Abstract
The centromeres of budding yeast are ∼120 bp in size and contain three functional elements: an AT-rich region flanked by binding sites for Cbf1 and CBF3. A specialized nucleosome containing the H3 variant Cse4 (CenH3) is formed at the centromere. Our genome-wide paired-end sequencing of nucleosomal DNA reveals that the centromeric nucleosome contains a micrococcal nuclease-resistant kernel of 123–135 bp, depending on the centromere, and is therefore significantly shorter than the canonical nucleosome. Unlike canonical nucleosomes, the centromeric nucleosome is essentially perfectly positioned. The entire centromere is included, together with at least 1 bp of DNA upstream of the Cbf1 site and at least 4 bp downstream of the CBF3 site. The fact that the binding sites for Cbf1 and CBF3 are included within the centromeric nucleosome has important implications for models of the centromeric nucleosome and for kinetochore function.
Keywords: chromatin, histone, Saccharomyces cerevisiae
The centromere is a DNA sequence that is required to anchor the chromosome to the kinetochore during mitosis. It is essential for proper segregation of the chromosomes. Budding yeast (Saccharomyces cerevisiae) has served as a very tractable model for the study of centromere and kinetochore function because its centromeres are very short (∼120 bp), relatively simple, and yet remain basically similar to those of other eukaryotes, which are much larger and contain repetitive sequences. Yeast centromeres (CENs) have been divided into three functional elements, termed CDEI, CDEII, and CDEIII (1, 2), which appear in this order on each of the 16 chromosomes. CDEI corresponds to a binding site for the transcription factor Cbf1 (3), CDEII is an extremely AT-rich region, and CDEIII is a binding site for the CBF3 complex, which is composed of Cep3, Ndc10, Ctf13, and Skp1. Cep3 binds specifically to CDEIII (4) and Ndc10 binds specifically to CDEII and CDEIII (5). The CBF3 complex is an essential component of the kinetochore, which is a large multisubunit assembly responsible for attaching the chromosome to the spindle microtubules.
The budding yeast centromere is organized into a single, specialized nucleosome containing Cse4, a centromere-specific variant of histone H3 (6–8), which is also found in other eukaryotes where it is variously referred to as CENP-A or CenH3. The canonical nucleosome contains 145–147 bp of DNA wrapped in ∼1.7 negative superhelical turns around a central octamer of the core histones, containing two molecules each of H2A, H2B, H3, and H4 (9). In contrast, the composition and structure of the centromeric nucleosome is highly controversial. Various models for the centromeric nucleosome have been proposed, based on data from yeast, Drosophila and other organisms, including: (i) A (CenH3-H4)2 tetramer bound with two molecules of Scm3, a centromeric protein capable of replacing H2A and H2B in the core histone octamer (10); in this model the only conventional histone is H4. It now seems likely that Scm3 is a chaperone rather than an integral component of the centromeric nucleosome (11). (ii) A canonical octamer containing CenH3 instead of H3 (12). (iii) A “hemisome” composed of only one molecule each of CenH3, H4 , H2A, and H2B (13, 14). In addition, there is an active debate over whether the DNA in CenH3-containing nucleosomes is wrapped around the histones in the opposite sense to the DNA in the canonical nucleosome (15–18). A critical issue is the role of the sequence-specific DNA-binding proteins (Cbf1, Cep3, and Ndc10) in the structure of the CEN nucleosome. An important piece of information, lacking until now, is the position of the CEN nucleosome with respect to the centromere—are the binding sites for these proteins included within the CEN nucleosome and how precisely is it positioned?
Here, we describe the results of a genome-wide paired-end sequencing study of yeast nucleosomes. The advantage of paired-end sequencing is that the length of each nucleosome can be deduced from alignment of the paired reads, corresponding to each end of the nucleosomal DNA fragment, to the yeast genome. We show that the centromeric nucleosome is smaller than the canonical nucleosome and that, in striking contrast to canonical nucleosomes, it is essentially perfectly positioned, such that it covers the entire centromere.
Results
CEN Nucleosomes Are Significantly Shorter than Canonical Nucleosomes.
Digestion of chromatin by MNase involves initial cleavage in the linker DNA between nucleosomes, most rapidly at favored sites determined by DNA sequence, followed by exonucleolytic trimming of the linker DNA until MNase is halted at the border of the nucleosome core (reviewed in ref. 19). The result is the nucleosome core particle, which contains ∼147 bp of DNA. However, the kinetics of native chromatin digestion are such that it is difficult to obtain homogeneous core particle preparations. Thus, mononucleosomal DNA is a mixture of core particles, incompletely trimmed core particles (containing residual linker DNA), and overdigested core particles (cut internally), as shown by the length distribution of nucleosomal DNA fragments (Fig. 1). Below, we show data for control cells and cells treated with 3-aminotriazole (3AT), which are part of a wider study of 3AT-induced genes; 3AT is an inducer of Gcn4-dependent genes and is not expected to have any effect on centromeric nucleosomes.
