Abstract
GPR56, an orphan G protein-coupled receptor (GPCR) from the family of adhesion GPCRs, plays an indispensable role in cortical development and lamination. Mutations in the GPR56 gene cause a malformed cerebral cortex in both humans and mice that resembles cobblestone lissencephaly, which is characterized by overmigration of neurons beyond the pial basement membrane. However, the molecular mechanisms through which GPR56 regulates cortical development remain elusive due to the unknown status of its ligand. Here we identify collagen, type III, alpha-1 (gene symbol Col3a1) as the ligand of GPR56 through an in vitro biotinylation/proteomics approach. Further studies demonstrated that Col3a1 null mutant mice exhibit overmigration of neurons beyond the pial basement membrane and a cobblestone-like cortical malformation similar to the phenotype seen in Gpr56 null mutant mice. Functional studies suggest that the interaction of collagen III with its receptor GPR56 inhibits neural migration in vitro. As for intracellular signaling, GPR56 couples to the Gα12/13 family of G proteins and activates RhoA pathway upon ligand binding. Thus, collagen III regulates the proper lamination of the cerebral cortex by acting as the major ligand of GPR56 in the developing brain.
Keywords: brain malformation, meningeal fibroblasts
During cerebral cortical development, most cortical neurons are born deep in specialized proliferative regions near the lining of the lateral ventricles and migrate out to take up residence in the surface layer of the brain, eventually forming a highly folded sheet of six neuronal layers (1–5). Disruptions in normal neuronal migration and positioning lead to cortical disorders, one of which is cobblestone lissencephaly (6). Cobblestone lissencephaly is typically seen in three distinct human congenital muscular dystrophy syndromes: muscle-eye-brain disease, Fukuyama-type congenital muscular dystrophy, and Walker–Warburg syndrome (6). Mutant mice with deletions in some members of the integrin pathway molecules and the extracellular matrix (ECM) constituents also exhibit cortical migration defects with deficiencies in basal lamina integrity and cortical ectopias, features that resemble the human cobblestone malformation (7–12). The suggested mechanism leading to cobblestone lissencephaly has been a defective pial basement membrane (BM) (6). However, recent literature demonstrates that abnormal neuronal migration may account partially for the improper formation of the cobblestone-like cortex (13–16).
GPR56 is an orphan G protein-coupled receptor (GPCR) from the family of adhesion GPCRs. Mutations in GPR56 cause a specific human brain malformation called bilateral frontoparietal polymicrogyria (BFPP), an autosomal recessively inherited developmental disorder of the brain (17–20). Magnetic resonance imaging of BFPP brains shows numerous (poly) small (micro) gyri, which extend diffusely across the frontal and parietal lobes with a decreasing anterior-to-posterior gradient of severity (17–20). Further studies in Gpr56 knockout mice and postmortem human BFPP brain specimen revealed that the histopathology of BFPP is a cobblestone-like cortical malformation (21, 22). However, the molecular and cellular mechanisms of how GPR56 regulates brain development remain largely unknown.
GPR56 is cleaved into an N- and a C-terminal fragment through a GPCR proteolytic site-mediated autoproteolytic process (23–25). Previously, we used the soluble N-terminal fragment of GPR56 (GPR56N) as a probe to reveal the expression of the putative ligand of GPR56 in the meninges and pial BM of the developing brain (21). Here we identify the ligand of GPR56 as collagen III. We further reveal that collagen III is one of the key constituents of the pial BM, in addition to being a major collagen in the connective tissues. Moreover, we demonstrate that the interaction of GPR56 with its ligand collagen III inhibits neuronal migration by coupling to Gα12/13 and activating the RhoA pathway. Our data confirms a unique receptor-ligand pair that regulates cerebral cortical development.
Results
Meningeal Fibroblasts Express the Putative Ligand of GPR56.
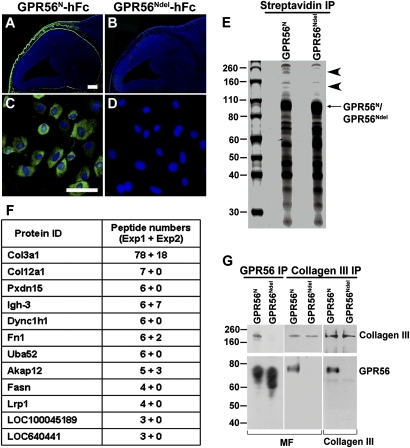
Using human Fc-tagged GPR56N (GPR56N-hFc), we revealed the presence of the putative ligand of GPR56 in the meninges and pial BM of the developing brain (Fig. 1A and Fig. S1) (21). To demonstrate the specificity of the ligand binding assay, we engineered a deletion mutant of GPR56N (GPR56Ndel) by deleting amino acids 93–143. We previously showed that GPR56Ndel was normally glycosylated and secreted into the conditioned media but lost the binding capacity to the putative ligand, allowing it to serve as a negative control (21).
Fig. 1.
Identification of GPR56 ligand. (A–D) GPR56N-hFc binds specifically the putative ligand of GPR56 (green) in meninges and pial BM (A) and MFs (C). GPR56Ndel-hFc was used as a negative control (B and D). A and B are composite images combined into a single image. Nuclear counterstain was performed by Hoechst 33342 (blue). (Scale bars: A and B, 200 μm; C and D, 100 μm.) (E) SDS/PAGE analysis of purified GPR56 ligand(s) eluted from streptavidin affinity resin. The putative ligand of GPR56 is a large protein with a molecular weight of 160–260 kDa (arrowheads). (F) Summary of MS results from two independent purifications. Total peptides sequenced and proteins specifically identified by GPR56N are indicated. Collagen III appeared as the top candidate based on the high number of matched peptides. (G) The association of collagen III and GPR56N was confirmed by co-IP. Collagen III was specifically pulled down by GPR56N, but not GPR56Ndel. Conversely, only GPR56N was pulled down by anti-collagen III antibody, but not GPR56Ndel.
