Abstract
Astroviruses are single-stranded, plus-sense RNA viruses that infect both mammals and birds, causing gastroenteritis and other extraintestinal diseases. Clinical studies have established astroviruses as the second leading cause of viral diarrhea in young children. Here we report the crystal structure of the human astrovirus dimeric surface spike determined to 1.8-Å resolution. The overall structure of each spike/projection domain has a unique three-layered β-sandwiches fold, with a core, six-stranded β-barrel structure that is also found in the hepatitis E virus capsid protrusions, suggesting a closer phylogenetic relationship between these two viruses than previously acknowledged. Based on a hepatitis E virus capsid model, we performed homology modeling and produced a complete, T = 3 astrovirus capsid model with features remarkably similar to those observed in a cryoelectron microscopy reconstruction image of a human astrovirus. Mapping conserved residues onto the astrovirus projection domain revealed a putative receptor binding site with amino acid compositions characteristic for polysaccharide recognition. Our results will have an important impact on future characterization of astrovirus structure and function, and will likely have practical applications in the development of vaccines and antivirals.
Keywords: structural protein, naked virus, attachment domain
Astroviruses are small, nonenveloped, icosahedral viruses with a positive-sense, single-stranded RNA genome of approximately 7 kb in size. With members infecting mammals and avian species, the family of Astroviridae consists of two genera, Mammastrovirus and Avastrovirus (1, 2). Human astroviruses, eight serotypes in total, are one of the leading causes of gastroenteritis in young children, elderly people, and immunocompromised adults. Sporadic astrovirus infections as well as large-scale outbreaks in susceptible populations have been described (2). In addition to humans, astroviruses can infect a wide range of wild and domestic animals, causing gastroenteritis in most mammals and both intestinal and extraintestinal diseases in birds (1).
The genomic RNA of astroviruses is polyadenylated with three ORFs. ORF1a and the downstream overlapping ORF1b encode two nonstructural polyproteins, nsp1a and nsp1ab. The nsp1ab protein is produced from both ORF1a and ORF1b through a translational frame-shift mechanism (3). Polyproteins nsp1a and nsp1ab are processed by a viral 3C-like serine protease and other cellular proteases into several proteins (e.g., the RNA-dependent RNA polymerase and the serine protease) that are likely to function in the replication of the viral genome (4). ORF2 at the 3′ end of the viral genome encodes the viral capsid protein (CP) (5) and is found in both genomic and subgenomic RNAs. It is unclear whether the astrovirus RNA contains a 5′ cap. Viral capping enzymes such as guanylyltranferase and methyltransferases have not been identified in the astrovirus genome.
When overexpressed in eukaryotic hosts, the 796-aa CP of the serotype 2 astrovirus was able to self-assemble into virus-like particles in the absence of other viral proteins and viral RNAs (6). By comparing the protein sequences of the CPs from the eight human astrovirus serotypes, astrovirus CP can be divided into three domains: a highly conserved N-terminal domain, a hypervariable domain, and a highly acidic C-terminal domain (7). The acidic C-terminal domain is eliminated from the virion by host caspases during viral maturation (8, 9). Once released from the infected cell, the astrovirus capsid is further processed by host extracellular proteases. The infectious virion of the astrovirus contains three predominant protein species with molecular weights of approximately 34 (VP34), 27/29 (VP27/29), and 25/26 (VP25/26) kDa, respectively (9–13). VP34 is derived from the highly conserved N-terminal region, whereas VP27/29 and VP25/26 are both from the hypervariable region with a different N terminus (14). Antigenicity studies indicate that neutralizing monoclonal antibodies recognize protein species derived from the hypervariable region (i.e., VP27/29 and VP25/26), suggesting that the hypervariable domain is involved in heterotypic immunity, virus neutralization, and viral attachment to target cells (13, 15).
Among the four small, nonenveloped positive-sense RNA viruses (i.e., picornavirus, calicivirus. astrovirus, and hepevirus) that infect animals, astrovirus is the only one whose atomic structure is not yet known. Electron microscopy of negatively stained astrovirus showed pentagonal and hexagonal contours, possibly corresponding to projections of icosahedral particles, with well-defined spikes/projections protruding from the viral surface (16). A low-resolution cryoelectron microscopy (cryo-EM) reconstruction image of astrovirus reveals a rippled, solid capsid shell of 33 nm in diameter that is decorated with 30 dimeric spikes extending 5 nm from the surface (1). It has been hypothesized that VP34 builds up the continuous capsid shell, while VP27/29 and VP25/26 form the dimeric projections seen in the cryo-EM reconstruction image.
To further explore the structural and functional properties of the astrovirus capsid, we determined the crystal structure of the projection domain of the human astrovirus CP to 1.8-Å resolution. Each projection domain, consisting of residues 415–646, folds into a unique, triple-layered β-sandwich structure. The core of the β-sandwich is a squashed β-barrel that bears resemblance to the P2 domain of the hepatitis E virus (HEV) CP, suggesting a phylogenetic relationship between these two viruses. Like in HEV, the astrovirus projection domain forms stable dimers in both solution and in crystal, and this interaction may play an important role in stabilizing viral particles. Mapping the conserved sequences of the eight astrovirus serotypes onto the crystal structure reveals a putative receptor binding site. The stereochemical feature of the receptor binding site indicates that a di/trisaccharide moiety may function as an astrovirus cell receptor.