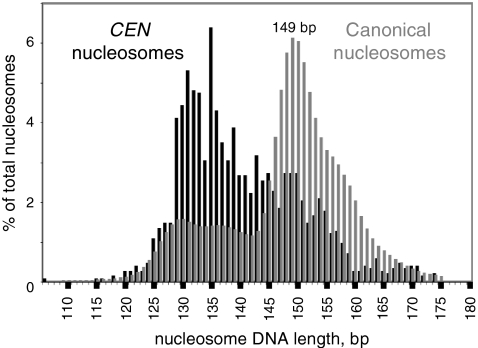
Fig. 1.
CEN nucleosomes are smaller than canonical nucleosomes. Size length distributions of nucleosomal DNA sequences from control cells, expressed as a percentage of the total number of nucleosome sequences. Gray histogram: all nucleosomes (∼16 million sequences). Black histogram: all nucleosomes containing at least 1 bp of any of the 16 CEN sequences (1,587 nucleosome sequences).
The length distribution of centromeric nucleosomes was compared with that of all canonical nucleosomes (Fig. 1). A centromeric nucleosome was defined as any nucleosome containing at least 1 bp of CEN sequence. Control cells yielded 1,587 CEN nucleosomes out of ∼16 million nucleosome sequences. There was an obvious difference between the size distributions of CEN and canonical nucleosomes (Fig. 1). The length distribution of CEN nucleosomes was approximately biphasic, indicating the presence of at least two different particles. The major peak ranged from 129–145 bp and accounted for just over half of the CEN nucleosomes. The minor peak ranged from 146–160 bp and accounted for about a quarter of the CEN nucleosomes. Thus, most CEN nucleosomes are significantly shorter than canonical nucleosomes, but the minor fraction is similar in size to the canonical core particle (which ranged from 146–153 bp and peaked at 149 bp; Fig. 1). By analogy with the canonical nucleosome, it is proposed that the minor fraction of CEN nucleosomes represents particles that were incompletely trimmed by MNase (i.e., they contain some undigested linker DNA) and that the major fraction represents the actual CEN nucleosome core particle, which contains significantly less DNA than the canonical core particle.
CEN Nucleosomes Are Perfectly Positioned.
Yeast centromeres are labeled CEN1-CEN16 to indicate which chromosome they belong to. Nucleosome occupancy profiles for the individual CEN nucleosomes were examined. The occupancy profile is a plot of the number of nucleosome sequences including a particular nucleotide against the chromosome coordinate and is effectively a plot of the probability of a nucleotide being present in a nucleosome. The profiles in Fig. 2 show the centromeres in selected chromosomes with 1 kb of flanking sequence on both sides. Generally, the data for control and 3AT-treated cells were very similar. In all cases, the CEN nucleosome was immediately obvious as a square peak located directly over the centromere. A square peak is observed when nearly all of the sequences begin within a few nucleotides of one another and end within a few nucleotides of one another, and is indicative of a unique position (19). Thus, CEN nucleosomes are perfectly positioned—i.e., each CEN nucleosome is in essentially the same position in all cells.
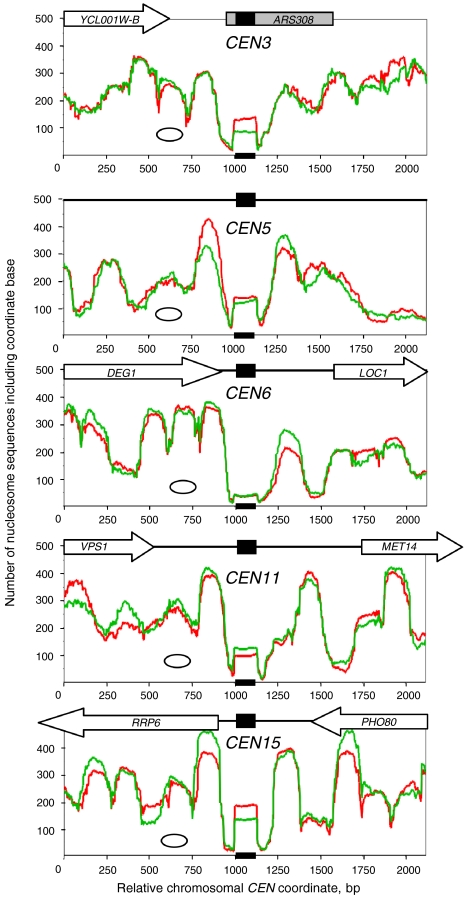
Fig. 2.
Perfect positioning of CEN nucleosomes. Nucleosome occupancy profiles for selected centromeres. The scale has been set arbitrarily such that the first nucleotide of CDEI in the CEN sequence as defined by the Saccharomyces Genome Database (black box) is at coordinate 1,001. The oval indicates the size of a canonical nucleosome. Red trace: control cells. Green trace: 3AT-treated cells. All nucleosome sequences were included. Raw data are shown. The data for 3AT-treated cells were multiplied by 1.27 to adjust for the different total number of nucleosomes sequenced for the two samples. Occupancy profiles for all of the other centromeres are presented in Fig. S1.