Because meningeal fibroblasts (MFs) contribute to the pial BM by secreting and organizing the majority of basal lamina constituents, we investigated whether they express the ligand(s) of GPR56. We performed a putative ligand binding assay on primary MFs. An abundance of specific putative ligand binding in MFs was detected, providing a source for the subsequent ligand search (Fig. 1C).
GPR56N Binds Collagen III.
To identify the ligand of GPR56, we used an in vitro biotinylation/proteomics approach (Fig. S2). A single-step capture on streptavidin beads was performed to purify the binding partner(s) of GPR56, using MFs as the ligand source and the biotinylated GPR56N as a probe, and the biotinylated GPR56Ndel served as a negative control (Fig. S2). SDS/PAGE revealed the putative ligand(s) to be a large protein with a molecular weight of 160–260 kDa (Fig. 1E). The specific protein bands, together with the corresponding regions of the negative control, were excised and subjected to MS analysis. Results from two independent pull-down assays and MS analyses identified collagen III as the top candidate (Fig. 1F).
We next validated the interaction between collagen III and GPR56 by performing a series of coimmunoprecipitation (co-IP) experiments. To remove the background created by mouse IgG Fc, we engineered GPR56N and GPR56Ndel constructs by replacing the mFc-biotag with a Flag tag and an in vivo biotinylation signal peptide (Fig. S3). In vivo biotinylated GPR56N and GPR56Ndel proteins were used in the following co-IP experiments. Collagen III was found to specifically associate with GPR56N, but not GPR56Ndel (Fig. 1G). Conversely, we identified GPR56N, but not GPR56Ndel, in the collagen III IP complex by reverse co-IP using anti-collagen III antibody (Fig. 1G). We thus confirmed collagen III as a true binding partner of GPR56, indicating an as-yet-unrecognized function for collagen III in brain development.
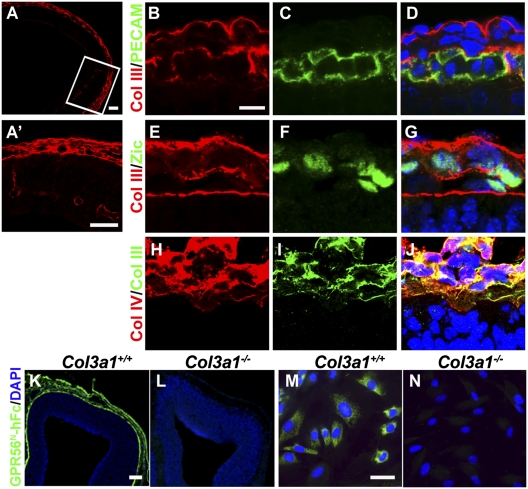
Collagen III is expressed in the meninges and blood vessels in the developing brain. Collagen III is a major collagen in connective tissues, with integrins α1β1 and α2β1 serving as its typical receptors (26, 27). There has been only scant literature suggesting the presence of collagen III in the brain through microarray analysis (28). To establish the expression pattern of collagen III in the brain, we performed collagen III immunohistochemistry (IHC) on embryonic mouse brains. The specificity of anti-collagen III antibody was confirmed by the absence of collagen III immunostaining on Col3a1−/− brain sections (Fig. S4). We detected an abundance of collagen III immunostaining in the meninges, pial BM, and blood vessels at embryonic day (E)12.5 (Fig. 2 A and A′). Double IHC of collagen III and either platelet endothelial cell adhesion molecule (PECAM) or Zic on E11.5 mouse brain sections demonstrated that only MFs, and not endothelial cells, express collagen III (Fig. 2 B–G). Further double IHC of collagen III and a known ECM constituent of the pial BM, collagen IV, confirmed that collagen III is an ECM component in the developing brain, and its location is consistent with its role as a ligand for GPR56 (Fig. 2 H–J).
Fig. 2.
Collagen III is the major binding partner of GPR56 in the mouse developing brain. (A) Collagen III immunostaining in E12.5 developing cortex. (A′) Higher-magnification views of the boxed region. (B–G) Double IHC of collagen III/PECAM and collagen III/Zic on E11.5 wild-type mouse brains. (H–J) Double IHC of collagen III (green) and collagen IV (red) in E12.5 neocortex. (K and L) GPR56N-hFc (green) detected a strong signal in the meninges and pial BM of E12.5 Col3a1+/+ brain but failed to detect any signals in the Col3a1−/− brain. Nuclear counterstain was performed by Hoechst 33342 (blue). (M and N) Abundant GPR56N-hFc–specific (green) binding was detected in primary MFs derived from the meninges of Col3a1+/+ but not Col3a1−/− mice. Nuclear counterstain was performed by Hoechst 33342 (blue). (Scale bars: A and K–N, 100 μm; A′, 50 μm; B–J, 10 μm.)
Collagen III Is the Major Binding Partner of GPR56 in the Developing Brain.
To examine whether collagen III is the major ligand of GPR56 in the developing brain, we performed a putative ligand binding assay on E12.5 brain sections of Col3a1 wild-type and null mutant embryos. We failed to detect any GPR56N-hFc binding signals in Col3a1 null mutant mouse brains, indicating that the deletion of Col3a1 eliminates the pial binding partner of GPR56N (Fig. 2L). This finding was further confirmed by the absence of GPR56N-hFc binding in the MFs derived from Col3a1−/− mice (Fig. 2N). Taken together, this data suggests that collagen III is the major binding partner of GPR56 in the mouse embryonic brain.
