Abstract
Autotransporters are bacterial virulence factors that consist of an N-terminal extracellular (“passenger”) domain and a C-terminal β barrel domain (“β domain”) that resides in the outer membrane. Here we used an in vivo site-specific photocrosslinking approach to gain insight into the mechanism by which the β domain is integrated into the outer membrane and the relationship between β domain assembly and passenger domain secretion. We found that periplasmic chaperones and specific components of the β barrel assembly machinery (Bam) complex interact with the β domain of the Escherichia coli O157:H7 autotransporter extracellular serine protease P (EspP) in a temporally and spatially regulated fashion. Although the chaperone Skp initially interacted with the entire β domain, BamA, BamB, and BamD subsequently interacted with discrete β domain regions. BamB and BamD remained bound to the β domain longer than BamA and therefore appeared to function at a later stage of assembly. Interestingly, we obtained evidence that the completion of β domain assembly is regulated by an intrinsic checkpoint mechanism that requires the completion of passenger domain secretion. In addition to leading to a detailed model of autotransporter biogenesis, our results suggest that the lipoprotein components of the Bam complex play a direct role in the membrane integration of β barrel proteins.
Keywords: membrane protein assembly, molecular chaperones, protein translocation
The outer membrane (OM) of Gram-negative bacteria is an unusual organelle whose inner leaflet consists of phospholipids and whose outer leaflet consists mainly of LPS. Most of the integral membrane proteins that reside in the OM fold into a characteristic β barrel structure that comprises multiple amphipathic β strands or contains a β barrel domain (β domain) that serves as a membrane anchor. Although it has long been known that OM proteins (OMPs) are transported into the periplasm via the Sec machinery, many aspects of OMP biogenesis are still poorly understood.
The transit of OMPs through the periplasm requires the activity of the molecular chaperones SurA, Skp, and DegP, which function in two parallel pathways. Escherichia coli that lack only one of the chaperones are viable, but cells that lack SurA and Skp or SurA and DegP are inviable (1). The double mutants display strong defects in the assembly of trimeric porins and OmpA, which are the major OMPs. Although the two pathways are redundant, the major OMPs preferentially use the SurA pathway (2). Skp appears to interact with OMPs at a very early stage of assembly, possibly even before their release from the Sec complex (3, 4). Although the chaperones play an essential role in OMP biogenesis, their exact function is unknown. It seems likely that they prevent OMPs from aggregating or misfolding, but they also may catalyze early steps in the folding process. It is also unclear if the SurA and Skp/DegP pathways are required for the biogenesis of all OMPs.
Recent studies have shown that the membrane integration of OMPs is catalyzed by the β barrel assembly machinery (Bam) or Omp85 complex, a heteroligomer comprised of an integral OMP (BamA) and four lipoproteins (BamB–E) (5–8). BamC/D/E exists as a discrete subcomplex that interacts with BamA through BamD (9). BamA and BamD are the only components of the complex that are essential for viability (6, 9). BamA belongs to a superfamily of proteins that catalyze the transport of other proteins into or across the bacterial, mitochondrial, and chloroplast OM and appears to recognize a conserved motif found at the C terminus of OMPs (10, 11). In addition to a β domain, BamA contains five N-terminal polypeptide transport-associated (POTRA) domains that are situated in the periplasm. The POTRA domains have been proposed to facilitate β barrel assembly through β augmentation (12). Structural data and molecular modeling suggest that a hinge between POTRA domains 2 and 3 allows the POTRA domains to adopt both a compact, fishhook-like conformation and an extended conformation (13). Although the deletion of POTRA domains 2, 3, 4, or 5 eliminates the binding of BamB to the complex, only the deletion of POTRA domain 5 eliminates the binding of BamC/D/E (9, 12). Interestingly, SurA appears to interact with the first POTRA domain of BamA (7, 14). This observation suggests that SurA might deliver OMPs to the Bam complex. BamB forms an eight-bladed β propeller structure, and available evidence suggests that residues that lie on one side of this structure mediate the binding of both BamA and ligands (15–18). BamD contains five copies of a structural motif (tetratricopeptide repeat) that often mediates protein–protein interactions (19). At present, however, neither the mechanism by which the Bam complex promotes the membrane integration of OMPs nor the function of the individual Bam complex subunits is known.
Autotransporters are a large superfamily of specialized OMPs whose assembly is especially enigmatic. In addition to containing a C-terminal ∼30-kDa β domain, these proteins contain an N-terminal extracellular (passenger) domain that often exceeds 100 kDa and that mediates a virulence function (reviewed in ref. 20). Although their sequences are highly diverse, passenger domains almost always form a β-helical structure and are transported across the OM in a C-terminal to N-terminal direction (21, 22). Many passenger domains are released from the cell surface by proteolytic cleavage after their transport across the OM (23). The sequence of autotransporter β domains is also conserved only weakly, but the four β domains that have been crystallized to date form nearly superimposable 12-stranded β barrels (24–27). Based on the observation that the deletion of the β barrel domain prevents passenger domain secretion, it was originally proposed that the β domain transports the covalently linked passenger domain across the OM (28). In apparent agreement with this proposal, the C terminus of the passenger domain protrudes from the β domain pore (24, 29). Recent experiments, however, have challenged the “autotransporter” hypothesis. Folded polypeptides that cannot fit through the ∼10-Å pore of a fully assembled β domain have been shown to be secreted efficiently via the autotransporter pathway (23, 30). Furthermore, it has been shown that the incorporation of the passenger domain C terminus into the β domain pore precedes rather than follows translocation, at least in the case of the E. coli O157:H7 autotransporter extracellular serine protease P (EspP) (31, 32). Finally, a passenger domain whose translocation is transiently stalled can be crosslinked to BamA when the reactive group is placed close to the stall point (22, 32). Taken together, these results suggest that the Bam complex not only promotes the membrane integration of the β domain but also may facilitate the secretion of the passenger domain.
In this study we used EspP as a model protein to gain insight into both OMP assembly and autotransporter biogenesis. The passenger domain of EspP is released in an autocatalytic intrabarrel cleavage reaction following its secretion (33). The status of the EspP β domain can be monitored both before and after its integration into the OM because the integration process occurs relatively slowly, especially at low temperature (31). In addition, because the initiation and completion of passenger domain translocation can be monitored as distinct events, it is also possible to examine the temporal relationship between β barrel assembly and passenger domain translocation. By following the interactions of newly synthesized EspP molecules with chaperones and components of the Bam complex using site-specific photocrosslinking, we identified several β domain assembly intermediates. Interestingly, we found that Skp is the only known chaperone that forms a stable interaction with the β domain, whereas SurA appears to play a specialized role in passenger domain secretion. We also obtained evidence that BamA, BamB, and BamD play a direct role in subsequent steps of β domain assembly by binding to the β barrel with a specific geometry. Furthermore, we found that the completion of β barrel assembly is linked obligately to the completion of passenger domain translocation. Based on our results, we propose a detailed model for autotransporter biogenesis and for the role of Bam complex subunits in OMP assembly.