The square peak due to the CEN nucleosome contrasted with the clear but rounded peaks due to canonical nucleosomes on adjacent sequences. A rounded peak corresponds to a cluster of mutually exclusive overlapping positions, usually including a dominant position; it indicates that this nucleosome is not positioned identically in all cells (19, 20). Unlike the CEN nucleosomes, which were flanked by troughs almost reaching the baseline, the troughs between the canonical nucleosome peaks generally did not approach the baseline, indicating the presence of substantial numbers of nucleosomes in alternative positions that include linker DNA between dominant positions. These nucleosomes are far from perfectly positioned; they have a position cluster organization instead (19, 20).
In most cases, the CEN nucleosome was flanked on both sides by one or more rounded peaks corresponding to canonical nucleosome position clusters including a dominant position, which might indicate a phasing effect of the CEN nucleosome (Fig. 2). This effect was particularly clear for CEN15, but interpretation is complicated by the unknown contributions of the neighboring RRP6 promoter which had a nucleosome on it, and the 3’-UTR of PHO80, which was somewhat nucleosome-depleted. The situation for CEN5 was clearer because the flanking sequences do not contain any genes: there was a relatively well positioned nucleosome on either side of the CEN5 nucleosome but, if this is a phasing effect, it is short range, because it did not extend to a second nucleosome.
The rate of digestion of CEN nucleosomes appeared to vary quite markedly. CEN1, CEN9, and CEN16 yielded very few reads, suggesting that these CEN nucleosomes had been destroyed (Fig. S1). The CEN6 nucleosome had almost disappeared, but the square peak was just visible (Fig. 2). All of the other CEN nucleosomes gave clear square peaks (Fig. 2, Fig. S1), but the overall number of reads appeared to be lower than the average for canonical nucleosomes (Fig. 2), suggesting that the CEN nucleosome might be somewhat less resistant to MNase than the canonical nucleosome. The variability in rate of digestion of individual CEN nucleosomes might indicate that CDEII, the central AT-rich region of the centromere, varies in its susceptibility to MNase, because CDEI and CDEIII are almost identical in every centromere.
The CEN Nucleosome Covers the Entire Centromere.
The position of the CEN nucleosome was equivalent for each centromere: the entire CEN sequence was contained within the CEN nucleosome in all cases (Fig. 2). The precise position of the CEN nucleosome with respect to the centromere was analyzed at nucleotide resolution by examining the nucleosome sequences more closely (Fig. 3). For each centromere, the number of nucleosome sequences beginning or ending at each chromosomal coordinate was plotted. The boundaries of each CEN nucleosome were generally sharp, as expected from the occupancy profiles. Particularly sharp boundaries were observed at CEN15, where both boundaries were defined within a few base pairs. The relatively small number of nucleosome sequences which initiated or terminated within a CEN sequence (Fig. 3) were attributed to background cleavage within the CEN nucleosome by MNase.
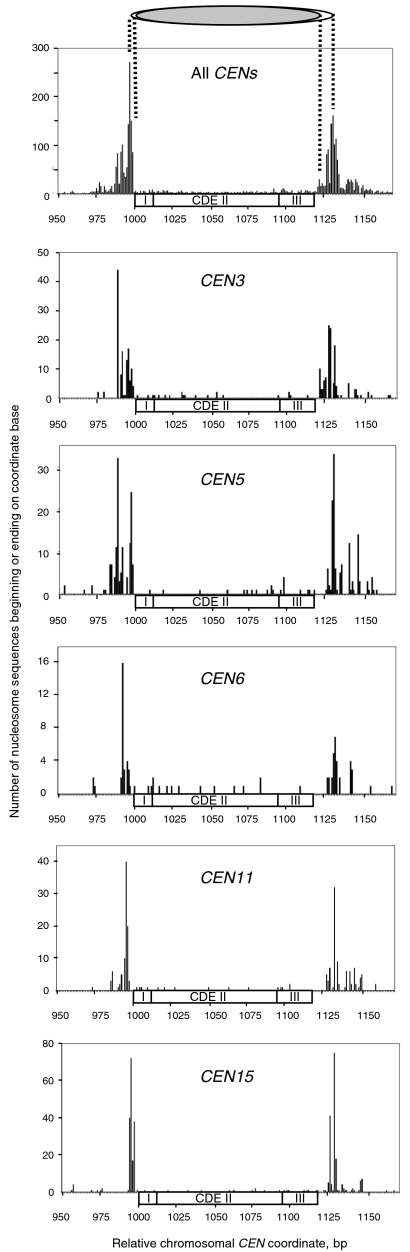
Fig. 3.