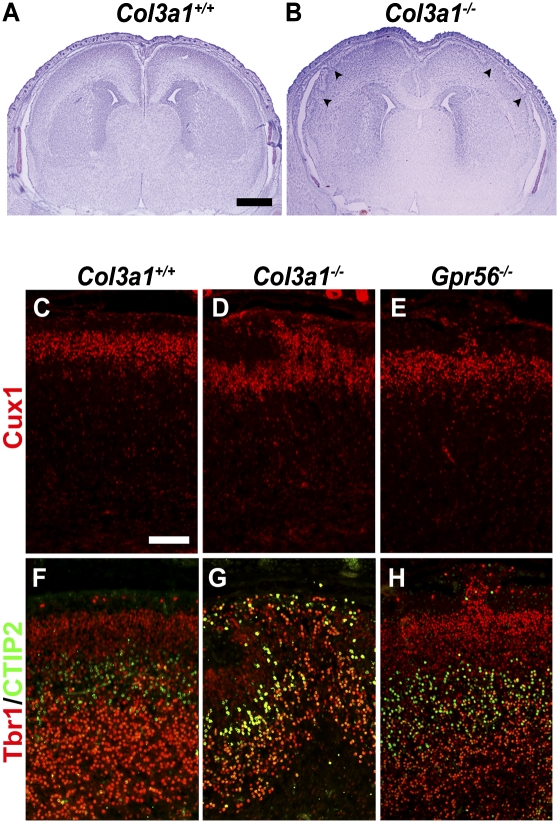
Deleting the mouse Col3a1 results in cobblestone lissencephaly as seen in Gpr56 null mutant mice. The brain phenotype of Col3a1 mutant mice has not been described previously. Examination of brain histology of Col3a1 mutant mice revealed frequent subpial neuronal ectopias (Fig. 3B). Only homozygous mutant embryos were phenotypically affected, whereas heterozygotes appeared to have a normal neocortex (Fig. S5). Notably, subpial ectopias occurred exclusively in the frontal regions bilaterally, resembling the anatomical distribution observed in Gpr56 null mutant mice (Fig. 3B and Fig. S5) (21).
Fig. 3.
Deletion of the Col3a1 gene results in neuronal ectopias and abnormal cortical lamination. (A and B) Nissl staining of E18.5 Col3a1+/+ and Col3a1−/− brains. Neuronal ectopias (arrowheads) were seen in Col3a1−/− mouse brain (B). (C–H) Immunostaining of Cux1 (red in C–E), Tbr1 (red in F–H), and CTIP2 (green in F–H) in E18.5 Col3a1+/+, Col3a1−/−, and Gpr56−/− brains. Neurons positive for Cux1, Tbr1, and CTIP2 were detected in the ectopias of Col3a1−/− and Gpr56−/− brains (D, E, G, and H). (Scale bars: A and B, 500 μm; C–H, 100 μm.)
During corticogenesis the cortical plate is expanded in an inside-out manner, in which progressively newer neurons migrate past the early-born neurons to occupy the more superficial layers of the cortex to eventually form a six-layered cortex (2–5, 29, 30). To determine the neuronal composition of the ectopias, we performed immunostaining with three cortical layer-specific markers: Cux1 for layers II–IV; Tbr1 for layers II–III and VI; and CTIP2 for layer V (31–33). We used E18.5 embryos for the study because over 95% of Col3a1−/− mice were lethal upon birth (34). Neurons positive for Cux1, Tbr1, and CTIP2 were detected in the ectopias, suggesting that the ectopic cells in the Col3a1−/− cortex were neurons from both deep and superficial cortical layers, as observed in Gpr56 null mutant mice (Fig. 3 D, E, G, and H) (21). Taken together, our data provides genetic evidence supporting the conclusion that collagen III functions as the ligand of GPR56 in regulating cortical development.
Collagen III Inhibits Neural Migration in a GPR56-Dependent Fashion.
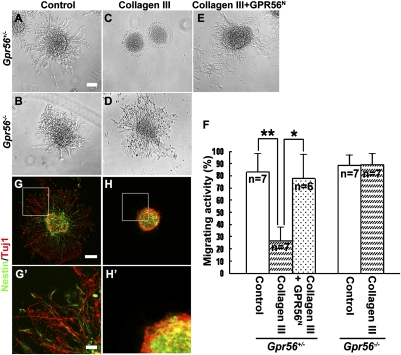
To study the biological function of the interaction of GPR56 with its ligand collagen III, we performed a neurosphere migration assay to evaluate the effect of collagen III on neural migration. Neurospheres established from E13.5 cortices of either Gpr56+/− or Gpr56−/− mice were cultured in neuron culture medium containing 84 nM recombinant collagen III, with or without 90 nM GPR56N, or carrier solution (acetic acid) as a control. After 2 d in culture, we observed robust neural migration from both Gpr56+/− and Gpr56−/− neurospheres in the control medium (Fig. 4 A and B). In contrast, the presence of recombinant collagen III significantly inhibited neural migration in Gpr56+/− neurospheres, but not in Gpr56−/− neurospheres, suggesting that the inhibition requires the expression of GPR56 (Fig. 4 C, D, and F). To further demonstrate the specificity of collagen III-mediated inhibition, we performed a competition assay. As expected, blocking collagen III activity by exogenous GPR56N significantly rescued the Gpr56+/− neurospheres from migration inhibition (Fig. 4 E and F).
Fig. 4.