Results
Sequential Interaction of Skp and SurA/BamA with the EspP Passenger Domain.
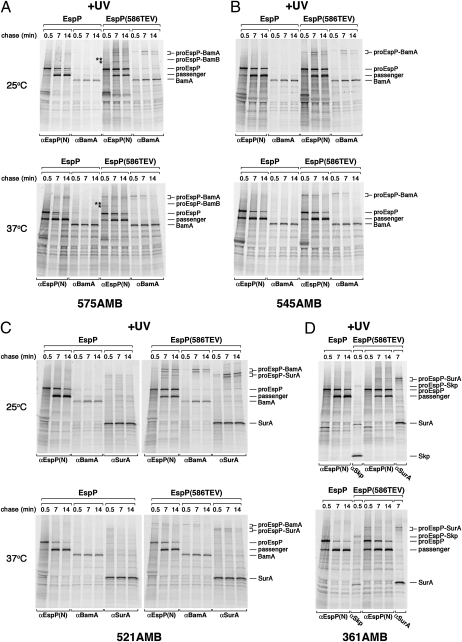
We used a site-specific photocrosslinking method to identify cellular factors that interact with EspP during its assembly. This technique employs the coexpression of an amber suppressor tRNA and an amino acyl-tRNA synthetase from Methanococcus jannaschii that are orthogonal to the E. coli translation machinery to incorporate the photoactivable amino acid analog p-benzoyl-l-phenylalanine (Bpa) at amber (AMB) codons engineered into a protein of interest (34). Bpa can be crosslinked only to molecules that are within ∼4 Å of the polypeptide backbone. We introduced an amber codon at one of four positions in the passenger domain and at eight surface-exposed positions in the β domain of wild-type EspP and an EspP mutant that contains a short linker insertion at residue 586 [EspP(586TEV)]. It was shown previously that translocation of the EspP(586TEV) passenger domain stalls transiently near the point of the insertion and that consequently the cleavage of the passenger domain is delayed (22). AD202 cells were transformed with two plasmids that encode an amber mutant and the amber suppression system (pDULEBpa) (35). Isopropyl-β-d-thio-galactoside (IPTG) was added to induce espP expression, and cells were subjected to pulse-chase labeling. Two equal samples were removed from radiolabeled cultures at various time points, and one sample from each time point was exposed to UV light. Immunoprecipitations were conducted using antisera generated against N- and C-terminal EspP peptides and various cellular proteins to isolate EspP-containing polypeptides and to identify crosslinking products. Unlike previous experiments in which we performed only a 1-min chase at 37 °C, in these experiments we conducted a longer chase and radiolabeled cells at both 37 °C and 25 °C. The lower temperature was used to slow EspP biogenesis and to facilitate the identification of assembly intermediates (31). The secretion and cleavage of 90% of newly synthesized EspP passenger domains requires ∼2 min at 37 °C but requires ∼15 min at 25 °C (Fig. S1) (22). Likewise, the stalling of EspP(586TEV) passenger domain secretion dissipates after 2 min at 37 °C but still is detectable after 15 min at 25 °C.
Consistent with previous results (22), we found that the EspP passenger domain could crosslink to BamA, but only when translocation stalled. When Bpa was introduced into EspP(586TEV) mutants at residues that are relatively close to the site of stalling (residues 545 and 575) and cells were incubated at 25 °C, high molecular weight adducts that could be immunoprecipitated with both anti–N-terminal EspP and BamA antisera were observed in UV-irradiated cells (Fig. 1A and B, Upper gels). These adducts were not produced when Bpa was introduced into the same positions in wild-type EspP. Based on previous results (21) and the size of the adducts, they very likely correspond to BamA crosslinked to proEspP, the precursor form of the protein that contains covalently linked passenger and β domains. Because the assembly of the protein was relatively slow at 25 °C, crosslinking was seen predominantly at the middle (7 min) and late (14 min) time points. Curiously, two discrete adducts were produced. Multiple bands have been observed previously in site-specific crosslinking experiments and have been attributed to the interaction of the photoprobe with multiple sites in the target protein (36). When the cells were incubated at 37 °C, a similar crosslinking pattern was observed (Fig. 1 A and B, Lower gels). However, presumably because EspP assembly is faster at the higher temperature, the proEspP-BamA adduct was seen primarily at the early (0.5 min) and middle time points. At both temperatures, residue 575 of the pro form of EspP(586TEV) also was crosslinked to an unidentified ∼30- to 40-kDa protein that is neither BamB nor BamC (single asterisks in Fig. 1A and in Fig. S2). Furthermore, a weak crosslink between residue 575 and BamB was observed (double asterisks in Fig. 1A and in Fig. S2).
Fig. 1.
Crosslinking of passenger domain residues of an EspP secretion intermediate to BamA, SurA, and Skp. AD202 cells were transformed with pDULEBpa and a derivative of pRI22 (Plac-espP) or pRI23 [Plac-espP(586TEV)] harboring an amber codon at residue 575 (A), 545 (B), 521 (C), or 361 (D). Cells were subjected to pulse-chase labeling at 25 °C or 37 °C after the addition of 200 μM IPTG. Half of each sample was UV irradiated, and equal portions were used for immunoprecipitations with the indicated antisera. Only the UV-irradiated samples are shown. In A, adducts that result from the crosslinking of residue 575 to an unknown protein (asterisk) and BamB (double asterisk) are denoted.
Interactions between the passenger domain and molecular chaperones were observed when the photoprobe was introduced closer to the N terminus. At both 25 °C and 37 °C, EspP(586TEV/521AMB) was crosslinked to BamA and SurA (Fig. 1C). At the lower temperature, crosslinking was observed primarily at the later time points. The finding that the level of the EspP(586TEV)-BamA adducts was comparable to the level of the EspP(586TEV)-SurA adducts at each time point indicates that the interaction of the secretion intermediate with both factors followed a similar time course. The data suggest that residue 521 was in proximity to both BamA and SurA when translocation stalled and therefore could be crosslinked to either factor. EspP(586TEV/361AMB) was crosslinked to both Skp and SurA (Fig. 1D). Interestingly, crosslinking of residue 361 to Skp was observed at the 0.5 min time point at 25 °C and clearly preceded crosslinking to SurA. Consistent with the notion that SurA interacts with this residue only when translocation stalls (and a segment of the passenger domain is exposed on the cell surface), the treatment of intact cells with PK led to the quantitative cleavage of the proEspP-SurA adduct (Fig. S3A). A significant fraction of the proEspP-Skp adduct was protease resistant, however, and therefore presumably was located in the periplasm. The two chaperones are not interchangeable, because crosslinking to SurA was not observed at the early time point in a Δskp strain (Fig. S3B). In addition, although none of the residues of wild-type EspP that we examined were crosslinked to BamA or SurA, residue 361 was crosslinked to Skp (Fig. 1D). Taken together, these results strongly suggest that the two chaperones interact with the passenger domain at distinct stages of EspP biogenesis: Skp binds to the passenger domain before the initiation of translocation, whereas SurA binds during the translocation reaction. Furthermore, the crosslinking of SurA to EspP(586TEV) residues 361 and 521 but not to residues 202, 545, or 575 (Fig. 1) (22) suggests that it interacts selectively with segments that are at a specific distance from the site of transfer.