Results and Discussion
Structure of the Astrovirus Projection Domain.
Assuming that the variable portion (excluding the highly acidic C-terminal domain) of the astrovirus CP forms the viral surface spikes, we made several constructs in an attempt to express just the projection/spike domain. These five constructs contain residues 394–646, 501–658, 415–628, 501–628, and 415–646, respectively. Of these five constructs, only the last one, P2415–646, produced soluble proteins in Escherichia coli. The ability of P2415–646 to stay soluble is because this construct encompasses an intact spike domain, and its two ends both terminate in flexible domain linkers (see discussion below). P2415–646 exhibited an apparent molecular mass (MM) of approximately 60 kDa as judged by gel filtration chromatography (Fig. S1), consistent with the calculated MM of a dimer (ca. 54.2 kDa).
Purified P2415–646 dimer was crystallized in two different crystal forms (Table S1). The I213 crystals, grown at pH 4.6, diffracted to only 2.8 Å, possibly due to its high solvent content (82.8%). The P3221 crystals, which were grown at pH 9.5, diffracted far better (solvent content = 41.0%). Using the P3221 crystals, the structure of the P2415–646 dimer has been determined to 1.8-Å resolution by multiwavelength anomalous dispersion (MAD) (Fig. 1). Our final model contains amino acid residues 430–645 that fold into a β-structure with a total of 11 β-strands. Although there is only one molecule in each crystal asymmetric unit, two P2415–646 molecules related by the twofold crystal symmetry interact to form a compact dimer. We have also determined the structure of the I213 crystal and found that the two structures differ by an rmsd of only 0.548 Å, despite the fact that these two crystals were grown at very different pH values and had different packing interactions. For simplicity, the P3221 structure will be used in all of our discussions below.
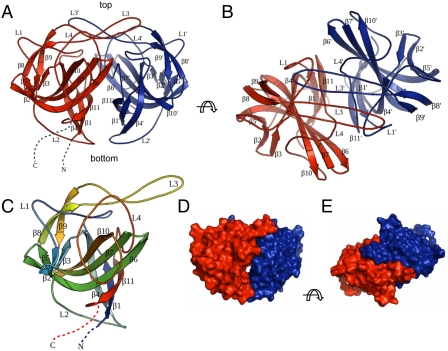
Fig. 1.
Crystal structure of astrovirus P2415–646. (A) Side view of the P2415–646 dimer. The two subunits are colored in red and blue, respectively. The N and C termini of the red subunit are highlighted by dotted lines. The viral capsid lies at the bottom the dimeric spike. (B) Top view of the P2415–646 dimer. (C) Structure of one P2415–646 molecule. The polypeptide is rainbow-colored with the N terminus in blue and the C terminus in red. (D and E) Surface representations of the P2415–646 dimer for comparison with the cryo-EM reconstruction image. Molecules in D and E are viewed from the same orientation as in A and B, respectively. The figures were made with PyMOL (39).
Close inspection of the folding topology reveals that each P2415–646 molecule contains a core, six-stranded β-barrel (β1, β4, β6, β7, β10, and β11) with a tightly packed hydrophobic core (Figs. 1C and 2A). Another three-stranded β-sheet, consisting of β2, β3, and β5, is packed against the outside of the β-barrel away from the dimer interface. Therefore, the structure of P2415–646 can be formalized as a triple-layered β-sandwich, in which the three orthogonal β-sheets are connected by two highly bent β-strands (β4 and β6). Two long loops, L1 and L4 (L1, residues 439–466; L4, residues 597–632), span across the top of the β-barrel, while a shorter loop, L2 (residues 491–506), runs across the bottom of the barrel (Fig. 1C). A β-hairpin motif, consisting of β8, β9, and L3 (residues 554–574), sits on top of L1 and L4, and may help to reinforce the structural conformation of these two long loops.
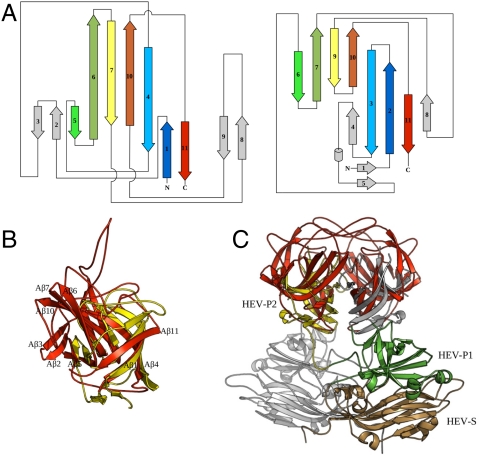
Fig. 2.