Collagen III inhibits neuronal migration. (A–E) NPC migration assay. In contrast to the robust NPC migration observed in the control media, recombinant collagen III inhibited NPC migration from Gpr56+/− neurospheres (C), but not from Gpr56−/− neurospheres (D). The addition of exogenous GPR56N significantly blocked collagen III-mediated migration inhibition (E). (F) The degree of collagen III-mediated migration inhibition was quantified as a percentage of the migrating neurospheres. Data are presented as mean ± SD; n = 7 for all groups except for the competition experiment, where n = 6. *P = 0.0032, **P = 0.00046, Student t test. (G–H) Double IHC of Nestin and Tuj1 of the cultured neurospheres. (G′–H′) Higher-magnification views of the boxed regions in G and H. (Scale bars: A–E, G, and H, 100 μm; G′–H′, 25 μm.)
To reveal the identity of the migrated cells from the neurospheres, we performed double IHC of Nestin and Tuj1. Interestingly, both Nestin+ and Tuj1+ cells were among the migrated cells, suggesting collagen III-mediated migration inhibition affects both neural progenitor cells (NPCs) and migrating neurons (Fig. 4 G and H).
Migrating Neurons Express GPR56.
We have previously shown that GPR56 is expressed in radial glial cells, but it was not clear whether migrating neurons expressed GPR56 (21). To investigate the potential function of GPR56 in neuronal migration, we examined whether migrating neurons express GPR56 using a monoclonal antibody against GPR56N (SI Materials and Methods). Surprisingly, we found that GPR56 is expressed in Tuj1+ migrating neurons, in addition to its expression in radial glial cells (Fig. 5 A–I) (21).
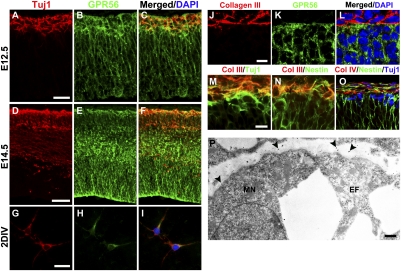
Fig. 5.
Migrating neurons express GPR56. (A–I) Immunostaining of Tuj1 (red) and GPR56 (green) in E12.5 (A–C), E14.5 (D–F) neocortex, and 2-d in vitro-cultured (2DIV) primary cortical neurons (G–I). GPR56 was expressed in the Tuj1+ cell population. (J–L) Double IHC of collagen III/GPR56 in E12.5 neocortex. (M and N) Double IHC of collagen III/Tuj1 and collagen III/Nestin in E12.5 neocortex. (O) Triple IHC of collagen IV, Nestin, and Tuj1 in E11.5 neocortex. (P) Immunoelectron microscopy of collagen III. Immunogold-positive collagen III (arrowheads) is expressed in the pial BM. Both migrating neuron (MN) and radial glial endfeet (EF) have direct contact to the collagen III-containing pial BM. (Scale bars: A–F, 50 μm; G–I, 25 μm; J–O, 10 μm; P, 500 nm.)
To reveal the spatial relationship of collagen III and GPR56-expressing cells, we performed double IHC of GPR56 and collagen III. As expected, the main interaction site for GPR56+ cells and collagen III is at the pia surface/marginal zone (Fig. 5 J–L).
It is traditionally thought that migrating neurons have no direct contact to the pial BM. To investigate the relationship between migrating neurons and the pial BM, we performed double and triple IHC on floating brain sections. As we suspected, both radial glial endfeet and migrating neurons directly contact the pial BM during early developmental stages, thus providing them access to the ligand of GPR56 in the pial BM (Fig. 5 M–O). This observation was further verified by immunoelectron microscopy analysis against collagen III in E12.5 brain sections (Fig. 5P).
Cajal–Retzius (CR) cells and interneurons are the major neurons in the preplate and marginal zone of the developing brain, and we investigated whether they also express GPR56. We performed double IHC of GPR56 and Reelin as well as GPR56 and Calbindin on E12.5 and E14.5 brains, respectively. CR cells indeed express GPR56, and likely account for some of the Tuj1+ neurons in the preplate at E12.5 (Fig. S6 A–C). Interestingly, only some of the interneurons in the marginal zone express GPR56, whereas interneurons in the deeper layers are GPR56− (Fig. S7). Furthermore, we observed an ectopic cluster of CR cells in E11.5 Col3a1−/− neocortex, suggesting a potential role of GPR56 signaling in the regulation of CR cell laminar position (Fig. S6 F and G).
Interaction of GPR56 and Its Ligand Collagen III Activates RhoA Pathway.
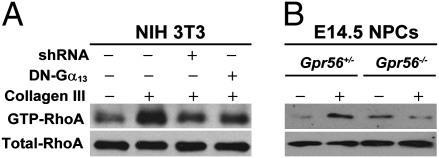
RhoA activation has been shown to regulate cell migration (35). Given the potential function of GPR56 and its ligand collagen III on neural migration, it is conceivable that RhoA is downstream of GPR56 signaling. To test this hypothesis, we performed a GTP-RhoA pull-down assay using NIH 3T3 as host cells that express an abundance of endogenous GPR56 (Fig. S8). Addition of recombinant collagen III to the cells resulted in an increased level of GTP-RhoA in comparison with the control (Fig. 6A). To determine whether collagen III-mediated RhoA activation is through GPR56, we tested the effect of GPR56 knockdown on collagen III-triggered RhoA activation. As expected, Gpr56 shRNA transformed cells failed to manifest RhoA activation upon addition of recombinant collagen III (Fig. 6A). To further confirm that collagen III-mediated RhoA activation is through GPR56, we performed a GTP-RhoA pull-down assay using NPCs derived from E14.5 brains of Gpr56+/− and Gpr56−/− mice. RhoA was strongly activated upon addition of recombinant collagen III in NPCs derived from Gpr56+/− mice but not from Gpr56−/− mice (Fig. 6B).
Fig. 6.