Sequential Interaction of Skp and Specific Bam Complex Subunits with the EspP β Domain.
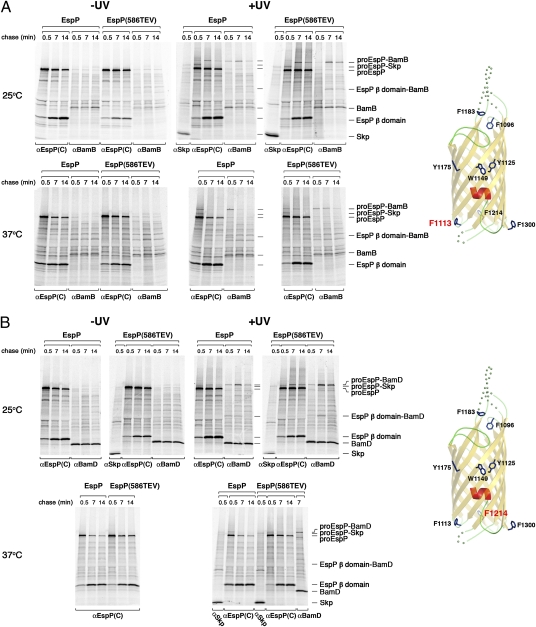
A rather different crosslinking pattern was observed when Bpa was incorporated at residue 1113, which is situated on the periplasmic side of the fully folded EspP β barrel. When cells that produced either EspP(1113AMB) or EspP(586TEV/1113AMB) were incubated at 25 °C, several bands that appeared only in UV-irradiated samples were immunoprecipitated with the anti-EspP C-terminal antiserum (Fig. 2A, Upper gels). At the 0.5 min time point an ∼150-kDa adduct that corresponds to proEspP crosslinked to Skp was observed. Because an adduct of about the same size was observed in Δskp cells, it appears that proEspP interacts with other ∼15- to 20-kDa proteins at early time points (Fig. S4). At later time points the ∼150-kDa adducts disappeared, and a larger adduct became more prominent. Consistent with previous results (22), this band corresponds to proEspP crosslinked to BamB. Although the proEspP-BamB adduct clearly peaked at the 7 min time point in samples that contained wild-type EspP, a strong signal also was observed at the 14 min time point in samples that contained EspP(586TEV). Indeed the interaction between EspP(586TEV) residue 1113 and BamB appeared to coincide with the interaction between passenger domain residues and BamA (compare Figs. 1 A–C and 2A). Taken together, the results show that Skp and BamB interact sequentially with residue 1113. Unlike the interaction between the passenger domain and BamA, however, the interaction between residue 1113 and BamB did not depend on the stalling of translocation but was prolonged by a mutation that slows passenger-domain secretion and cleavage. An ∼70-kDa band that could be immunoprecipitated with both anti-EspP C-terminal and BamB antisera was also observed in parallel with the proEspP-BamB adduct. Based on its size, this polypeptide almost certainly corresponds to the cleaved EspP β domain crosslinked to BamB. The isolation of this adduct demonstrates that the interaction of BamB with the β barrel persists even after passenger domain cleavage. When cells were incubated at 37 °C, the same overall crosslinking pattern was observed but was accelerated considerably (Fig. 2A, Lower gels).
Fig. 2.
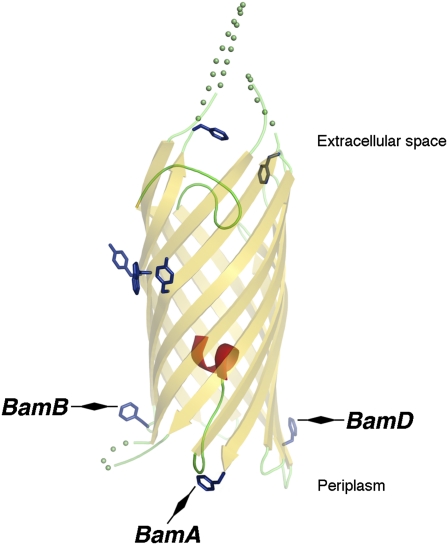
Crosslinking of EspP β domain residues 1113 and 1214 to Skp, BamB, and BamD. AD202 cells were transformed with pDULEBpa and a derivative of pRI22 or pRI23 harboring an amber codon at residue 1113 (A) or 1214 (B). Cells were subjected to pulse-chase labeling at 25 °C or 37 °C after the addition of IPTG, and samples were processed as described in the legend of Fig. 1. The crystal structure of the β domain (25) and the location of each residue are shown in the diagram on the right.
Interestingly, sequential interactions between EspP and Skp and another Bam complex lipoprotein, BamD, were observed when the photoprobe was incorporated at residue 1214. This residue also resides on the periplasmic side of the OM, but it is situated ∼120° away from residue 1113 on the circumference of the fully folded β domain (diagram in Fig. 2B). Like EspP(1113AMB) and EspP(TEV586/1113AMB), both EspP(1214AMB) and EspP(TEV586/1214AMB) were crosslinked to Skp at the 0.5 min time point at both 25 °C and 37 °C (Fig. 2B). At the same time points that EspP(1113AMB) and EspP(TEV586/1113AMB) were crosslinked to BamB, however, EspP(1214AMB) and EspP(TEV586/1214AMB) were crosslinked to BamD. Furthermore, an adduct that corresponds to the cleaved EspP β domain crosslinked to BamD was also observed when Bpa was incorporated into residue 1214 of either wild-type EspP or EspP(586TEV). Thus, the interaction between BamD and residue 1214, like the interaction between BamB and residue 1113, persists after passenger domain cleavage.