Structural comparison of P2415–646 with HEV capsid P2 domain. (A) Topology diagrams for the astrovirus P2415–646 (Left) and the HEV P2 domain (Right). Related β-strands in the two structures are rendered in matching colors, and unrelated structural elements are shaded in gray. (B) Superposition of the astrovirus P2415–646 (red) and the HEV P2 domain (yellow). Structure elements in astrovirus P2415–646 are labeled as in Fig. 1C with a letter “A.” (C) Superposition of the atrovirus P2415–646 dimer with the HEV CP dimer. The atrovirus P2415–646 dimer is in red. For HEV CP, one subunit is in gray, and the other is colored by domains: S in gold, P1 in green, and P2 in yellow.
Structure of the Astrovirus Projection Domain Dimer.
Crystal packing analysis indicates that one P2415–646 molecule and its twofold crystal symmetry related partner form a dimer (Fig. 1 A and B), consistent with our gel filtration chromatography results showing that P2415–646 forms dimers in solution. Structural analysis using the PISA server (17) confirms the dimeric configuration of P2415–646. The buried surface area of the P2415–646 dimer is approximately 3,490 Å2, which counts for 20.2% of the solvent-accessible area of the complex. The calculated free energy of dimer dissociation from the PISA server is 15.6 kcal/mol, suggesting that the dissociation constant is within the low nanomolar range. Therefore, the dimer interaction mediated by the projection domain is likely to play an important role in stabilizing the viral capsid. Because the astrovirus projection domain is connected to the capsid shell at its N terminus, the thinner end of the projection dimer (where both the N and C termini are located) will be referred to as the bottom, whereas the opposite, flat end will be referred to as the top (Fig. 1A).
The projection dimer interface is primarily mediated by loops L3 and L4 and strands β1 and β11 (Fig. 1 A and B). In particular, two L3 loops from the two subunits of a dimer reach over to the opposing subunit like two crossing arms, making extensive interactions along their entire length. Sitting on top of the projection domain dimer, L3 also makes close contacts with the L1 and L4 loops from both subunits. Therefore, loop L3 is likely to play a pivotal role in not only CP dimer formation, but also the overall folding of the astrovirus projection domain. It is also likely that folding of the projection domain is closely coupled to its dimerization. Indeed, approximately 90% of the overexpressed P2415–646 proteins were insoluble, possibly due to misfolding because proper dimerization of the projection domain had not occurred.
A total of 51 residues from each subunit are observed at the dimer interface, among which 29 of the residues are highly conserved. Among the nonconserved interface residues, 12 residues interact with the other subunit via backbone atoms. Except for the protruding L3 loop, residues at the dimer interface create a nearly flat surface in between the two molecules that is mostly hydrophobic in nature. The protruding L3 loop, however, contains mostly polar/charged residues and thus predominantly makes hydrogen bonds at the dimer interface. For example, 20 of the 26 intermolecular hydrogen bonds involve residues from the protruding L3 loop. Intermolecular hydrogen bonds are mediated by either main-chain or side-chain atoms of highly conserved residues (i.e., Gln450, Tyr555, Arg558, and Arg572). The highly conserved nature of these intersubunit hydrogen bonds indicates an important functional role and suggests that they may determine the specificity of dimer interaction.
When visually compared to a cryo-EM reconstruction image of an astrovirus (1), the overall morphology of the P2415–646 dimer matches that of the dimeric surface spikes. For instance, the dimensions of our P2415–646 dimer are approximately 60 × 25 × 46 Å3, which are in the same range as our visual estimate of the surface spikes observed in the cryo-EM image. Furthermore, the top view of the P2415–646 dimer crystal structure shows a rectangular shape with two small knobs protruding sideways, whereas the side view of the dimer shows a bowl-like shape with a small hole in the middle (Fig. 1 D and E), again consistent with the cryo-EM reconstruction. The comparison also confirms the orientation of the P2415–646 dimer relative to the protein shell: the twofold axis of the dimer is normal to the viral capsid, with the two L3 loops located on the top end of the spike distal from the viral surface.
After it was reported that astrovirus CP of serotype 8 is cleaved by a host caspase after Asp657 during viral particle assembly (9), we expressed and crystallized the P2415–657 protein. The structure of P2415–657 is essentially identical to that of P2415–646, with residues from 646 to 657 being disordered. Although the last ordered residue (Pro645) from our final model is shielded under the surface spikes, we expect that the caspase cleavage site Asp657, which is 12 residues away from Pro645, should be accessible to the caspase in intact particles.
Structure Comparison.
The amino acid sequence of the astrovirus CP shows no significant homology to any other known viral proteins. Using the structure of the astrovirus CP projection domain as a reference, we searched for structural homologs using the DALI server (18) against all Protein Data Bank (PDB) entries. The hit with the highest Z score is the projection domain of the HEV CP (Z score = 3.9). The others are nonviral proteins including archaea riboflavin kinases (Z score = 3.5), a siderophore-interacting protein (Z score = 3.0), ferredoxin-NADP reductases (Z score = 2.0–2.7), a flavodoxin reductase (Z score = 2.5), and a dihydroorotate dehydrogenase (chain b, Z score = 2.5). Except for the HEV CP P2 domain, other hits have an α/β- rather than an all-β-structure. Thus we speculate that their structure similarity to astrovirus P2415–646, which appears to be insignificant by visual inspection, may be due to evolutionary convergence.