The CEN nucleosome covers the entire centromere. The positions of the individual CEN nucleosomes shown in Fig. 2 are shown at nucleotide resolution, together with a plot including all of the CEN nucleosomes. Each plot shows the number of nucleosome sequences from control cells beginning or ending at each nucleotide coordinate in the vicinity of the CEN sequence. Each CEN is shown with its CDEI, II and III elements. The position and length of the CEN nucleosome was defined by the distance between the most prominent peaks on each side (white oval). The kernel of the CEN nucleosome was defined as the MNase-resistant region between the peak clusters (gray oval).
In all cases, the CEN nucleosome profile exhibited a cluster of peaks at both borders of the nucleosome, separated by a region that was essentially resistant to MNase (Fig. 3). The coordinates of both major boundary peaks for each CEN nucleosome are recorded in Table 1. Using this measure, the size of the CEN nucleosome ranged from 131 to 143 bp. The average size was 138 ± 4 bp, corresponding to the majority population in the length distribution (Fig. 1) and is significantly smaller than the canonical nucleosome core particle. The peak clusters probably represent different degrees of trimming of the CEN nucleosome by MNase and the pattern is somewhat variable from one CEN nucleosome to the next. Therefore, we focused on the MNase-resistant region, which we term the “kernel” of the CEN nucleosome. The kernel was defined as the distance between the clusters of peaks, and corresponds to the minimum size of the CEN nucleosome. The kernel ranged from 123–135 bp in length (Table 1; average = 130 ± 3 bp). Thus, the kernel was ∼8 bp shorter than the CEN nucleosome as defined by the most prominent peaks.
Table 1.
Positions of the CEN nucleosomes
| Centromere | CEN length (bp)* | CEN nucleosome length (bp)† | CEN nucleosome position‡ | CEN kernel length (bp)§ | CEN kernel position¶ |
| CEN1 | 118 | too few reads | - | - | - |
| CEN2 | 117 | 131 | 998–1,128 | 127 | 999–1,125 |
| CEN3 | 117 | 139 | 989–1,127 | 123 | 999–1,121 |
| CEN4 | 111 | 137 | 992–1,128 | 126 | 997–1,122 |
| CEN5 | 118 | 143 | 989–1,131 | 129 | 999–1,127 |
| CEN6 | 118 | 140 | 993–1,132 | 135 | 997–1,131 |
| CEN7 | 119 | 134 | 997–1,130 | 131 | 999–1,129 |
| CEN8 | 118 | 141 | 991–1,131 | 133 | 999–1,131 |
| CEN9 | 117 | too few reads | - | - | - |
| CEN10 | 119 | 136 | 997–1,132 | 134 | 998–1,131 |
| CEN11 | 118 | 137 | 997–1,133 | 131 | 998–1,128 |
| CEN12 | 120 | 138 | 997–1,134 | 133 | 998–1,130 |
| CEN13 | 119 | 143 | 991–1,133 | 130 | 999–1,128 |
| CEN14 | 118 | 138 | 997–1,134 | 129 | 998–1,126 |
| CEN15 | 119 | 134 | 997–1,130 | 129 | 999–1,127 |
| CEN16 | 117 | too few reads | - | - | - |
*Length of the centromere given by the Saccharomyces Genome database.
†Length of the CEN nucleosome, defined as the distance between the most prominent peaks in the analysis shown in Fig. 3.
‡Positions of the CEN nucleosomes; the first nucleotide of CDEI was arbitrarily given a coordinate of 1,001 (as in Fig. 2).
§Length of the CEN kernel, defined as the MNase-resistant region between the most prominent peaks in the analysis shown in Fig. 3.
¶Position of the CEN kernel; the first nucleotide of CDEI was defined as coordinate 1,001.
The kernel included all of the CEN sequence in every case. The CEN sequences are all between 117 and 120 bp in length, except CEN4, which is only 111 bp long (Table 1). The CEN kernel was 6 to 19 bp longer than the CEN sequence that it contains. The noncentromeric DNA in the CEN kernel was apportioned such that 1 to 4 bp flanking CDEI and 4 to 14 bp flanking CDEIII were included within it.
The sequences of the MNase-resistant kernels of the CEN nucleosomes were aligned according to their presumed dyad axes (Fig. 4A). The Cbf1 binding site (RTCATGTG) was very close to the edge of the CEN nucleosome kernel, but was still included within it. Consequently, Cbf1 might define this border of the CEN nucleosome. The probable binding site of Cep3, a DNA sequence-specific subunit of the CBF3 complex, was located well within the CEN nucleosome.
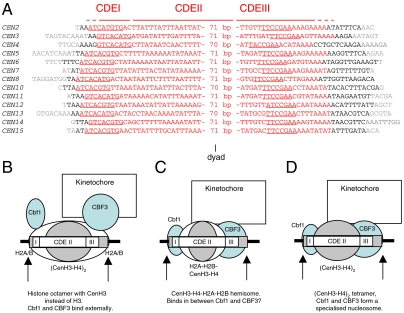
Fig. 4.