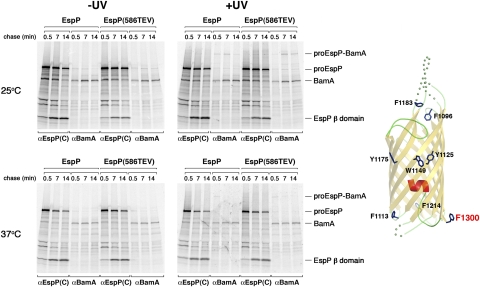
The periplasmic side of the EspP β barrel was crosslinked to BamA only when Bpa was incorporated at residue 1300, the C-terminal amino acid of the protein. This residue is located ∼120° away from both residues 1113 and 1214 on the circumference of the fully folded β domain (diagram in Fig. 3). Like the crosslinking of residues 1113 and 1214 to BamB and BamD, the crosslinking of residue 1300 to BamA was observed when Bpa was incorporated into either EspP or EspP(586TEV) (Fig. 3). In addition, the time course of the appearance and disappearance of the proEspP-BamA adduct was temperature dependent and was similar to that of the proEspP-BamB and proEspP-BamD adducts described above. We did not detect an ∼110-kDa adduct that corresponds to the free β domain crosslinked to BamA. The results suggest that the interaction of residue 1300 with BamA is terminated sooner than the interactions of residues 1113 and 1214 with BamB and BamD, respectively. In any case, the crosslinking of residue 1300 to BamA is noteworthy because it is consistent with the results of in vitro experiments that suggest that BamA specifically recognizes the sequence motif found at the C terminus of β barrel proteins (11). In this regard, it is also interesting that residue 1300 was the only amino acid in the EspP β domain that we examined that was not crosslinked to Skp or other ∼15- to 20-kDa proteins. The data raise the possibility that chaperones are excluded from binding to the C terminus of EspP to facilitate its recognition by BamA.
Fig. 3.
Crosslinking of EspP β domain residue 1300 to BamA. AD202 cells were transformed with pDULEBpa and a derivative of pRI22 or pRI23 harboring an amber codon at residue 1300. Cells were subjected to pulse-chase labeling at 25 °C or 37 °C after the addition of IPTG, and samples were processed as described in the legend of Fig. 1. The crystal structure of the β domain and the location of residue 1300 are shown in the diagram on the right.
By showing that each of three residues that are spatially separated on the base of the EspP β barrel interacts with a different component of the Bam complex, our crosslinking results suggest that BamA, BamB, and BamD at least partially surround the β barrel during its assembly. It is unlikely that the components function together as a simple channel, however, because elimination of BamB does not significantly affect EspP biogenesis or lead to the crosslinking of residue 1113 to a surrogate protein. Although the exact position of the three residues before the completion of β barrel assembly is unknown, the finding that the EspP β domain undergoes substantial folding before its integration into the OM (31) and the results of experiments described below suggest that the residues are relatively close to their ultimate positions when they interact with components of the Bam complex.
Crosslinking experiments in which Bpa was incorporated at other positions strongly suggested that most of the middle and outer segments of the EspP β barrel do not maintain a prolonged interaction with the Bam complex during β barrel assembly. When the photoprobe was incorporated into extracellular loop residues 1096 and 1183 or midbarrel residues 1125 and 1175 in either EspP or EspP(586TEV), an early interaction with Skp was observed, but crosslinks to Bam complex components were not detected (Figs. S5 and S6). A relatively weak crosslink between the midbarrel residue 1149 and BamB was observed, however, and a very weak crosslink to BamA was seen when EspP(586TEV/1149AMB) was examined (Fig. S7). The crosslinking data are consistent with the location of residue 1149 about halfway between residue 1113 and residue 1300 on the cylindrical surface of the fully assembled EspP β barrel. Taken together, these results provide evidence for the existence of a β barrel assembly intermediate in which the bulk of the barrel is located within the space of the OM (and therefore far from the Bam complex lipoproteins that reside in the periplasm).
Passenger Domain Secretion Is Initiated Before β Domain Assembly Is Complete.
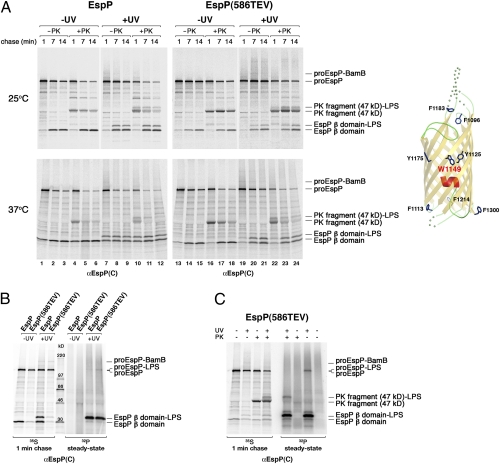
Although we did not detect strong crosslinking between midbarrel residues and Bam complex subunits, we did observe an extremely prominent crosslinking adduct that migrated at ∼33 kDa (lanes 7–12 and 19–24 in Fig. 4A and Figs. S6 and S7). This adduct appeared with the same kinetics as the cleaved β domain, and its level increased over time. Because the adduct is only ∼3 kDa larger than the β domain, we conjectured that it might represent the β domain crosslinked to a lipid, most likely LPS. To test this idea, we conducted pulse-chase labeling with 35S in parallel with 32P-labeling of cells that produced EspP(1149AMB) or EspP(TEV586/1149AMB) and then irradiated portions of each sample. As we predicted, the ∼33-kDa adduct was labeled with 32P and therefore contains a lipid moiety (Fig. 4B). Furthermore, we could detect the ∼33-kDa adduct on a Western blot probed with an anti-LPS antiserum (Fig. S8). Indeed the crosslinking of residues 1125, 1149, and 1175 to LPS is not surprising, because their midbarrel location implies that they reside in or very close to the outer leaflet of the OM when the β domain is fully assembled. The observation that residue 1150, which projects into the β barrel pore, was not crosslinked to lipid (Fig. S9) confirmed that the β domain was in a relatively native conformation when the ∼33-kDa adduct appeared.
Fig. 4.
Residue 1149 is exposed to lipid after the initiation of passenger domain secretion. (A) AD202 cells were transformed with pDULEBpa and a derivative of pRI22 or pRI23 harboring an amber codon at residue 1149. Cells were subjected to pulse-chase labeling at 25 °C or 37 °C after the addition of 200 μM IPTG, and half of each sample was UV irradiated. Both UV-irradiated and nonirradiated samples were divided in half, and one half was treated with PK. Equal portions of all the samples then were used for immunoprecipitations with an anti-EspP C-terminal antiserum. The crystal structure of the β domain and the location of residue 1149 are shown in the diagram on the right. (B and C) The experiment in A was repeated, except that a culture containing KH232PO4 was grown in parallel. UV irradiation (B) or both UV irradiation and PK treatment (C) of the 35S- and 32P-labeled cells was conducted as described above. Equal portions of all the samples were used for immunoprecipitations with an anti-EspP C-terminal antiserum.
Further analysis of the crosslinking of residue 1149 to LPS yielded strong evidence that passenger domain translocation begins before β barrel assembly is complete. The finding that residue 1149 of the pro form of EspP(586TEV) was crosslinked to a 32P-labeled molecule provided evidence that it was exposed to LPS before passenger domain cleavage (Fig. 4B). To determine whether the passenger domain emerges on the cell surface before this residue is exposed to LPS, we designed an experiment based on the observation that the folding of an ∼17-kDa C-terminal fragment of the EspP passenger domain is an early step in secretion (22, 32). Treatment of intact cells with PK leads to the generation of an ∼47-kDa C-terminal fragment that is derived from proEspP molecules whose passenger domains have been exposed on the cell surface but not yet cleaved. Because the cleavage of the EspP(586TEV) passenger domain is delayed, the ∼47-kDa C-terminal fragment persists longer than that of the wild-type protein (22).