Like astrovirus, HEV is a small, nonenveloped, single-stranded, positive-sense RNA virus with a genome size of about 7.2 kb. At present, it is the sole member of the Hepeviridae family. The HEV capsid has a T = 3 icosahedral symmetry (19), but recombinant CP of HEV forms T = 1 icosahedral particles (20, 21). The structure of each HEV CP contains three domains, S, P1, and P2. The S domain adopts a typical jelly-roll β-barrel fold commonly found in other positive-stranded RNA viruses. The P1 domain has a structure of six-stranded β-barrel decorated with four short α-helices (20, 21). The HEV CP P2 domain makes up the dimeric spikes on the viral surface and is responsible for receptor binding and antigenicity determination. The structural alignment generated by the DALI server shows that the HEV capsid P2 domain (20–22) has a core six-stranded β-barrel structure similar that of the astrovirus P2415–646 (Fig. 2). Indeed, the matching β-strands in these two β-barrel structures (β1, β4, β6, β7, β10, and β11 for astrovirus, and β2, β3, β7, β9, β10, and β11 for HEV) have very similar spatial positions, although their lengths may be different (Fig. 2B). The strand β5 of astrovirus P2415–646, although not part of the core six-stranded β-barrel, is also related to the strand β6 of HEV P2. In both structures, this β-strand is packed on the outer surface of the core β-barrel.
In addition to the similar structural folds, we have also noted several major structural differences between the two viral spikes (Fig. 2 A and B). First, the loop L2 that runs across the bottom of the core β-barrel of astrovirus P2415–646 is replaced by two β-strands (β4 and β5) and a one-turn α-helix in HEV. Second, the long β-hairpin β6 and β7 in astrovirus P2415–646 is replaced with two short strands (β7 and β9) with a loop in between that contains an extra β-strand (β8) in HEV. Third, there are two extra β-hairpin insertions in astrovirus, one between β1 and β4 and the other between β7 and β10. Fourth, β6, β7, and β10 in astrovirus CP are much longer than their counterparts in HEV P2. In astrovirus, these three β-strands form a small protruding knob on each side of the surface spikes seen in the cryo-EM image. Because of these insertions and also because of its longer β-strands, the astrovirus P2 domain is substantially larger in size compared to the HEV P2 (232 vs. 143 residues).
The P2 dimer interaction is mediated by similar structural elements in both astrovirus and HEV, as structure superposition based on P2 monomers also placed the P2 dimers on top of each other, with the two dimer axes differing by only about 14°. Therefore, the inclination angle of the β-barrel with respect to the icosahedral twofold symmetry axis must be similar in astrovirus and HEV, but azimuthal rotation of the dimeric spike about the icosahedral twofold axis may or may not be the same in these two viruses. The superposition of the astrovirus and HEV P2 dimers also confirms our earlier top/bottom assignment to the astrovirus P2 dimer (Figs. 1A and 2C). Like in the HEV CP, both N and C termini of the astrovirus P2 are located at the bottom of the spike close to the particle surface.
The unexpected structural similarity between the astrovirus and HEV surface spikes suggests that the two viruses may have a much closer phylogenetic relationship than previously expected based on sequence analyses (23). On the other hand, the caliciviruses, another group of small, nonenveloped, positive-sense RNA viruses also having dimeric surface spikes, are likely to be more distant compared to these two, because the structural organization of its CP projection domain P2 is different. Whereas the three domains of the HEV CP are organized in a linear fashion, the polypeptide of the calicivirus CP winds through the three domains in the order of S → P1 → P2 → P1 (24). Therefore, the surface spike of calicivirus is formed effectively by a polypeptide insertion in the P1 domain. In contract, the spike of both HEV and astrovirus is situated on the CP polypeptide after the P1 domain (Fig. 3A). It is important to note that all three viruses have a homologous spike structure, with the astrovirus spike being more closely related to HEV (Z = 3.9) than calicivirus (Z = 1.7) according to DALI pairwise comparison. Further studies on these small, nonenveloped RNA viruses, both on their capsid and their replication, would be necessary to establish their phylogenetic relationships, which, in turn, would greatly facilitate further structural and functional characterization of these viruses.
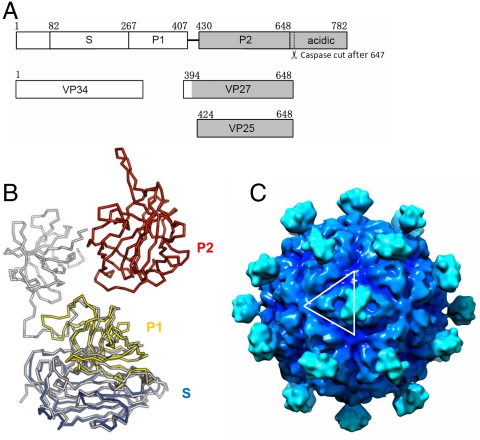
Fig. 3.