The position of the CEN nucleosome and implications for models of the CEN nucleosome. A. Alignment of the CEN nucleosome kernels according to their dyad axes (i.e., the centers of the kernel sequences). Nucleotides within the CEN sequence are indicated in red, those outside CEN but within the kernel are in black, and those that are outside the kernel are in gray. Nucleotides underlined in CDEI and CDEIII represent the specific binding sites of Cbf1 and CBF3 (Cep3), respectively. The complete sequences are given in Fig. S2. B–D. Models proposed for centromeric nucleosomes are shown positioned as determined in this work: B. The CenH3 histone octamer model (12) in which CenH3 replaces H3 in an otherwise canonical histone octamer. Because the octamer covers both CDEI and CDEIII, Cbf1 and CBF3 would have to bind to DNA on the outer surface of the CEN nucleosome. C. The hemisome model proposes a tetramer or half-nucleosome composed of CenH3, H4, H2A, and H2B (13). D. A (CenH3-H4)2 tetramer (equivalent to the canonical (H3-H4)2 tetramer) forms an atypical nucleosome with Cbf1 and CBF3 (10). Cbf1 and CBF3 bind specifically to CDEI and CDEIII respectively, and so could fix the positions of the other components. In all of these models, the position of the CEN nucleosome requires that the histone component interacts primarily with CDEII, the extremely AT-rich region between the Cbf1 and CBF3 sites.
Discussion
We have shown that the CEN nucleosome is significantly shorter than the canonical nucleosome, ranging from 131 to 143 bp, with a nuclease-resistant kernel of 123 to 135 bp. The CEN nucleosome is perfectly positioned, such that the kernel includes the entire CEN sequence together with ∼2 bp of noncentromeric DNA flanking CDEI and ∼10 bp flanking CDEIII (Fig. 4A).
Chromatin Structure of the Budding Yeast Centromere.
Early indirect end-labeling and restriction enzyme mapping studies indicated that there is a ∼200 bp MNase-resistant region at the budding yeast centromere (21, 22). In an elegant set of experiments, Furuyama and Biggins (8) demonstrated that there is only one Cse4-containing nucleosome at the centromere by probing Southern blots of immuno-purified tagged Cse4 nucleosomes with CEN probes. Their data demonstrate that CEN DNA is present in a Cse4-containing particle of mononucleosome size. The slightly smaller size of the CEN nucleosome which we have observed would not have been apparent in these experiments, because the agarose gels required for blotting did not have the resolution necessary. It is emphasized that although these data show clearly that the CEN sequence is at least partly contained within a single nucleosome, the position of the CEN nucleosome cannot be deduced.
It has also been noted (21) that ordered arrays of several positioned nucleosomes are formed on either side of CEN3 and CEN11. It has been suggested that such positioning is a general feature of CEN chromatin. However, although our occupancy profiles for CEN3 and CEN11 (Fig. 2) do show some relatively well positioned nucleosomes, the chromatin structures of sequences adjacent to the CEN nucleosome do not seem obviously more ordered than other regions (Fig. 2). The degree of phasing might depend more on whether the genes neighboring the centromere are active.
The CEN Nucleosome Contains less DNA than the Canonical Nucleosome.
The CEN nucleosome is shorter than the canonical nucleosome, indicating that its structure is significantly different. The outer 13 bp on each side of the canonical nucleosome are bound by the N-terminal α-helix of H3 (9). CenH3 would be expected to have an analogous role in determining the boundaries of the CEN nucleosome, as suggested by the crystal structure of the (CENP-A-H4)2 tetramer (17). Perhaps CenH3 binds less tightly than H3 to the outer DNA turns, affording less protection from MNase.
The size of the CEN nucleosome depends on the centromere. The size range seems larger than might be expected from the error in the measurement, assuming an identical structure (131–143 bp; range: 13 bp). Incomplete trimming seems an unlikely explanation because the nuclease-resistant kernels show a similar range (123–135 bp) and their termini are not unusually GC-rich (Fig. 4A), which might prevent MNase from removing residual linker DNA. All but one of the CEN sequences fall within the range 117–120 bp (Table 1), which seems too small a variation to account for the kernel size range. The exception is CEN4, which is only 111 bp. Surprisingly, the CEN4 nucleosome kernel is larger (126 bp) than that of CEN3 (123 bp), even though CEN3 is 117 bp. The limited sequence preference of MNase (23) is unlikely to be a major problem in determining the CEN nucleosome boundaries, because it is most evident in the early, endonucleolytic phase of digestion, typically used in the indirect end-labeling technique (19). The specificity is very weak in the later, exonucleolytic (trimming) phase of digestion, which is required to prepare core particles: a genome-wide analysis of nucleosomal DNA ends indicates that they are generated predominantly by cleavage of TT, AA, or TA, which will be found in almost any short DNA sequence (24).