We subjected cells that produced EspP(1149AMB) and EspP(586TEV/1149AMB) to pulse-chase labeling and UV irradiation as described above and then treated a portion of each sample with PK. As expected, a prominent ∼47-kDa C-terminal fragment was seen in samples that were PK treated but not UV irradiated (lanes 4–6, 10–12, 16–18, and 22–24 in Fig. 4A). Upon UV irradiation, an additional ∼50-kDa band that could be labeled with 32P and that presumably corresponds to an ∼47-kDa PK fragment-LPS crosslinking adduct was observed (lanes 10–12 and 22–24 in Fig. 4A, and Fig. 4C). Interestingly, the ratio of the ∼50-kDa fragment to the ∼47 kDa fragment increased over time. This effect was particularly clear when cells that produced EspP(586TEV/1149AMB) were incubated at 25 °C (lanes 22–24 in Fig. 4A, Upper gels). At this temperature, a significant amount of the ∼50-kDa band was not observed until 7 min. The data indicate that the transient stalling of passenger domain translocation results in a delayed exposure of residue 1149 to LPS and strongly suggest that the C terminus of the passenger domain is secreted before the β domain reaches its final position in the OM. Consistent with this interpretation, the 32P-labeled proEspP(586TEV)-LPS band was quantitatively converted to the ∼50-kDa band when cells were treated with PK (Fig. 4C). This result shows that the passenger domain of the entire population of proEspP(586TEV) that is exposed on the cell surface is also in contact with LPS. If exposure of residue 1149 to LPS occurred first, we would have expected a portion of the proEspP-LPS band to be resistant to PK digestion. Taken together, the results imply that after the onset of passenger domain secretion either the β barrel undergoes a conformational change or the segment of the β barrel containing residue 1149 is released from a proteinaceous environment.
Additional information about the disposition of the β domain during the early stages of passenger domain secretion emerged from experiments in which a linker was inserted into the third extracellular loop of the EspP β barrel. AD202 cells transformed with a plasmid that encodes EspP, EspP containing the linker insertion [EspP(L3ins)], or EspP(586TEV) containing the linker insertion [EspP(586TEV/L3ins)] were subjected to pulse-chase labeling at 37 °C after the addition of IPTG, and a portion of each sample was treated with PK. Although the β domain of EspP was insensitive to exogenously added PK, an ∼18-kDa C-terminal fragment that resulted from the cleavage of the extended loop of EspP(L3ins) and EspP(586TEV/L3ins) began to appear at early time points (Fig. S10). Based on the susceptibility of the pro form of the protein to PK digestion, the appearance of this fragment seemed to coincide with the onset of passenger domain secretion. In any case, because the peptide insertion at residue 586 prolongs the translocation reaction, it was clear that the ∼18-kDa fragment could be generated long before the EspP(586TEV/L3ins) passenger domain was completely secreted. Taken together, the results strongly suggest that even though the β barrel is incompletely assembled at the onset of passenger domain secretion, it is positioned so that its loops penetrate the OM.
Discussion
In this study we used site-specific crosslinking in combination with pulse-chase labeling to identify intermediates in the assembly of a model OMP, the EspP β domain. Consistent with previous results that suggest that Skp functions at an early stage of OMP biogenesis, we found that widely dispersed hydrophobic residues that ultimately reside on the exterior surface of the folded β barrel initially interact with this chaperone. Although Skp appears to maintain OmpA in an unfolded state (37), it is unclear whether Skp binds to an unfolded form of the EspP β domain or to a more structured intermediate that has been inferred from biochemical studies (31). Furthermore, because Skp forms a trimer whose binding cavity appears to be too small to accommodate the entire β domain (37), multiple Skp trimers may interact with the ∼30-kDa fragment. Based on recent in vitro experiments, it is possible that the highly basic Skp protein targets the β domain directly to the negatively charged inner leaflet of the OM (38). However, it also is conceivable that other chaperones that have been implicated in OMP biogenesis (SurA and DegP) bind to the EspP β domain but that these interactions are too transient to be detected by our methods. At later time points we observed interactions between residues that are located on the periplasmic side of the β domain and BamA, BamB, and BamD that likely occur after much of the β barrel traverses the OM. Strikingly, the data showed that each Bam complex subunit contacts a different side of the β barrel and suggested that the complex might form a channel-like structure. Although stable interactions between the Bam complex and the β domain of wild-type EspP were seen, the finding that a mutation that stalls passenger domain secretion prolonged the interactions demonstrated a link between passenger domain secretion and β domain assembly. The link between the two processes serves as a valuable checkpoint to ensure that passenger domain cleavage does not occur until the entire domain is extruded across the OM. Furthermore, the observation that BamB and BamD bind to the cleaved β domain suggests that they function at a later step of assembly than BamA.
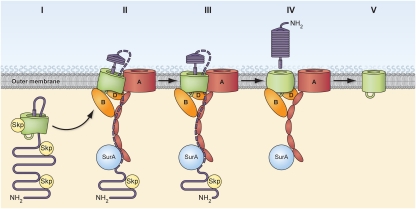
Based on the results of this and previous studies, we propose an integrated model for the biogenesis of both autotransporter domains. In our model, Skp binds to both domains following (or even during) the translocation of EspP into the periplasm, presumably by recognizing the amphipathic β strands that are present throughout the protein. The β domain then undergoes substantial folding, and an α-helical segment that encompasses the passenger domain–β domain junction is incorporated into the cavity of the resulting “protobarrel” structure (Fig. 5, step I) (31). Subsequently EspP is targeted to the OM, at least in part through the interaction of BamA with the C terminus of the β domain. The β domain then penetrates the OM through the combined action of multiple Bam complex subunits, including BamA, BamB, and BamD. BamA concurrently facilitates the initiation of passenger domain secretion (Fig. 5, step II). At this stage Skp dissociates from the β domain but remains associated with periplasmically disposed segments of the passenger domain. During passenger domain secretion, an additional step(s) in β domain assembly exposes midbarrel residues (e.g., W1149) to LPS and presumably moves the β barrel to its final position (Fig. 5, step III). The secretion and processing of the passenger domain is completed, at which point only BamB and BamD remain bound to the β domain (Fig. 5, step IV). Shortly after the passenger domain is released, the Bam complex lipoproteins dissociate, presumably after promoting a final step in β domain assembly (Fig. 5, step V). Although one might imagine that the Bam complex promotes a conformational change in the β barrel that is required to align the residues that catalyze passenger-domain cleavage, the role of the Bam complex after the release of the passenger domain is unclear. However, because EspP cleavage mutants are not toxic and therefore do not appear to sequester the Bam complex, it is unlikely that the dissociation of the Bam complex from the β domain requires the acquisition of a specific postcleavage conformation. In any case, our data imply that the interaction of the β domain with the Bam complex (which we observed under all conditions) is more prolonged than the interaction of the passenger domain with BamA (which can be detected only when translocation stalls).
