Abstract
DNA topoisomerases are believed to promote transcription by removing excessive DNA supercoils produced during elongation. However, it is unclear how topoisomerases in eukaryotes are recruited and function in the transcription pathway in the context of nucleosomes. To address this problem we present high-resolution genome-wide maps of one of the major eukaryotic topoisomerases, Topoisomerase II (Top2) and nucleosomes in the budding yeast, Saccharomyces cerevisiae. Our data indicate that at promoters Top2 binds primarily to DNA that is nucleosome-free. However, although nucleosome loss enables Top2 occupancy, the opposite is not the case and the loss of Top2 has little effect on nucleosome density. We also find that Top2 is involved in transcription. Not only is Top2 enriched at highly transcribed genes, but Top2 is required redundantly with Top1 for optimal recruitment of RNA polymerase II at their promoters. These findings and the examination of candidate-activated genes suggest that nucleosome loss induced by nucleosome remodeling factors during gene activation enables Top2 binding, which in turn acts redundantly with Top1 to enhance recruitment of RNA polymerase II.
Keywords: histone eviction, relaxase, gene regulation
Topoisomerase II (Top2) produces a transient double-stranded DNA break allowing it to transport a second duplex through the break in order to regulate DNA supercoiling (1). Thus, Top2 acts to decatenate DNA during replication (2, 3) and to regulate chromosome condensation during mitosis (4). Preventing excess supercoiling during transcription is also a proposed function for Top2. Studies in prokaryotes, later performed in budding yeast, led to the development of the twin-supercoiled-domain model, in which positive supercoils ahead of RNA polymerase and negative supercoils behind it need to be relaxed by topoisomerases for efficient transcription elongation to occur (5–7). However, it is not clear how topoisomerases are recruited to active genes in eukaryotes especially in the context of chromatin structure.
Several conflicting results have been reported with regard to the effect of topoisomerase activity on nucleosomes in vitro, and the relationship between topoisomerase activity and chromatin architecture in vivo is unclear. Chromatin assembly extracts from budding yeast show that topoisomerase activity is required for nucleosome assembly as measured by plasmid supercoiling and micrococcal nuclease digestion (8). Yet, chemical inhibition of Top2 in Xenopus extracts leads to nucleosome repositioning, but not to reduced nucleosome occupancy arguing that Top2 may not be required for nucleosome assembly (9). In contrast, relaxed DNA is not a favorable substrate for nucleosome formation (10), suggesting that the release of DNA supercoils by Top2 may inhibit nucleosome assembly. In support of this hypothesis, it has been shown that Top2 can facilitate histone eviction in vitro and that loss of topoisomerase activity leads to decreased nucleosome disassembly in vivo in Schizosaccharomyces pombe (11, 12).
Top2 may also be important for releasing supercoils generated during Pol II transcription in vivo. In Saccharomyces cerevisiae topoisomerase I (Top1) and Top2 can both relax positively and negatively supercoiled DNA, but Top2 is the main relaxase on chromatin templates (13). In top1Δtop2-ts mutant strains in which both topoisomerase activities are defective ribosomal RNA transcription is significantly decreased (2, 14). Surprisingly, global polyA+ RNA synthesis was reduced by only approximately 3-fold, whereas mRNA synthesis of specific target genes was not affected (2). Other studies have found that Pol II transcription on a chromatin template in vitro requires topoisomerase activity only in long genes (15, 16), suggesting its involvement in transcription elongation. This is also the case in S. pombe, where removal of Top1 and Top2 activity leads to decreased transcription by Pol II at long genes in vivo (11). Despite this association between Top2 and Pol II transcription, it has been reported that although Top2 binds to intergenic regions in S. cerevisiae in a pattern correlated with transcriptional activity, Top2 is dispensable for Pol II transcription, at least in S phase (17).
In light of these contradictory findings, we wished to further characterize the role of Top2 on nucleosomal DNA in vivo and its role in Pol II transcription. Using high-density microarrays and genetic analysis, we address the relationship between nucleosome presence and Top2 binding. In addition, we address the relationship between Top2 and Pol II transcription. Our data suggest that nucleosome remodeling during gene activity displaces nucleosomes to allow Top2 binding, which functions redundantly with Top1 to enhance Pol II recruitment at highly transcribed genes.
Results
Top2 Binds Primarily to DNA That Is Nucleosome-Free.