Thus, the size of the CEN nucleosome is not determined exclusively by the length of the CEN sequence in base pairs. An important factor here might be the variable length of CDEII, which accounts for the differences in CEN sequence length. Even though CDEII sequences are ∼90% AT, there is surprisingly little homology between them (Fig. S2). Centromeric DNA is naturally curved, a property attributed to CDEII, and the extent of curvature is different for different centromeres (25). Differences in curvature might compensate for differences in the number of base pairs, resulting in the same path length (measured by distance) and therefore essentially the same structure. Alternatively, the double helix could be over- or undertwisted to accommodate DNA with different numbers of base pairs. Such stretching to accommodate DNA of a slightly different length has been observed in the structure of a canonical nucleosome (26).
Another apparent difference between the CEN nucleosome and the average canonical nucleosome is the absence of some important DNA sequence signals. In canonical nucleosomes, the distribution of AA/AT/TA/TT dinucleotides has a ∼10-bp periodicity which is exactly out of phase with the ∼10-bp periodicity exhibited by GG/GC/CG/CC dinucleotides; the former are preferentially located for bending into the minor groove and the latter for bending into the major groove (27, 28). The fact that CDEII is 90% AT and accounts for most of the DNA in the CEN nucleosome indicates that the G/C pattern is not present. In addition, the much remarked exclusion of runs of A or T from canonical nucleosomes (24, 29) is clearly not true for CEN nucleosomes (Fig. S2). These differences may reflect the presence of CenH3 in the CEN nucleosome, which probably interacts with CDEII (see below).
Models for the CEN Nucleosome.
The precise position of the CEN nucleosome with respect to the CEN sequence is revealing. Because it covers the entire centromere, the proteins which bind to CEN DNA must either be part of the inner core of the CEN nucleosome, or must bind on the outside of the nucleosome. By analogy with the canonical nucleosome, the position of the CEN nucleosome predicts that CDEI and CDEIII might each interact with an H2A-H2B dimer and that CDEII would interact with the (CenH3-H4)2 tetramer. The AT-rich CDEII element is ∼80 bp, similar to the length of DNA bound by the canonical (H3-H4)2 tetramer (9). However, there is controversy over whether the CEN nucleosome contains an octamer or a tetramer and over whether it contains H2A-H2B (10, 12, 13).
If the CEN nucleosome contains two H2A-H2B dimers and a central CenH3-H4 tetramer (12), Cbf1 and CBF3 would have to bind to DNA on the outside of the nucleosome (Fig. 4B). Most DNA-binding proteins bind to nucleosomal sites with much lower affinity, but there are some exceptions, including the pioneer transcription factors, which prefer nucleosomal sites (30). Different affinities for nucleosomal and free DNA sites are to be expected because binding to a site on the outside of the nucleosome requires recognition of DNA that is highly bent away from the protein and partly obscured by the histones. The bending of CEN DNA by Cbf1 and CBF3 (31, 32) might facilitate or inhibit binding to the CEN nucleosome, depending on whether the induced bend is similar to the bent DNA in the CEN nucleosome. A potential problem for the model is that CBF3 binds to both sides of the DNA helix (33) and so may clash with one of the two H2A-H2B dimers. Support for this model comes from the fact that nucleosomes can be reconstituted with CenH3, H4, H2A, and H2B, and that they protect less DNA than a canonical nucleosome from MNase (15, 17, 34), indicating that they are similar and perhaps identical to the CEN nucleosome. The fact that such nucleosomes can be reconstituted on noncentromeric DNA argues against a critical role for CenH3 in determining the position of the CEN nucleosome, although CenH3 might have a significant preference for binding to CEN DNA.
In the hemisome model (13), the CEN nucleosome contains only one molecule each of CenH3, H4, H2A, and H2B (Fig. 4C). It seems unlikely that such a tetramer could protect ∼135 bp by itself. However, if the hemisome binds to CDEII and fits snugly between Cbf1 and the CBF3 complex, such that MNase cannot cut in between, then such protection can be envisaged. Similarly, if the CEN nucleosome contains a central CenH3-H4 tetramer but no H2A-H2B (10), then Cbf1 and/or CBF3 might form part of the CEN nucleosome core, binding to the CEN sequence instead of H2A-H2B (Fig. 4D). Thus, Cbf1 and/or CBF3 might be built into the CEN nucleosome. If so, Cbf1 and CBF3 might be expected to interact directly with CenH3 or H4, as well as with CEN DNA. There is evidence that Cbf1 and CBF3 interact directly on CEN DNA in vitro, promoting looping (35). The model can explain the perfect positioning of the CEN nucleosome, because the position would be fixed by the sequence-specific interactions of Cbf1 and/or CBF3 with CEN DNA. Of these, CBF3 seems most likely because mutations in CDEIII result in the complete loss of nuclease resistance at the centromere, whereas mutations in CDEI have less drastic effects (36).