Fig. 5.
Model of EspP biogenesis. After EspP is transported across the inner membrane, both the passenger domain (purple) and the β domain (green) interact with Skp. The β domain rapidly begins to fold, and a hairpin that includes a peptide that spans the passenger domain–β domain junction and a more N-terminal segment of the passenger domain is incorporated into the cavity of the partially folded protobarrel structure (step I). EspP then is targeted to the OM, where the C terminus of the β domain is recognized by BamA. Skp dissociates from the β domain, and BamA, BamB, and BamD (perhaps in concert with other factors) initiate the integration of the β domain into the OM by interacting with distinct surfaces of the client protein (step II). BamA concomitantly promotes the initiation of passenger domain secretion. The path taken by the passenger domain across the OM is unknown, but it may move progressively from SurA to the BamA POTRA domains and then traverse the OM through a channel formed between the EspP β domain and BamA (dotted purple line). After the initiation of passenger domain secretion, midbarrel residues in the EspP β domain are exposed to LPS (step III). Once the passenger domain is fully secreted and cleaved, only BamB and BamD remain associated with the β domain (step IV). Finally, assembly of the β domain is completed, and BamB and BamD dissociate (step V). For clarity, the BamC and BamE subunits of the Bam complex are not shown.
The identity of the channel that facilitates passenger-domain secretion and the reason why the completion of β barrel assembly is contingent upon the completion of passenger domain secretion remain to be determined. Although it is unknown whether the crosslinking of the EspP(586TEV) passenger domain to BamA is caused by an interaction with the BamA β barrel the with POTRA domains, or with both, it is unclear how the passenger domain would enter the pore of the BamA β barrel if, as our results suggest, the β domain does not pass through the same channel. In this regard, it is interesting that although passenger domain residues that are relatively close to the stall point are crosslinked to BamA but not SurA, amino acids that are located ∼80 residues away are crosslinked to both BamA and SurA (Fig. 1C and ref. 31). Given that SurA appears to interact with the first POTRA domain of BamA (15), the data suggest that passenger domain secretion might involve a stepwise transfer of polypeptide segments from SurA to the POTRA domains (Fig. 5, step II, dotted purple line) and subsequent transport through an OM channel. One attractive possibility is that the passenger domain is secreted through a heterooligomeric channel formed partly by the β domain and partly by BamA, Bam lipoproteins, and perhaps other factors that simultaneously envelop the β domain (Fig. 5, step II). The crosslinking of EspP(586TEV) residues 575 and 600, which are close to the point of passenger domain stalling, to BamB and other unidentified proteins (Fig. 1A and ref. 21) is consistent with this hypothesis. In the proposed scenario, the β domain protobarrel that forms in the periplasmic space would accommodate the C-terminal α-helical segment of the passenger domain (31) as well as a more N-terminal segment and would be partially open (Fig. 5, step I). The passenger domain would be extruded across the OM through a β domain pore that is held open on one side by BamA. A major strength of this hypothesis is that it would account for the obligate coupling of passenger domain secretion and β domain assembly by implying that the β domain cannot adopt a native conformation until after passenger domain secretion is terminated. In addition, if the β domain channel can be held open to different degrees by BamA, this hypothesis also might account for the secretion of folded polypeptides via the autotransporter pathway (23, 30).
Our analysis of the stages of assembly of the EspP β domain may provide significant insight into the biogenesis of the broader class of bacterial β barrel proteins. Most notably, our results suggest that the Bam complex lipoproteins, whose function has been mysterious, contribute directly to the assembly of β barrel proteins. It might be expected that BamA plays a predominant role in the assembly process because it is the only conserved subunit. Sam50, a mitochondrial ortholog that promotes the assembly of mitochondrial OM proteins, forms a complex with proteins that are completely unrelated to the Bam complex lipoproteins. Our results suggest that the membrane integration of β barrel proteins involves residues on the outer surface of BamA (instead of the BamA pore) and raise the possibility that the function of BamA is limited to recognizing the conserved C-terminal motif of client proteins. The observation that BamB and BamD are structurally related to proteins that have ligand binding activity (16–19) is consistent with the notion that they interact directly with β barrel proteins. Recent NMR studies also have shown that BamE binds phosphatidylglycerol in a pocket that resides near the site of interaction with BamD (39). The lipid binding activity might help disrupt the OM locally in the vicinity of BamD or might help expose the outer surface of β barrel proteins to the lipid bilayer. Furthermore, our results suggest that periplasmic chaperones do not play the same role in the biogenesis of all OMPs. Although the assembly competence of the major OMPs in the periplasm is maintained primarily by SurA (2), we did not detect an interaction between SurA and the EspP β domain. Instead, the β domain interacted stably with Skp, whereas SurA appeared to have a specialized role in transferring segments of the passenger domain to BamA. One possible explanation of the data is that Skp interacts with OMPs as they emerge on the periplasmic side of the inner membrane and then typically transfers them to SurA. SurA then binds to BamA and facilitates β barrel assembly in concert with the POTRA domains. Indeed there is evidence that SurA either directly or indirectly facilitates β barrel folding (40). In the case of autotransporter β domains, however, the presence of an intrabarrel segment catalyzes folding by a different pathway, and the role of SurA is limited to presenting the β-helical passenger domain to the secretion machinery in an appropriate conformation.
Materials and Methods
Strains, Antibiotics, and Antisera.
The strains used in this study were AD202 (MC4100 ompT::kan (41), HDB131 (AD202 Δskp), and HDB133 (AD202 bamB::kan). Antisera generated against EspP N- and C-terminal peptides and purified BamA have been described (22, 42). Anti-SurA and anti-BamB antisera were provided by Rajeev Misra (Arizona State University, Tempe, AZ) and Dan Kahne (Harvard University, Cambridge, MA), respectively, and anti-Skp, anti-BamC, anti-BamD, and anti-LPS antisera were provided by Natacha Ruiz and Tom Silhavy (Princeton University, Princeton, NJ). The anti-LPS antiserum was originally obtained from HyCult Biotech. Ampicillin (100 μg/mL) and tetracycline (5 μg/mL) were added as needed.
Plasmid Construction.