Astrovirus capsid structure model. (A) Domain structure of astrovirus CP of serotype 8. Astrovirus CP has a conserved region (in white), a variable region and an acidic C-terminal region (in gray). The S and P1 domain of astrovirus as defined in this paper can be roughly mapped to the conserved region. The P2 domain, however, is mapped to the variable region. The coverage of VP34, VP27, and VP24 is also indicated. Although the N-terminal residues of these three polypeptides have been previously mapped, it is not clear exactly where they end. Our crystal structure of the surface spike strongly argues that the C-terminal residues of both VP27 and VP25 are Arg648, which is the first trypsin cleavage site after the last ordered residues (Pro645) in the spike. This residue is also the only trypsin susceptible site that would explain the size of VP28 and VP27, because these two proteins have the same N-terminal residue (12). The C terminus of VP34 remains ambiguous. (B) Astrovirus CP superimposed onto HEV CP. HEV CP is shown in gray, whereas the astrovirus CP is colored by domains with S in blue, P1 in yellow, and P2 in red. (C) T = 3 astrovirus capsid model rendered at 15-Å resolution. The white lines highlight one asymmetric unit. The contour level was chosen so that the volume is consistent with 10 Å3 per non-hydrogen atom. The isosurfaces were drawn using Chimera (40).
Structure Modeling of the Human Astrovirus CP.
Considering its well-conserved nature, the N-terminal portion of the astrovirus CP should have a structure that perhaps bears even more similarity to the HEV CP than the hypervariable P2 domain. Indeed, using the astrovirus CP sequence alone, homology structure recognition using the HHPRED server (25) indicates that the HEV CP is the best structural homolog (among all PDB entries). Specifically, residues 82–266 and 267–407 of the human astrovirus (serotype 8) CP are matched to the S (amino acids 118–313) and P1 (amino acids 314–453) domains of the HEV CP, respectively (Fig. 3A). Based on this alignment, a homology model of the human astrovirus CP (excluding the P2 domain) was created using the program MODELLER (26) with the HEV CP structure as a template (Fig. 3B). The crystal structure of astrovirus CP P2 domain was then added to the structure model of the S and P1 domain, based on how the astrovirus CP P2 is superimposed to the HEV CP P2 dimer. By aligning the S and P1 domains of our astrovirus CP model onto the three quasi-equivalent HEV CP molecules from the T = 3 EM model (19), we were able to generate a T = 3 astrovirus capsid structure. Surface spikes made of P2 dimers were shown only on icosahedral twofolds, to be consistent with the cryo-EM reconstruction image of astrovirus (1). Some manual adjustments of the position and the azimuthal rotation of the P2 spikes around the icosahedral twofold were necessary to fit with the cryo-EM reconstruction. As a result, the two P2 domains of HEV and astrovirus do not superimpose because they assume different orientations in these two viruses (Fig. 3B).
Our astrovirus capsid model shows stunning consistency with the cryo-EM reconstruction of astrovirus (1) (Fig. 3C). The diameter of our model is approximately 440 Å including the spikes, and the distance between adjacent spikes is approximately 110 Å, similar to those measured from the cryo-EM reconstruction. Most importantly, the landscape variation of the continuous capsid shell (excluding the spikes) is also similar in both the astrovirus capsid model and the cryo-EM reconstruction. For instance, three bumps are found around quasi-threefold axes, large depressions are seen on quasi-sixfold axes, and a five-pointed, star-shaped ridge is observed on each fivefold axis (Fig. 3C). The close resemblance of the two structures not only provides support for our T = 3 astrovirus capsid model, but also further confirms the notion that astroviruses are phylogenetically related to HEV. Our models should be useful for further analyses of astrovirus structure and assembly.
During maturation, the immature viral particles are processed by extracellular proteases to produce smaller peptides species that are observed in infectious particles. VP34 is derived from the conserved region, whereas VP27 and VP25 are from the hypervariable region of the astrovirus CP (1) (Fig. 3). The N terminus of VP27 and VP25 starts from Gln394 and Ser424, respectively (12). Based on our structural model, we have mapped the N-terminal cleavage site of VP27 (Gln394) to the unstructured loop right before the last β-strand in the P1 domain. Therefore, VP27 should be able to remain associated with the viral particle through main-chain H bonds mediated by this last β-strand in the P1 domain. The N-terminal cleavage site of VP25, however, is located in an extended linker (the P1–P2 linker) between the P1 and the P2 domain. Therefore, VP25 may need to dimerize with a VP27 to remain stably bound to the viral particle.
It is interesting why spikes are observed only on the icosahedral twofolds in mature virion (1). Previously published SDS-PAGE gels show that the amount of VP34 present in infectious particles was comparable to that of VP27 and VP25 (10, 12), indicating that most of the VP27 and VP25, although covalently independent, remain associated with the particle in a disordered manner. The loss of the connection of VP25 to the capsid shell, however, may allow VP25/VP27 spikes to wobble around, thus resulting in orientation disorder and their disappearance from the EM reconstruction on quasi-twofold axes. The spikes located on the icosahedral twofolds are likely to be VP27 homodimers, which have restricted movement just like the spikes in unprocessed capsid and thus can be visualized in a population-averaged reconstruction. We did not model the structure of the P1–P2 linker due to significant sequence variation from that of the hepatitis E virus CP, but we speculate that proteolytic cleavage at Ser424 (that produces VP25) is probably unfavored in icosahedral twofold spikes due to their particular conformation and environment.