The interaction of GPR56 and its ligand collagen III activates RhoA by coupling to Gα12/13. (A) RhoA activation in NIH 3T3 cells. The addition of collagen III caused an increased level of GTP-RhoA, which was dependent on the presence of GPR56 and Gα13. (B) Collagen III induced the elevation of GTP-RhoA level in NPCs derived from Gpr56+/− but not Gpr56−/− brains. Total RhoA expression in the cell lysate served as a loading control.
The RhoA activation assay requires serum starvation to decrease the basal level of activated RhoA. To evaluate the effect of serum starvation on collagen III-mediated migration inhibition, we performed a modified neurosphere migration assay. A similar result was observed as in Fig. 4, although there were more processes and fewer cells in the migrating zone, indicating some degree of differentiation caused by serum starvation (Fig. S9).
GPR56 Activates RhoA Pathway by Coupling to Gα12/13.
GPCR-mediated RhoA activation is primarily coupled to the Gα12/13 family of G proteins (36). To investigate whether GPR56 also signals through Gα12/13 upon ligand binding, we tested the effect of a catalytically inactive dominant-negative Gα13 (DN-Gα13) on RhoA activation. Expression of DN-Gα13 blunted the activation of RhoA by recombinant collagen III (Fig. 6A). Thus, our data suggests that GPR56 signals through the Gα12/13 and RhoA pathway, consistent with the literature that Gα12 and Gα13 double-mutant mice exhibit a cobblestone-like brain malformation (13).
Discussion
Here we demonstrated that collagen III regulates cortical development by functioning as the major ligand of GPR56. Mutations in one allele of the human COL3A1 cause type IV Ehlers–Danlos syndrome, an autosomal dominantly inherited disorder of the connective tissues in which affected individuals present with excessive bleeding, bruising, and vascular problems, including aortic dissection (37–41). Deleting both alleles of the mouse Col3a1 results in perinatal lethality of unknown etiology in the majority of the homozygous mutant mice, although losing one allele of the gene is not associated with any obvious defects in mice (34). The phenotype associated with the homozygous null mutation of the human COL3A1 is unknown. Given that the homozygous deletion of the mouse Col3a1 causes cobblestone lissencephaly and perinatal lethality in the majority of the null mutant mice, it is possible that its null allele homozygous mutation in humans might be associated with a lethal form of cobblestone lissencephaly.
GPR56 undoubtedly plays an important role in radial glial cells, given its abundant expression in that cell population. However, considering the fact that the anatomic distribution of the cerebellar defect seen in Gpr56−/− mice highly correlates with the restricted expression pattern of GPR56 in the developing granule cells but not the glial cells, our data supports a possible intriguing role of GPR56 in migrating neurons (42). We have previously shown that the pial BM is well organized at E12.5 in Gpr56 null mutant embryonic brains, and that the regional breakdown of the pial BM develops concurrently with the overmigrating neurons (21). Here we further demonstrated that the interaction of GPR56 and its ligand collagen III inhibits neuronal migration. Therefore, it is conceivable to suggest that GPR56 signaling may regulate BM transmigration, a cellular trafficking across BM barriers that occurs in the course of development, inflammation, and tumor metastasis (43). This possibility is certainly consistent with previous observations that down-regulation of GPR56 expression renders a high metastatic potential in melanoma cells (25). GPR56 mRNA is found to be significantly down-regulated in several highly metastatic melanoma cell lines compared with poorly metastatic cells (44). The role of GPR56 in tumor metastasis was further confirmed by a series of elegant assays where overexpression of GPR56 in highly metastatic melanoma cells suppressed tumor growth and their metastatic ability, whereas down-regulation of GPR56 by siRNA enhanced tumor growth and metastasis (25).
GPR56 has been shown to associate with the tetraspanin molecules CD9 and CD81, whose complex then associates with Gαq/11 and Gβγ subunits (45). More recently, a polyclonal antibody against GPR56N was shown to activate RhoA, presumably by mimicking ligand binding (46). In this study, we confirm that GPR56 is indeed signaling through Gα12/13 by demonstrating that (i) collagen III activates RhoA in a GPR56-dependent manner and (ii) the expression of DN-Gα13 dampened the activation. The Gα12/13 pathway has been previously shown to provide positioning cues for cortical neurons during brain development. Conditional ablation of the genes encoding the α-subunits of G12 and G13 in the nervous system results in a cobblestone-like cortical malformation (13). Furthermore, the embryonic cortical neurons derived from those conditional knockout mice were unable to retract their neurites in response to lysophosphatidic acid and sphingosine-1-phosphate, suggesting that they have lost the ability to respond to repulsive mediators acting via GPCRs (13).
We have thus defined a unique ECM receptor-ligand pair—GPR56 and collagen III—that is essential in regulating cortical development. Our data overwhelmingly supports the notion that collagen III is the major ligand of GPR56 in the developing brain and that the interaction of GPR56 with its ligand collagen III occurs at the level of the pial BM involving radial glial cells and neurons, which in turn regulates the integrity of the pial BM and cortical lamination. Our discovery of GPR56 and collagen III as an ECM receptor-ligand pair opens up avenues for further study of the functions of GPR56 in brain development as well as in tumor growth and metastasis.
Materials and Methods
GPR56 Fusion Proteins.
Three different types of fusion proteins were generated for different experiments. Human IgG Fc-tagged GPR56N/GPR56Ndel fusion proteins were used for the putative ligand binding assay (Fig. S1). GPR56N/GPR56Ndel-mFc-biotag fusion proteins were used for the putative ligand pull-down (Fig. S2). GPR56N/GPR56Ndel-Flag-biotag fusion proteins were used for the co-IP experiments (Fig. S3).
Mice.