Plasmids pRLS5 (pTRC99a-EspP), pJH97 [pTRC99a-EspP(586TEV)], pRI22 (Plac-10His-EspP), and pRI23 [Plac-10His-EspP(586TEV)] have been described (22, 42). Amber mutations were introduced into pRI22 and pRI23 by site-directed mutagenesis using the QuikChange mutagenesis kit (Stratagene). To insert linkers into extracellular loop 3 of the EspP β domain, a Pst I site first was introduced into pRLS5 and pJH97 using the oligonucleotide 5′-CACTGCAACCTTTGCCCTGCAGGGACTCGGAACCCG-3′ and its complement. Tandem TEV sites then were introduced into the Pst I site using the oligonucleotides PstTev(+) and PstTev(−) (22) to generate plasmids pPT17 and pPT18.
Pulse-Chase Labeling and Photocrosslinking.
Cells were grown at 37 °C in M9 medium containing 0.2% glycerol and all the l-amino acids (40 μg/mL) except methionine and cysteine. Overnight cultures were washed and diluted into fresh M9 medium at OD550 = 0.02–0.03. When cultures reached OD550 = 0.2, EspP synthesis was induced by the addition of 200 μM IPTG unless otherwise noted. After 30 min, cells were subjected to pulse-chase labeling as described (22). In experiments performed at low temperature, cultures were shifted to 25 °C 5 min before pulse labeling. In photocrosslinking experiments, 1 mM Bpa was added along with the inducer. At each time point, duplicate 4-mL aliquots were pipetted over ice into a 15-mL tube (untreated sample) or into a six-well plate and were UV irradiated for 4 min as described (22). Cells were concentrated by centrifugation (3,500 × g, 10 min, 4 °C) and resuspended in 1 mL M9 medium. Proteins in all samples were collected by trichloroacetic acid (TCA) precipitation. In some experiments, the resuspended cells were divided in half, and one half was treated with PK as described (22) before TCA precipitation. Immunoprecipitations were performed as described (31), and proteins were resolved by SDS/PAGE on 8–16% minigels (Invitrogen).
32P Labeling.
Overnight cultures grown in modified G56 medium [45 mM MES (pH 7.0), 10 mM KCl, 10 mM MgCl2, 5 mM NaCl, 15 mM (NH4)2SO4, 5 μg/L thiamine, 0.2% glycerol, and 40 μg/mL l-amino acids (except methionine and cysteine)] (43) containing 0.13 mM KH2PO4.were washed twice with medium without phosphate and diluted to OD550 = 0.03 in medium containing either 0.13 mM KH2PO4 or 133 μCi/mL KH232PO4 (900–1,100 mCi/mM; PerkinElmer) to start two parallel cultures. When the cultures reached OD550 = 0.3, 1 mM Bpa and 200 μM IPTG were added. Pulse-chase labeling and UV irradiation of cells grown in medium containing KH2PO4 was performed as described above. Subsequently, two 6-mL aliquots from the culture containing KH232PO4 were pipetted into a 15-mL tube and chilled on ice (untreated sample) or placed in a six-well plate on ice and UV irradiated for 7 min as described (22). Cells were concentrated by centrifugation (3,500 × g, 10 min, 4 °C) and resuspended in M9 before PK treatment or TCA precipitation.
Supplementary Material
Acknowledgments
We thank Susan Buchanan, Jim Fairman, and Olga Pavlova for critical reading of the manuscript. We thank Travis Barnard for providing illustrations of the crystal structure of the EspP β domain and the National Institutes of Health Division of Medical Arts for helping to construct Fig. 5. This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Author Summary on page 12577.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1103827108/-/DCSupplemental.
References
- 1.Rizzitello AE, Harper JR, Silhavy TJ. Genetic evidence for parallel pathways of chaperone activity in the periplasm of Escherichia coli. J Bacteriol. 2001;183:6794–6800. doi: 10.1128/JB.183.23.6794-6800.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sklar JG, Wu T, Kahne D, Silhavy TJ. Defining the roles of the periplasmic chaperones SurA, Skp, and DegP in Escherichia coli. Genes Dev. 2007;21:2473–2484. doi: 10.1101/gad.1581007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schäfer U, Beck K, Müller M. Skp, a molecular chaperone of gram-negative bacteria, is required for the formation of soluble periplasmic intermediates of outer membrane proteins. J Biol Chem. 1999;274:24567–24574. doi: 10.1074/jbc.274.35.24567. [DOI] [PubMed] [Google Scholar]
- 4.Harms N, et al. The early interaction of the outer membrane protein phoe with the periplasmic chaperone Skp occurs at the cytoplasmic membrane. J Biol Chem. 2001;276:18804–18811. doi: 10.1074/jbc.M011194200. [DOI] [PubMed] [Google Scholar]
- 5.Voulhoux R, Bos MP, Geurtsen J, Mols M, Tommassen J. Role of a highly conserved bacterial protein in outer membrane protein assembly. Science. 2003;299:262–265. doi: 10.1126/science.1078973. [DOI] [PubMed] [Google Scholar]
- 6.Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Sklar JG, et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci USA. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hagan CL, Kim S, Kahne D. Reconstitution of outer membrane protein assembly from purified components. Science. 2010;328:890–892. doi: 10.1126/science.1188919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malinverni JC, et al. YfiO stabilizes the YaeT complex and is essential for outer membrane protein assembly in Escherichia coli. Mol Microbiol. 2006;61:151–164. doi: 10.1111/j.1365-2958.2006.05211.x. [DOI] [PubMed] [Google Scholar]
- 10.Gentle IE, Burri L, Lithgow T. Molecular architecture and function of the Omp85 family of proteins. Mol Microbiol. 2005;58:1216–1225. doi: 10.1111/j.1365-2958.2005.04906.x. [DOI] [PubMed] [Google Scholar]
- 11.Robert V, et al. Assembly factor Omp85 recognizes its outer membrane protein substrates by a species-specific C-terminal motif. PLoS Biol. 2006;4:e377. doi: 10.1371/journal.pbio.0040377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim S, et al. Structure and function of an essential component of the outer membrane protein assembly machine. Science. 2007;317:961–964. doi: 10.1126/science.1143993. [DOI] [PubMed] [Google Scholar]