Putative Receptor Binding Site(s).
Little is known about astrovirus receptor(s) and virus-receptor binding, although previous antigenicity and sequence analysis implicate the P2 domain in cell attachment and receptor binding (13–15). Considering that amino acid residues lining up the receptor binding site on viral capsids are generally highly conserved (27, 28), we should be able to locate a receptor binding site(s) that is common for all human astrovirus types by mapping conserved amino acid residues onto the surface of the astrovirus P2. It is likely that all human astroviruses use a common receptor for cell internalization, even different astrovirus serotypes might use different cell surface molecules for initial attachment (29).
Aligning the human astrovirus P2415–646 domain of serotype 8 to that of the other seven serotypes gave pairwise percentage identities ranging from 47% to 57%. With such high sequence identities, it is reasonable to assume that the P2 domain of different human astroviruses has similar structure and shape. We then calculated the amino acid conservation scores for the P2 domain using the ConSurf server (30) and the sequences of all eight human astrovirus serotypes. When the amino acid conservation scores were mapped to astrovirus P2, a few conserved regions were observed. The most interesting one, which we call the P site, is located in a shallow groove on top of the P2 spike (Fig. 4). This region has a large number of conserved residues (Lys455, Ser 554, Thr575, and Glu610 strictly conserved in all eight serotypes), all with their side chains exposed to solvent. The location of this site also makes it highly accessible to cell receptors without steric hindrance. Another conserved surface patch, the S site, is located on the side of the P2 spike. This site contains three strictly conserved aromatic residues (Tyr475, Trp604, and Trp606), but two of these (Trp604 and Trp606) have their side chains tucked inside and thus may have been retained for the folding of the protein rather than for ligand recognition. Interestingly, an exposed β-turn (Thr529-Asn530-Asn531-Arg532) between strands β6 and β7 is also well conserved. As discussed above, this β-turn forms a knob on each side of the P2 spike. This β-turn and its adjacent regions have the highest B factors among the entire structure. The exact function of this region is unclear, but we suspect that it may be involved in interactions with other viral or host proteins. A search for potential translational modification sites using PROSITE (31) for this β-turn returned negative results.
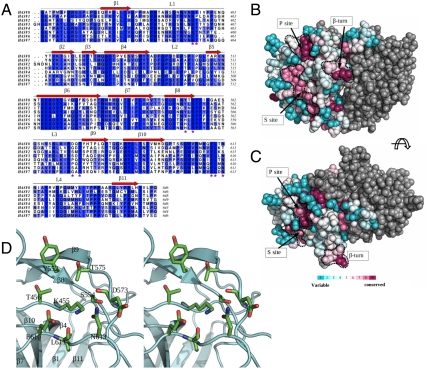
Fig. 4.
Conserved surfaces on the human astrovirus projection domain. (A) Multiple sequence alignment of the projection domains (residues 415–646 for serotype 8) from human astroviruses of the eight different serotypes. The level of conservation is color-coded by different shades of white (least conserved) and blue (strictly conserved). The multiple sequence alignment was computed using ALINE (41). (B and C) Astrovirus projection domain, viewed from the side and the top, respectively. Residues in one subunit are colored according to their levels of conservation. The other subunit is colored in gray. Three highly conserved sites are delineated by dotted black curves. (D) Stereo view of the putative receptor-binding site (P site).
Close inspection of the P site shows that it is largely hydrophilic in nature, and is abundant in hydrogen bond donors and acceptors (Fig. 4D). In addition to the strictly conserved residues Lys455, Ser 554, Thr575, and Glu610, other residues found in the vicinity are Thr456 (Pro in serotype 1), Tyr552 (His in serotypes 5, 6, and 7), and Asn613 (Thr in serotype 1, Ser in serotype 2), together with main-chain carbonyls from Asp573 and Leu611. These residues mainly come from the three loops L1, L3, and L4 (Fig. 4D). The polar residue composition, especially the positively/negatively charged pair Lys455 and Glu610, is also observed in the receptor binding sites of several viruses that use oligosaccharide moieties as receptors (e.g., influenza A virus and norovirus) (32, 33), suggesting that human astroviruses are likely to use a polysaccharide molecule as a cell receptor. Further molecular modeling and docking indicate that the P site may be able to accommodate a di- or trisaccharide moiety.
Consistent with our structural analysis, our biochemical data show that the addition of heparin, heparan sulfate, or dextran sulfate was able to partially block HAstV-8 infectivity in a consistent manner (Fig. S2). Among the three polysaccharides used, heparin had the most pronounced effect. This result thus confirms our hypothesis that a sugar molecule may function as a cell receptor or is at least used for the initial recognition of astrovirus on the cell. It is also worth mentioning that the addition of glycophorin (a protein rich in sialic acid) or the sialidase treatment of host cells had no effect on infectivity, suggesting that sugar molecules other than sialic acid may function as the cell receptor.
In summary, the crystal structure of the human astrovirus P2415–646 provides a first glimpse into the astrovirus capsid structure at the atomic level. The P2415–646 dimer constitutes the astrovirus surface spikes that mediate receptor binding as well as viral antigenicity. The structure reveals a surprising phylogenetic relationship between astroviruses and HEV. Putative receptor binding site(s) have also been mapped based on sequence conservation.