Col3a1 mice were obtained from the Jackson Laboratory with the strain name C.129S4 (B6)-Col3a1tm1Jae/J in a BALB/c background. The Gpr56 knockout mice, kindly provided by Genentech, were produced in collaboration between Genentech and Lexicon Genetics to analyze the function of ∼500 secreted and transmembrane proteins. All animals were treated according to the guidelines of the Animal Care and Use Committee of Children's Hospital Boston.
Purification of GPR56 Immunocomplexes and Tandem MS and Sequencing.
Purified GPR56N-mFc-biotag and GPR56Ndel-mFc-biotag proteins were biotinylated in vitro with BirA enzyme as described previously (47). Biotinylated GPR56N and GPR56Ndel proteins were separately incubated with primary MF lysates in lysis buffer [20 mM Hepes (pH 7.3), 150 mM NaCl, and 5 mM MgCl2] with protein inhibitor mixture (Roche Molecular Biochemicals) and 1% Brij 96 (Fluka). Mouse primary MFs were established from the meninges of E14.5 mice, and amplified in DMEM with 10% FBS. Immunocomplexes were affinity purified by streptavidin beads (Sigma). GPR56-associated proteins were eluted in 2× SDS loading buffer, subjected to SDS/PAGE and Coomassie blue stain.
The specific protein bands, together with the corresponding regions of the negative control, were excised and prepared for MS by the MS core facility at Children's Hospital Boston. MS data were searched against the mouse International Protein Index (IPI mouse 339) database using the protein identification software Mascot (v2.2.04, Matrix Sciences).
Co-IP and Immunoblotting.
Co-IP and immunoblotting were done as previously described, using mouse primary MF lysates and recombinant collagen III (23). The GPR56N/Ndel and collagen III proteins were detected by Western blot by using anti-GPR56 antibody (2b) or collagen III antibody, respectively.
Histology and IHC.
Histology analysis was carried out as previously described (21). Frozen sections were collected on a cryostat. Free-floating sections were collected by a vibratome (Leica). Sections were processed for immunostaining using standard procedures. Primary antibodies were visualized by appropriate fluorophore-conjugated secondary antibodies. Equivalent amounts of fusion proteins were used as probes to examine the distribution of the putative ligand of GPR56 in mouse brains and meningeal fibroblasts. Images were captured using a Nikon 80i upright microscope or a confocal FluoView Laser System (FluoView FV1000; Olympus). Representative photographs were obtained with the same exposure setting for control and mutant.
Migration Assay.
Neurospheres were prepared from E13.5 mouse cerebral cortex as described previously and detailed in SI Materials and Methods (46, 48). The neurospheres were cultured in neural culture medium with 84 nM collagen III (Abcam) with or without 90 nM GPR56N, or with carrier solution (acetic acid) as control. Forty-eight hours later, the neurospheres were imaged and the number of migrating neurospheres was quantified. The number of migrating neurospheres was determined by meeting one of the following two criteria: (i) the distance from the edge of the neurosphere to the leading cell of outgrowth was more than the diameter of the neurosphere, or (ii) the neurosphere was completely dispersed. The migration activity was assessed by the percentage of the number of migrating neurospheres out of the total number of neurospheres. The results were summarized from seven independent experiments (six for the competition assay) and statistically analyzed with Student t test.
GTP-Rho Pull-Down Assay.
The GTP-Rho pull-down assay was done as previously described, using either NPCs or NIH 3T3 cells (46, 49). The NPCs used for the GTP-Rho pull-down assay was prepared as described in the migration assay. NIH 3T3 cells were transfected with empty vector, Gpr56 shRNA, or DN-Gα13 plasmid (a gift from A. Irving, University of Dundee, Nethergate, Dundee, Scotland) (50). Twenty-four hours after transfection, the cells were subject to serum starvation for 36 h followed by the addition of recombinant collagen III 84 nM or acetic acid as control for 5 min.
Supplementary Material
Acknowledgments
We thank Drs. Christopher A. Walsh, Rosalind A. Segal, Robert Hevner, Andrew Irving, Hsi-Hsien Lin, and Alan Cantor for providing antibodies and plasmids; Maria Ericsson at the Harvard Medical School Electron Microscope facility for the assistance in immunoelectron microscopy analysis; and the Intellectual and Developmental Disabilities Research Center proteomics center at the Children's Hospital Boston for assistance with mass spectrometry analysis. This research was supported in part by National Institute of Neurological Disorders and Stroke Grant R01 NS057536 (to X.P.), the William Randolph Hearst Fund Award (S.-J.J.), the Leonard and Isabelle Goldenson Research fellowship (R.L.), and a Flight Attendant Medical Research Institute Young Clinical Scientist Award (Z.J.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104821108/-/DCSupplemental.