- 13.Gatzeva-Topalova PZ, Warner LR, Pardi A, Sousa MC. Structure and flexibility of the complete periplasmic domain of BamA: The protein insertion machine of the outer membrane. Structure. 2010;18:1492–1501. doi: 10.1016/j.str.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bennion D, Charlson ES, Coon E, Misra R. Dissection of β barrel outer membrane protein assembly pathways through characterizing BamA POTRA 1 mutants of Escherichia coli. Mol Microbiol. 2010;77:1153–1171. doi: 10.1111/j.1365-2958.2010.07280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vuong P, Bennion D, Mantei J, Frost D, Misra R. Analysis of YfgL and YaeT interactions through bioinformatics, mutagenesis, and biochemistry. J Bacteriol. 2008;190:1507–1517. doi: 10.1128/JB.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heuck A, Schleiffer A, Clausen T. Augmenting β-augmentation: Structural basis of how BamB binds BamA and may support folding of outer membrane proteins. J Mol Biol. 2011;406:659–666. doi: 10.1016/j.jmb.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 17.Kim KH, Paetzel M. Crystal structure of Escherichia coli BamB, a lipoprotein component of the β barrel assembly machinery complex. J Mol Biol. 2011;406:667–678. doi: 10.1016/j.jmb.2010.12.020. [DOI] [PubMed] [Google Scholar]
- 18.Noinaj N, Fairman JW, Buchanan SK. The crystal structure of BamB suggests interactions with BamA and its role within the BAM complex. J Mol Biol. 2011;407:248–260. doi: 10.1016/j.jmb.2011.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandoval CM, Baker SL, Jansen K, Metzner SI, Sousa MC. Crystal structure of BamD: An essential component of the β barrel assembly machinery of Gram-negative bacteria. J Mol Biol. 2011 doi: 10.1016/j.jmb.2011.03.035. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein HD. In: EcoSal—Escherichia coli and Salmonella: Cellular and Molecular Biology. Böck A, et al., editors. Washington: ASM; 2010. http://www.ecosal.org, Chap 4.3.6. [Google Scholar]
- 21.Junker M, Besingi RN, Clark PL. Vectorial transport and folding of an autotransporter virulence protein during outer membrane secretion. Mol Microbiol. 2009;71:1323–1332. doi: 10.1111/j.1365-2958.2009.06607.x. [DOI] [PubMed] [Google Scholar]
- 22.Ieva R, Bernstein HD. Interaction of an autotransporter passenger domain with BamA during its translocation across the bacterial outer membrane. Proc Natl Acad Sci USA. 2009;106:19120–19125. doi: 10.1073/pnas.0907912106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Skillman KM, Barnard TJ, Peterson JH, Ghirlando R, Bernstein HD. Efficient secretion of a folded protein domain by a monomeric bacterial autotransporter. Mol Microbiol. 2005;58:945–958. doi: 10.1111/j.1365-2958.2005.04885.x. [DOI] [PubMed] [Google Scholar]
- 24.Oomen CJ, et al. Structure of the translocator domain of a bacterial autotransporter. EMBO J. 2004;23:1257–1266. doi: 10.1038/sj.emboj.7600148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barnard TJ, Dautin N, Lukacik P, Bernstein HD, Buchanan SK. Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nat Struct Mol Biol. 2007;14:1214–1220. doi: 10.1038/nsmb1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Berg B. Crystal structure of a full-length autotransporter. J Mol Biol. 2010;396:627–633. doi: 10.1016/j.jmb.2009.12.061. [DOI] [PubMed] [Google Scholar]
- 27.Zhai Y, et al. Autotransporter passenger domain secretion requires a hydrophobic cavity at the extracellular entrance of the β-domain pore. Biochem J. 2011;435:577–587. doi: 10.1042/BJ20101548. [DOI] [PubMed] [Google Scholar]
- 28.Pohlner J, Halter R, Beyreuther K, Meyer TF. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature. 1987;325:458–462. doi: 10.1038/325458a0. [DOI] [PubMed] [Google Scholar]
- 29.Tajima N, Kawai F, Park SY, Tame JR. A novel intein-like autoproteolytic mechanism in autotransporter proteins. J Mol Biol. 2010;402:645–656. doi: 10.1016/j.jmb.2010.06.068. [DOI] [PubMed] [Google Scholar]
- 30.Jong WS, et al. Limited tolerance towards folded elements during secretion of the autotransporter Hbp. Mol Microbiol. 2007;63:1524–1536. doi: 10.1111/j.1365-2958.2007.05605.x. [DOI] [PubMed] [Google Scholar]
- 31.Ieva R, Skillman KM, Bernstein HD. Incorporation of a polypeptide segment into the β-domain pore during the assembly of a bacterial autotransporter. Mol Microbiol. 2008;67:188–201. doi: 10.1111/j.1365-2958.2007.06048.x. [DOI] [PubMed] [Google Scholar]
- 32.Peterson JH, Tian P, Ieva R, Dautin N, Bernstein HD. Secretion of a bacterial virulence factor is driven by the folding of a C-terminal segment. Proc Natl Acad Sci USA. 2010;107:17739–17744. doi: 10.1073/pnas.1009491107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dautin N, Barnard TJ, Anderson DE, Bernstein HD. Cleavage of a bacterial autotransporter by an evolutionarily convergent autocatalytic mechanism. EMBO J. 2007;26:1942–1952. doi: 10.1038/sj.emboj.7601638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc Natl Acad Sci USA. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Farrell IS, Toroney R, Hazen JL, Mehl RA, Chin JW. Photo-cross-linking interacting proteins with a genetically encoded benzophenone. Nat Methods. 2005;2:377–384. doi: 10.1038/nmeth0505-377. [DOI] [PubMed] [Google Scholar]
- 36.Plath K, Mothes W, Wilkinson BM, Stirling CJ, Rapoport TA. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- 37.Walton TA, Sandoval CM, Fowler CA, Pardi A, Sousa MC. The cavity-chaperone Skp protects its substrate from aggregation but allows independent folding of substrate domains. Proc Natl Acad Sci USA. 2009;106:1772–1777. doi: 10.1073/pnas.0809275106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patel GJ, Behrens-Kneip S, Holst O, Kleinschmidt JH. The periplasmic chaperone Skp facilitates targeting, insertion, and folding of OmpA into lipid membranes with a negative membrane surface potential. Biochemistry. 2009;48:10235–10245. doi: 10.1021/bi901403c. [DOI] [PubMed] [Google Scholar]
- 39.Knowles TJ, et al. Structure and function of BamE within the outer membrane and the β barrel assembly machine. EMBO Rep. 2011;12:123–128. doi: 10.1038/embor.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouvière PE, Gross CA. SurA, a periplasmic protein with peptidyl-prolyl isomerase activity, participates in the assembly of outer membrane porins. Genes Dev. 1996;10:3170–3182. doi: 10.1101/gad.10.24.3170. [DOI] [PubMed] [Google Scholar]
- 41.Akiyama Y, Ito K. SecY protein, a membrane-embedded secretion factor of E. coli, is cleaved by the ompT protease in vitro. Biochem Biophys Res Commun. 1990;167:711–715. doi: 10.1016/0006-291x(90)92083-c. [DOI] [PubMed] [Google Scholar]
- 42.Szabady RL, Peterson JH, Skillman KM, Bernstein HD. An unusual signal peptide facilitates late steps in the biogenesis of a bacterial autotransporter. Proc Natl Acad Sci USA. 2005;102:221–226. doi: 10.1073/pnas.0406055102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ganong BR, Leonard JM, Raetz CR. Phosphatidic acid accumulation in the membranes of Escherichia coli mutants defective in CDP-diglyceride synthetase. J Biol Chem. 1980;255:1623–1629. [PubMed] [Google Scholar]