A nucleosome-like structure containing Cbf1, CBF3, CenH3-H4, and potentially Scm3 (Fig. 4D) should stabilize the CEN complex. Generally speaking, sequence-specific factors bind their sites reversibly, facilitating the search process necessary to locate their specific binding sites. However, reversible binding of the kinetochore to centromeric DNA through Cbf1, Ndc10, and Cep3 would present a problem for chromosome segregation, because there would be a reasonable chance of dissociation during segregation. One of the characteristics of the canonical nucleosome is its extreme stability, in the absence of ATP-dependent remodeling machines. Thus, adoption of a nucleosome-like structure by the CEN complex might confer the necessary property of an extremely low probability of dissociation. A stable structure would be particularly critical in yeast because there is only one CEN nucleosome in each chromosome.
Materials and Methods
Preparation of Core Particle DNA.
YDC111 (MATa ade2-1 can1-100 HIS3 leu2-3,112 trp1-1 ura3-1 RAD5+) (20) was grown to late log phase in synthetic complete (SC) medium (control) or SC medium lacking histidine to which 3AT was added to 10 mM for 20 min just before harvesting (3AT-treated). Core particle DNA was prepared by MNase digestion of nuclei as described (37).
Paired-End Sequencing.
Samples were prepared essentially as suggested by the manufacturer (Illumina). Repaired core particle DNA was purified using Qiaquick PCR Purification columns (Qiagen 28104). The DNA was treated as follows, with purification using a Qiaquick column after each step: (i) a 5′-phosphate was added to the DNA using T4 polynucleotide kinase; (ii) dA was added to the 3′-ends using Klenow 3′–5′ exo-; (iii) Paired-end adaptors (Illumina) were added using the Quick Ligation kit (NEB); (iv) ligation products with an adapter at each end were purified from a 6% (40∶1) polyacrylamide gel stained with ethidium bromide; (v) core particle DNA with ligated adapters was amplified by 14 or 15 cycles of PCR using 25 μM paired-end sequencing primers 1.0 and 2.0 (Illumina) and Phusion polymerase (Finnzymes Oy); and (vi) the PCR product was gel-purified. The DNA concentration was adjusted to 100 nM and 30 μL was sent to the Tufts Core Facility for one lane of paired-end sequencing (Illumina Solexa). Control cells yielded 16,601,752 and 12,285,181 aligned paired reads of 40 nt each for the first and second experiments, respectively; 3AT-treated cells gave 13,120,871 and 13,548,103 aligned paired reads. Paired reads were aligned to the S. cerevisiae genome using ELAND. Reads with mis-matches were excluded from the analysis. The GEO accession number for the data presented here is GSE26493.
Supplementary Material
Acknowledgments.
We thank Munira Basrai, Steve Henikoff, and Carl Wu for helpful comments on the manuscript. We thank Kip Bodi and Michael Berne at the Tufts University Core Facility for paired-end sequencing. This work was supported by the Intramural Research Program of the National Institutes of Health (NIH), National Institute of Child Health and Human Development.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no GSE26493: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=hvsbfoqmqecaorq&acc=GSE26493).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104978108/-/DCSupplemental.
References
- 1.FitzGerald-Hayes M, Clarke L, Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982;29:235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- 2.Hieter P, et al. Functional selection and analysis of yeast centromeric DNA. Cell. 1985;42:913–921. doi: 10.1016/0092-8674(85)90287-9. [DOI] [PubMed] [Google Scholar]
- 3.Bram RJ, Kornberg RD. Isolation of a Saccharomyces cerevisiae centromere DNA binding protein, its human homologue, and its possible role as a transcription factor. Mol Cell Biol. 1987;7:403–409. doi: 10.1128/mcb.7.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lechner J, Carbon J. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 1991;64:717–725. doi: 10.1016/0092-8674(91)90501-o. [DOI] [PubMed] [Google Scholar]
- 5.Espelin CW, Simons KT, Harrison SC, Sorger PK. Binding of the essential Saccharomyces cerevisiae kinetochore protein Ndc10 to CDEII. Mol Biol Cell. 2003;14:4557–4568. doi: 10.1091/mbc.E02-08-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meluh PB, Yang P, Glowczewski L, Koshland D, Smith MM. Cse4p is a component of the core centromere of Saccharomyces cerevisiae. Cell. 1998;94:607–613. doi: 10.1016/s0092-8674(00)81602-5. [DOI] [PubMed] [Google Scholar]
- 7.Smith MM. Centromeres and variant histones: what, where, when and why? Curr Opin Cell Biol. 2002;14:279–285. doi: 10.1016/s0955-0674(02)00331-9. [DOI] [PubMed] [Google Scholar]