References
- 1.Gaitanis JN, Walsh CA. Genetics of disorders of cortical development. Neuroimaging Clin N Am. 2004;14:219–229. doi: 10.1016/j.nic.2004.03.007. viii. [DOI] [PubMed] [Google Scholar]
- 2.Ayala R, Shu T, Tsai LH. Trekking across the brain: The journey of neuronal migration. Cell. 2007;128:29–43. doi: 10.1016/j.cell.2006.12.021. [DOI] [PubMed] [Google Scholar]
- 3.Marín O, Rubenstein JL. Cell migration in the forebrain. Annu Rev Neurosci. 2003;26:441–483. doi: 10.1146/annurev.neuro.26.041002.131058. [DOI] [PubMed] [Google Scholar]
- 4.Caviness VS, Jr, Rakic P. Mechanisms of cortical development: A view from mutations in mice. Annu Rev Neurosci. 1978;1:297–326. doi: 10.1146/annurev.ne.01.030178.001501. [DOI] [PubMed] [Google Scholar]
- 5.Bystron I, Blakemore C, Rakic P. Development of the human cerebral cortex: Boulder Committee revisited. Nat Rev Neurosci. 2008;9:110–122. doi: 10.1038/nrn2252. [DOI] [PubMed] [Google Scholar]
- 6.Olson EC, Walsh CA. Smooth, rough and upside-down neocortical development. Curr Opin Genet Dev. 2002;12:320–327. doi: 10.1016/s0959-437x(02)00305-2. [DOI] [PubMed] [Google Scholar]
- 7.Georges-Labouesse E, Mark M, Messaddeq N, Gansmüller A. Essential role of alpha 6 integrins in cortical and retinal lamination. Curr Biol. 1998;8:983–986. doi: 10.1016/s0960-9822(98)70402-6. [DOI] [PubMed] [Google Scholar]
- 8.De Arcangelis A, Mark M, Kreidberg J, Sorokin L, Georges-Labouesse E. Synergistic activities of alpha3 and alpha6 integrins are required during apical ectodermal ridge formation and organogenesis in the mouse. Development. 1999;126:3957–3968. doi: 10.1242/dev.126.17.3957. [DOI] [PubMed] [Google Scholar]
- 9.Graus-Porta D, et al. Beta1-class integrins regulate the development of laminae and folia in the cerebral and cerebellar cortex. Neuron. 2001;31:367–379. doi: 10.1016/s0896-6273(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 10.Beggs HE, et al. FAK deficiency in cells contributing to the basal lamina results in cortical abnormalities resembling congenital muscular dystrophies. Neuron. 2003;40:501–514. doi: 10.1016/s0896-6273(03)00666-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niewmierzycka A, Mills J, St-Arnaud R, Dedhar S, Reichardt LF. Integrin-linked kinase deletion from mouse cortex results in cortical lamination defects resembling cobblestone lissencephaly. J Neurosci. 2005;25:7022–7031. doi: 10.1523/JNEUROSCI.1695-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costell M, et al. Perlecan maintains the integrity of cartilage and some basement membranes. J Cell Biol. 1999;147:1109–1122. doi: 10.1083/jcb.147.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moers A, Nürnberg A, Goebbels S, Wettschureck N, Offermanns S. Galpha12/Galpha13 deficiency causes localized overmigration of neurons in the developing cerebral and cerebellar cortices. Mol Cell Biol. 2008;28:1480–1488. doi: 10.1128/MCB.00651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwiatkowski AV, et al. Ena/VASP is required for neuritogenesis in the developing cortex. Neuron. 2007;56:441–455. doi: 10.1016/j.neuron.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Sarkisian MR, et al. MEKK4 signaling regulates filamin expression and neuronal migration. Neuron. 2006;52:789–801. doi: 10.1016/j.neuron.2006.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaglin XH, et al. Mutations in the beta-tubulin gene TUBB2B result in asymmetrical polymicrogyria. Nat Genet. 2009;41:746–752. doi: 10.1038/ng.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piao X, et al. An autosomal recessive form of bilateral frontoparietal polymicrogyria maps to chromosome 16q12.2-21. Am J Hum Genet. 2002;70:1028–1033. doi: 10.1086/339552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chang BS, et al. Bilateral frontoparietal polymicrogyria: Clinical and radiological features in 10 families with linkage to chromosome 16. Ann Neurol. 2003;53:596–606. doi: 10.1002/ana.10520. [DOI] [PubMed] [Google Scholar]
- 19.Piao X, et al. G protein-coupled receptor-dependent development of human frontal cortex. Science. 2004;303:2033–2036. doi: 10.1126/science.1092780. [DOI] [PubMed] [Google Scholar]
- 20.Piao X, et al. Genotype-phenotype analysis of human frontoparietal polymicrogyria syndromes. Ann Neurol. 2005;58:680–687. doi: 10.1002/ana.20616. [DOI] [PubMed] [Google Scholar]
- 21.Li S, et al. GPR56 regulates pial basement membrane integrity and cortical lamination. J Neurosci. 2008;28:5817–5826. doi: 10.1523/JNEUROSCI.0853-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bahi-Buisson N, et al. GPR56-related bilateral frontoparietal polymicrogyria: Further evidence for an overlap with the cobblestone complex. Brain. 2010;133:3194–3209. doi: 10.1093/brain/awq259. [DOI] [PubMed] [Google Scholar]
- 23.Jin Z, et al. Disease-associated mutations affect GPR56 protein trafficking and cell surface expression. Hum Mol Genet. 2007;16:1972–1985. doi: 10.1093/hmg/ddm144. [DOI] [PubMed] [Google Scholar]
- 24.Shashidhar S, et al. GPR56 is a GPCR that is overexpressed in gliomas and functions in tumor cell adhesion. Oncogene. 2005;24:1673–1682. doi: 10.1038/sj.onc.1208395. [DOI] [PubMed] [Google Scholar]
- 25.Xu L, Begum S, Hearn JD, Hynes RO. GPR56, an atypical G protein-coupled receptor, binds tissue transglutaminase, TG2, and inhibits melanoma tumor growth and metastasis. Proc Natl Acad Sci USA. 2006;103:9023–9028. doi: 10.1073/pnas.0602681103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JK, et al. A novel binding site in collagen type III for integrins alpha1beta1 and alpha2beta1. J Biol Chem. 2005;280:32512–32520. doi: 10.1074/jbc.M502431200. [DOI] [PubMed] [Google Scholar]