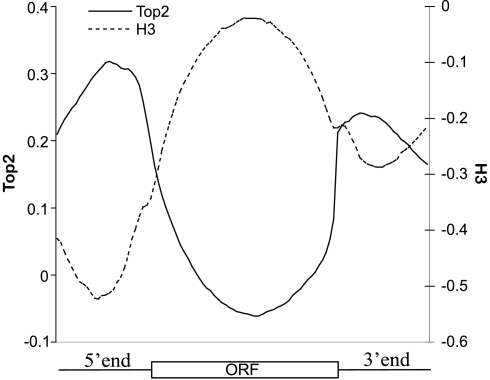
In order to map the location of Top2 in the yeast genome we first used chromatin immunoprecipitation and Affymetrix high-density microarrays, which tile the entire yeast genome at 5-base pair (bp) resolution (ChIP-chip) (18). ChIP DNA associated with Top2 was compared to input DNA using Affymetrix tiling analysis software (17, 19). As shown in Fig. 1, Top2 is enriched in intergenic regions that are coincident with the promoter nucleosome-free region (NFR) (20–22). In order to compare the relationship between Top2 and nucleosomes, we performed ChIP-chip using an antibody directed against the histone H3 C terminus to measure nucleosome occupancy. As shown in the composite profiles of Top2 binding and nucleosome density (Fig. 1) Top2 and nucleosomes are found in a mutually exclusive pattern across genes throughout the S. cerevisiae genome. Detailed maps of Top2 and histone binding at chromosome VI are shown in Fig. S1. We conclude that Top2 is primarily found at chromosomal regions, such as promoters, that are nucleosome-free. Therefore, we have chosen to focus this study on the role of Top2 within nucleosome-free promoter regions upstream of ORFs.
Fig. 1.
Exclusive distribution of Top2 and nucleosome occupancy. ChIP DNA of Top2 and H3 and input from wild-type cells were amplified, labeled, and hybridized to Affymetrix Tiling arrays. The average binding over 6,576 annotated genes and their adjacent 500-bp regions are shown. Top2 and H3 averages at intergenic and ORF regions were done using 25 and 50 equal size bins, respectively. Enrichment of Top2 or H3 ChIP DNA is shown as the log 2 ratios of IP versus input DNA.
Nucleosome Loss Enables Top2 Occupancy.
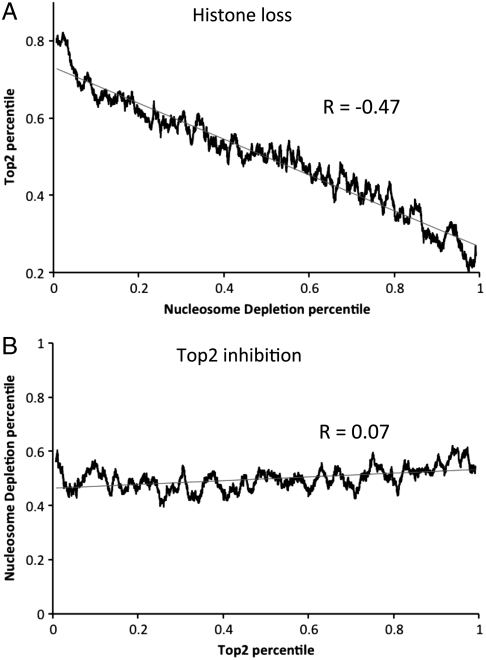
Given the mutually exclusive distributions of nucleosomes and Top2, we wished to know whether histones prevent Top2 binding or vice versa. To deplete histones from chromatin, we repressed histone H4 mRNA synthesis using a yeast strain containing a single copy of the H4 gene fused to the inducible GAL promoter (23). By shifting cells from galactose to glucose we were able to selectively shut off H4 synthesis, depleting cells of approximately 50% of histones because histone H4 synthesis is repressed while the cells progress through one final round of DNA replication (23). After repression of the histone H4 gene for 6 h, nucleosome density and Top2 binding were measured by ChIP-chip using Agilent tiling arrays (50-bp resolution). In these two-channel microarrays the data represent the relative change in DNA binding of histone H3 and Top2 between the mutant and wild-type strains. Paradoxically, we noted that a number of genes showed an apparent increase in H3 signal following nucleosome depletion. In order to determine whether this was an artifact of array normalization, where loci with relatively unchanged H3 levels appear to gain signal due to a genome-wide decrease, we examined genes that have an apparent increase in H3 level using semiquantitiative PCR. Fig. S2 shows the results of H3 ChIP at the ORFs of eight randomly selected genes, four from the top 100 depleted and four from the bottom 100 depleted in H3 occupancy following nucleosome depletion. We confirmed that histone depletion results only in the loss but not in any significant gain of histone H3. Because we find no absolute increase in H3 binding in the bottom 100 group upon histone depletion, we present the binding data on a relative scale as percentile rank in which 0 and 100th percentile correspond to the observed maximal and minimal decrease in H3 upon nucleosome depletion. H3 and Top2 binding at 6,368 coding regions were compared as a moving average with a window size of 100 genes and step size of one gene. As shown in Fig. 2A, levels of Top2 binding increase the most at sites where nucleosome depletion is the greatest (Pearson correlation = -0.47). We conclude that decreased nucleosome presence enables increased Top2 binding to DNA.
Fig. 2.