- 8.Furuyama S, Biggins S. Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc Natl Acad Sci USA. 2007;104:14706–14711. doi: 10.1073/pnas.0706985104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 10.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, Wu C. Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell. 2007;129:1153–1164. doi: 10.1016/j.cell.2007.04.026. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Z, et al. Structural basis for recognition of centromere histone variant CenH3 by the chaperone Scm3. Nature. 2011 doi: 10.1038/nature09854. doi: 10.1038/nature09854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Camahort R, et al. Cse4 is part of an octameric nucleosome in budding yeast. Mol Cell. 2009;35:794–805. doi: 10.1016/j.molcel.2009.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalal Y, Furuyama T, Vermaak D, Henikoff S. Structure, dynamics and evolution of centromeric nucleosomes. Proc Natl Acad Sci USA. 2007;104:15974–15981. doi: 10.1073/pnas.0707648104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y. Tetrameric organization of vertebrate centromeric nucleosomes. Proc Natl Acad Sci USA. 2010;107:20317–20322. doi: 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoda K, et al. Human centromere protein A (CENP-A) can replace histone H3 in nucleosome reconstitution in vitro. Proc Natl Acad Sci USA. 2000;97:7266–7271. doi: 10.1073/pnas.130189697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Furuyama T, Henikoff S. Centromeric nucleosomes induce positive DNA supercoils. Cell. 2009;138:104–113. doi: 10.1016/j.cell.2009.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)2 reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kingston IJ, Yung JSY, Singleton MR. Biophysical characterization of the centromere-specific nucleosome from budding yeast. J Biol Chem. 2010;286:4021–4026. doi: 10.1074/jbc.M110.189340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark DJ. Nucleosome positioning, nucleosome spacing and the nucleosome code. J Biomol Struc Dyn. 2010;27:781–793. doi: 10.1080/073911010010524945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim Y, McLaughlin N, Lindstrom K, Tsukiyama T, Clark DJ. Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene. Mol Cell Biol. 2006;26:8607–8622. doi: 10.1128/MCB.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bloom KS, Carbon J. Yeast centromere DNA is in a unique and highly ordered structure in chromosomes and small circular minichromosomes. Cell. 1982;29:305–317. doi: 10.1016/0092-8674(82)90147-7. [DOI] [PubMed] [Google Scholar]
- 22.Funk M, Hegemann JH, Philippsen P. Chromatin digestion with restriction endonucleases reveals 150–160 bp of protected DNA in the centromere of chromosome XIV in Saccharomyces cerevisiae. Mol Gen Genet. 1989;219:153–160. doi: 10.1007/BF00261171. [DOI] [PubMed] [Google Scholar]
- 23.Hörz W, Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981;9:2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Field Y, et al. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comp Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bechert T, Heck S, Fleig U, Diekmann S, Hegemann JH. All 16 centromere DNAs from Saccharomyces cerevisiae show DNA curvature. Nucleic Acids Res. 1999;27:1444–1449. doi: 10.1093/nar/27.6.1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasudevan D, Chua EYD, Davey CA. Crystal structures of nucleosome core particles containing the “601” strong positioning sequence. J Mol Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 27.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191:651–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 28.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iyer V, Struhl K. Poly(dA∶dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 1995;14:2570–2579. doi: 10.1002/j.1460-2075.1995.tb07255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zaret KS, et al. Pioneer factors, genetic competence and inductive signaling: programming liver and pancreas progenitors from the endoderm. \Cold Spring Harb Sym. 2008;73:119–126. doi: 10.1101/sqb.2008.73.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Niedenthal RK, Sen-Gupta M, Wilmen A, Hegemann JH. Cpf1 protein induced bending of yeast centromeric DNA element I. Nucleic Acids Res. 1993;21:4726–4733. doi: 10.1093/nar/21.20.4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pietrasanta L, et al. Probing the Saccharomyces cerevisiae centromeric DNA (CEN DNA)-binding factor 3 (CBF3) kinetochore complex by using atomic force microscopy. Proc Natl Acad Sci USA. 1999;96:3757–3762. doi: 10.1073/pnas.96.7.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Espelin CW, Kaplan KB, Sorger PK. Probing the architecture of a single kinetochore using DNA-protein crosslinking. J Cell Biol. 1997;139:1383–1396. doi: 10.1083/jcb.139.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conde e Silva N, et al. CENP-A-containing nucleosomes: easier disassembly versus exclusive centromeric localization. J Mol Biol. 2007;370:555–573. doi: 10.1016/j.jmb.2007.04.064. [DOI] [PubMed] [Google Scholar]
- 35.Hemmerich P, et al. Interaction of yeast kinetochore proteins with centromere-protein/transcription factor Cbf1. Proc Natl Acad Sci USA. 2000;97:12583–12588. doi: 10.1073/pnas.97.23.12583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saunders M, FitzGerald-Hayes M, Bloom K. Chromatin structure of altered yeast centromeres. Proc Natl Acad Sci USA. 1988;85:175–179. doi: 10.1073/pnas.85.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gaykalova DA, et al. A polar barrier to transcription can be circumvented by remodeler-induced nucleosome translocation. Nucleic Acids Res. 2011;39:3520–3528. doi: 10.1093/nar/gkq1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.