- 27.Nykvist P, et al. Distinct recognition of collagen subtypes by alpha(1)beta(1) and alpha(2)beta(1) integrins. Alpha(1)beta(1) mediates cell adhesion to type XIII collagen. J Biol Chem. 2000;275:8255–8261. doi: 10.1074/jbc.275.11.8255. [DOI] [PubMed] [Google Scholar]
- 28.Semeralul MO, et al. Microarray analysis of the developing cortex. J Neurobiol. 2006;66:1646–1658. doi: 10.1002/neu.20302. [DOI] [PubMed] [Google Scholar]
- 29.LoTurco JJ, Bai J. The multipolar stage and disruptions in neuronal migration. Trends Neurosci. 2006;29:407–413. doi: 10.1016/j.tins.2006.05.006. [DOI] [PubMed] [Google Scholar]
- 30.Hatten ME. New directions in neuronal migration. Science. 2002;297:1660–1663. doi: 10.1126/science.1074572. [DOI] [PubMed] [Google Scholar]
- 31.Englund C, et al. Pax6, Tbr2, and Tbr1 are expressed sequentially by radial glia, intermediate progenitor cells, and postmitotic neurons in developing neocortex. J Neurosci. 2005;25:247–251. doi: 10.1523/JNEUROSCI.2899-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molyneaux BJ, Arlotta P, Hirata T, Hibi M, Macklis JD. Fezl is required for the birth and specification of corticospinal motor neurons. Neuron. 2005;47:817–831. doi: 10.1016/j.neuron.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 33.Nieto M, et al. Expression of Cux-1 and Cux-2 in the subventricular zone and upper layers II–IV of the cerebral cortex. J Comp Neurol. 2004;479:168–180. doi: 10.1002/cne.20322. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Wu H, Byrne M, Krane S, Jaenisch R. Type III collagen is crucial for collagen I fibrillogenesis and for normal cardiovascular development. Proc Natl Acad Sci USA. 1997;94:1852–1856. doi: 10.1073/pnas.94.5.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narumiya S, Tanji M, Ishizaki T. Rho signaling, ROCK and mDia1, in transformation, metastasis and invasion. Cancer Metastasis Rev. 2009;28:65–76. doi: 10.1007/s10555-008-9170-7. [DOI] [PubMed] [Google Scholar]
- 36.Worzfeld T, Wettschureck N, Offermanns S. G(12)/G(13)-mediated signalling in mammalian physiology and disease. Trends Pharmacol Sci. 2008;29:582–589. doi: 10.1016/j.tips.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 37.Germain DP. Ehlers–Danlos syndrome type IV. Orphanet J Rare Dis. 2007;2:32. doi: 10.1186/1750-1172-2-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kontusaari S, Tromp G, Kuivaniemi H, Romanic AM, Prockop DJ. A mutation in the gene for type III procollagen (COL3A1) in a family with aortic aneurysms. J Clin Invest. 1990;86:1465–1473. doi: 10.1172/JCI114863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuivaniemi H, Tromp G, Bergfeld WF, Kay M, Helm TN. Ehlers–Danlos syndrome type IV: A single base substitution of the last nucleotide of exon 34 in COL3A1 leads to exon skipping. J Invest Dermatol. 1995;105:352–356. doi: 10.1111/1523-1747.ep12320704. [DOI] [PubMed] [Google Scholar]
- 40.Prockop DJ, Kivirikko KI. Heritable diseases of collagen. N Engl J Med. 1984;311:376–386. doi: 10.1056/NEJM198408093110606. [DOI] [PubMed] [Google Scholar]
- 41.Schwarze U, et al. Haploinsufficiency for one COL3A1 allele of type III procollagen results in a phenotype similar to the vascular form of Ehlers–Danlos syndrome, Ehlers–Danlos syndrome type IV. Am J Hum Genet. 2001;69:989–1001. doi: 10.1086/324123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koirala S, Jin Z, Piao X, Corfas G. GPR56-regulated granule cell adhesion is essential for rostral cerebellar development. J Neurosci. 2009;29:7439–7449. doi: 10.1523/JNEUROSCI.1182-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowe RG, Weiss SJ. Breaching the basement membrane: Who, when and how? Trends Cell Biol. 2008;18:560–574. doi: 10.1016/j.tcb.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 44.Zendman AJ, Cornelissen IM, Weidle UH, Ruiter DJ, van Muijen GN. TM7XN1, a novel human EGF-TM7-like cDNA, detected with mRNA differential display using human melanoma cell lines with different metastatic potential. FEBS Lett. 1999;446:292–298. doi: 10.1016/s0014-5793(99)00230-6. [DOI] [PubMed] [Google Scholar]
- 45.Little KD, Hemler ME, Stipp CS. Dynamic regulation of a GPCR-tetraspanin-G protein complex on intact cells: Central role of CD81 in facilitating GPR56-Galpha q/11 association. Mol Biol Cell. 2004;15:2375–2387. doi: 10.1091/mbc.E03-12-0886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iguchi T, et al. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G alpha 12/13 and Rho pathway. J Biol Chem. 2008;283:14469–14478. doi: 10.1074/jbc.M708919200. [DOI] [PubMed] [Google Scholar]
- 47.Stacey M, et al. EMR4, a novel epidermal growth factor (EGF)-TM7 molecule up-regulated in activated mouse macrophages, binds to a putative cellular ligand on B lymphoma cell line A20. J Biol Chem. 2002;277:29283–29293. doi: 10.1074/jbc.M204306200. [DOI] [PubMed] [Google Scholar]
- 48.Bix GJ, Clark GD. Platelet-activating factor receptor stimulation disrupts neuronal migration In vitro. J Neurosci. 1998;18:307–318. doi: 10.1523/JNEUROSCI.18-01-00307.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Henstridge CM, et al. The GPR55 ligand L-alpha-lysophosphatidylinositol promotes RhoA-dependent Ca2+ signaling and NFAT activation. FASEB J. 2009;23:183–193. doi: 10.1096/fj.08-108670. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.