Materials and Methods
Cloning, Expression, and Purification.
The DNA sequence encoding residues 415–646 of astrovirus serotype 8 (Yuc8 strain) ORF2 and a C-terminal 6-histidine tag with a 2-glycine linker was amplified and cloned into the vector pET28 (EMD Chemicals). The E. coli strain Rosetta 2 (DE3) (EMD Chemicals) was transformed with the pET28 vector, and recombinant protein expression was induced with 1 mM IPTG at 15 °C for 16 h after the optical density reached 0.5. seleno-methionine-substituted protein was expressed in M9 minimal medium supplemented with seleno-methionine. Harvested cells were sonicated in a lysis buffer containing 20 mM Tris·HCl (pH 7.5), 300 mM NaCl, and 2 mM PMSF, and the lysate was clarified by centrifugation (20,000 × g, 1 h). The supernatant was applied onto a Ni-nitrilotriacetate (Qiagen) affinity column, and the recombinant protein was eluted with an elution buffer (the lysis buffer with 250 mM imidazole). The protein was further purified by size-exclusion chromatography using a superdex-200 column (GE Healthcare Life Sciences). The purified protein was concentrated to 6–7 mg/mL and stored in 20 mM Tris·HCl (pH 7.5), 200 mM NaCl, and 0.02% sodium azide at 4 °C.
Crystallization, Data Collection, and Data Processing.
Crystals were grown using the hanging drop, vapor diffusion method. Two crystal forms were obtained (Table S1). Crystals in the cubic space group I213 were grown in 0.1 M sodium acetate (pH 4.6), 0.2 M ammonium sulfate, and 16% PEG 4000, whereas crystals in the trigonal space group P3221 were grown in 0.1 M 2-(N-cyclohexylamino)ethanesulfonic acid (pH 9.5), 0.1 M glycine, and 28% PEG 3000. Crystals were flash frozen in liquid nitrogen using 20% xylitol (for the cubic crystals) or 22.5% sucrose (for the trigonal crystals) as cryoprotectants. Diffraction data were collected at the beamline 4.2.2 at the Advanced Light Source. The diffraction data were processed with HKL2000 (34).
Structure Determination.
The astrovirus P2415–646 structure was solved by MAD (Table S1). PHENIX (35) was used to determine the location of the six selenium sites and to autobuild an initial model. With the initial model, native data and the program PHASER (36) were used to obtain the native protein structures. The structures were refined and manually rebuilt with Refmac5 (37) and Coot (38), respectively. The final model for the trigonal crystal has an Rwork of 17.9% and an Rfree of 21.5%. A total of 137 water molecules are included in the final model. A Ramachandran plot of the final model shows that 212 residues (98.2%) are in the most favored regions and four residues (1.8%) in the additionally allowed regions. The model has been deposited in the Research Collaboratory for Structural Bioinformatics Protein Data Bank (PDB ID 3QSQ).
Supplementary Material
Acknowledgments.
We thank Kelly Dryden, Mariana Tihova, and Mark Yeager for sharing their electron microscopy reconstruction images with us for comparison. We thank Yangyang Dong, Max Nibert, and Hongmao Sun for helpful discussions. X-ray diffraction data were collected at Advanced Light Source and Advanced Photon Source. This work is supported by the Welch Foundation (C-1565 to Y.T.), the National Institutes of Health (AI067638 to Y.T.), DGAPA-UNAM (219910 to E.M.), CONACyT (79574 to E.M.), and the Kresge Science Initiative Endowment Fund at Rice University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3QSQ).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1104834108/-/DCSupplemental.
References
- 1.Mendez E, Arias CF. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 982–1000. [Google Scholar]
- 2.Walter JE, Mitchell DK. Astrovirus infection in children. Curr Opin Infect Dis. 2003;16:247–253. doi: 10.1097/00001432-200306000-00011. [DOI] [PubMed] [Google Scholar]
- 3.Lewis TL, Matsui SM. Astrovirus ribosomal frameshifting in an infection-transfection transient expression system. J Virol. 1996;70:2869–2875. doi: 10.1128/jvi.70.5.2869-2875.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mendez E, Salas-Ocampo MPE, Munguia ME, Arias CF. Protein products of the open reading frames encoding nonstructural proteins of human astrovirus serotype 8. J Virol. 2003;77:11378–11384. doi: 10.1128/JVI.77.21.11378-11384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geigenmuller U, Ginzton NH, Matsui SM. Studies on intracellular processing of the capsid protein of human astrovirus serotype 1 in infected cells. J Gen Virol. 2002;83:1691–1695. doi: 10.1099/0022-1317-83-7-1691. [DOI] [PubMed] [Google Scholar]
- 6.Dalton RM, Pastrana EP, Sanchez-Fauquier A. Vaccinia virus recombinant expressing an 87-kilodalton polyprotein that is sufficient to form astrovirus-like particles. J Virol. 2003;77:9094–9098. doi: 10.1128/JVI.77.16.9094-9098.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang QH, et al. Genetic analysis of the capsid region of astroviruses. J Med Virol. 2001;64:245–255. doi: 10.1002/jmv.1043. [DOI] [PubMed] [Google Scholar]
- 8.Mendez E, Salas-Ocampo E, Arias CF. Caspases mediate processing of the capsid precursor and cell release of human astroviruses. J Virol. 2004;78:8601–8608. doi: 10.1128/JVI.78.16.8601-8608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banos-Lara MdR, Mendez E. Role of individual caspases induced by astrovirus on the processing of its structural protein and its release from the cell through a non-lytic mechanism. Virology. 2010;401:322–332. doi: 10.1016/j.virol.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 10.Bass DM, Qiu S. Proteolytic processing of the astrovirus capsid. J Virol. 2000;74:1810–1814. doi: 10.1128/jvi.74.4.1810-1814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Monroe SS, et al. Temporal synthesis of proteins and RNAs during human astrovirus infection of cultured cells. J Virol. 1991;65:641–648. doi: 10.1128/jvi.65.2.641-648.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mendez E, Fernandez-Luna T, Lopez S, Mendez-Toss M, Arias CF. Proteolytic processing of a serotype 8 human astrovirus ORF2 polyprotein. J Virol. 2002;76:7996–8002. doi: 10.1128/JVI.76.16.7996-8002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez-Fauquier A, et al. Characterization of a human astrovirus serotype 2 structural protein (VP26) that contains an epitope involved in virus neutralization. Virology. 1994;201:312–320. doi: 10.1006/viro.1994.1296. [DOI] [PubMed] [Google Scholar]
- 14.Krishna NK. Identification of structural domains involved in astrovirus capsid biology. Viral Immunol. 2005;18:17–26. doi: 10.1089/vim.2005.18.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bass DM, Upadhyayula U. Characterization of human serotype 1 astrovirus-neutralizing epitopes. J Virol. 1997;71:8666–8671. doi: 10.1128/jvi.71.11.8666-8671.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Risco C, Carrascosa JL, Pedregosa AM, Humphrey CD, Sanchez-Fauquier A. Ultrastructure of human astrovirus serotype 2. J Gen Virol. 1995;76:2075–2080. doi: 10.1099/0022-1317-76-8-2075. [DOI] [PubMed] [Google Scholar]
- 17.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xing L, et al. Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J Biol Chem. 2010;285:33175–33183. doi: 10.1074/jbc.M110.106336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guu TSY, et al. Structure of the hepatitis E virus-like particle suggests mechanisms for virus assembly and receptor binding. Proc Natl Acad Sci USA. 2009;106:12992–12997. doi: 10.1073/pnas.0904848106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamashita T, et al. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc Natl Acad Sci USA. 2009;106:12986–12991. doi: 10.1073/pnas.0903699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li S, et al. Dimerization of hepatitis E virus capsid protein E2s domain is essential for virus-host interaction. PLoS Pathog. 2009;5:e1000537. doi: 10.1371/journal.ppat.1000537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koci MD, Seal BS, Schultz-Cherry S. Molecular characterization of an avian astrovirus. J Virol. 2000;74:6173–6177. doi: 10.1128/jvi.74.13.6173-6177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Prasad BV, et al. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286:287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- 25.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2004;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 26.Marti-Renom MA, et al. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 27.Bewley MC, Springer K, Zhang YB, Freimuth P, Flanagan JM. Structural analysis of the mechanism of adenovirus binding to its human cellular receptor, CAR. Science. 1999;286:1579–1583. doi: 10.1126/science.286.5444.1579. [DOI] [PubMed] [Google Scholar]
- 28.Chapman MS, Rossmann MG. Comparison of surface properties of picornaviruses: Strategies for hiding the receptor site from immune surveillance. Virology. 1993;195:745–756. doi: 10.1006/viro.1993.1425. [DOI] [PubMed] [Google Scholar]
- 29.Brinker JP, Blacklow NR, Herrmann JE. Human astrovirus isolation and propagation in multiple cell lines. Arch Virol. 2000;145:1847–1856. doi: 10.1007/s007050070060. [DOI] [PubMed] [Google Scholar]
- 30.Ashkenazy H, Erez E, Martz E, Pupko T, Ben-Tal N. ConSurf 2010: Calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 2010;38:W529–533. doi: 10.1093/nar/gkq399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Castro E, et al. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao S, et al. Structural basis for the recognition of blood group trisaccharides by norovirus. J Virol. 2007;81:5949–5957. doi: 10.1128/JVI.00219-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gamblin SJ, et al. The structure and receptor binding properties of the 1918 influenza hemagglutinin. Science. 2004;303:1838–1842. doi: 10.1126/science.1093155. [DOI] [PubMed] [Google Scholar]
- 34.Otwinowski Z, Minor W. Macromolecular Crystallography Part A Processing of X-Ray Diffraction Data Collected in Oscillation Mode. London: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 35.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 38.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 39.Schrödinger, LLC . New York: Schrödinger, LLC; 2010. The PyMOL Molecular Graphics System. Ver. 1.3. [Google Scholar]
- 40.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.Bond CS, Schuttelkopf AW. ALINE: A WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.