Top2 binding is enabled by nucleosome depletion, but Top2 loss does not alter nucleosome density. Top2 and H3 levels were measured by ChIP-chip following nucleosome depletion (strain UKY403) or topoisomerase inactivation (strain RS192). UKY403 (pGal-H4) cells were grown to log phase in medium containing galactose before shifting to glucose medium for 6 h. Topoisomerase mutant strains were grown to log phase at 25 °C and then shifted to 37 °C for 30 min prior to harvesting chromatin. (A) Data for 6,368 ORFs were percentile ranked to correspond to the nucleosome depletion (x axis) or Top2 increase (y axis) and the moving average (window size 100; moving step 1 gene) was plotted. (B) Top2 and H3 binding upon Top2 inhibition was compared as in A. R represents the Pearson correlation between the percentile-ranked samples. Percentile ranking was used to facilitate correlation of microarray datasets.
In order to further characterize the recruitment of Top2 to a promoter of a gene undergoing natural nucleosome loss, we wished to examine nucleosome depletion and Top2 binding during activation of a gene. We chose to examine the PHO5 gene, which is induced following phosphate starvation and is known to rapidly lose nucleosomes from the promoter in an ordered manner during activation (24–26). Shown in Fig. S3 is H3, Top2, and Pol II binding at the PHO5 gene as measured by standard ChIP following shift to low phosphate media. These data demonstrate that, during activation, nucleosomes are disassembled at the promoter and Top2 accumulates at this site.
Top2 Occupancy Is Not Required for Nucleosome Loss.
To determine whether Top2 loss enables nucleosome occupancy, we examined H3 levels in topoisomerase deficient strains. Nucleosome formation has been reported to be more efficient on supercoiled DNA (10), suggesting that the DNA relaxing activity of Top2 (or Top1) might act to prevent nucleosome assembly (1, 27). To address this possibility we examined nucleosome binding in a mutant strain (top1Δtop2-ts) lacking Top1 and Top2 topoisomerase activity and compared it to wild type. After shifting to the nonpermissive temperature for 30 min, Top2 and H3 ChIP-chip analysis were performed as above. We find that following shift to the nonpermissive temperature there is a dramatic decrease in Top2 binding throughout the genome in the top2-ts mutant strain (data are clustered according to Pol II binding level) (Fig. S4). Therefore, we conclude that shift to the restrictive temperature causes mutant Top2 to dissociate from the genome; however, we cannot exclude the possibility that temperature shift alters only the epitope recognized by our antibody. In order to address whether Top2 directly affects nucleosome density, we determined the correlation between the degree of Top2 loss and change in H3 density. Shown in Fig. 2B is a plot of the percentile rank change in H3 and Top2 binding. We find that there is little to no correlation (Pearson correlation = 0.07) between loss of Top2 following transition to the restrictive temperature and change in histone H3 density. Therefore, we conclude that the loss of topoisomerase activity has little effect on nucleosome density.
Top2 Is Enriched at the Promoters of Highly Transcribed Genes.
Transcription is generally associated with nucleosome loss (28). Therefore we wished to know whether Top2 targets are more highly transcribed. Average Top2 binding across the promoter and ORF was separated into five groups according to previously reported transcription frequency rates (29). Fig. S5 shows the average levels of Top2 (Fig. S5A) and histone H3 (Fig. S5B) across the ORFs and their adjacent DNA regions. Consistent with previous reports (20, 30), we find that nucleosome occupancy is negatively correlated with transcription rate, whereas Top2 binding shows a positive correlation. This finding is consistent with previously published reports in budding yeast (17). However, in S. pombe, only Top1 is selectively enriched at active genes (11). In order to compare the roles of the topoisomerases in the two yeasts, we analyzed genome-wide Top1 binding data from a recently published report (31). Shown in Fig. S6 is Top1 binding, separated into groups according to transcriptional frequency at the promoter, transcription start site, and the ORF. We conclude that in contrast to S. pombe, in exponentially growing S. cerevisiae cells, both Top1 and Top2 bind to promoter DNA in a manner that is positively correlated with transcription.
Top2 Is Not Recruited to Promoters by RNA Pol II.
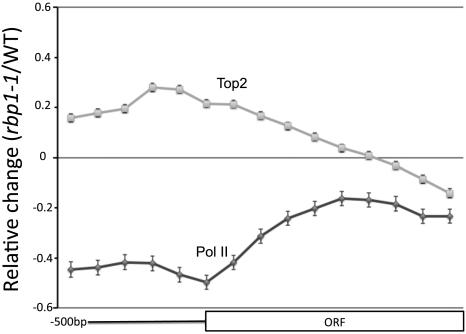
Many components of the transcriptional apparatus are recruited to genes by RNA polymerase itself, and we wondered whether this might be the case with Top2. We therefore asked whether Pol II activity determines Top2 binding using a temperature-sensitive allele of Pol II (rpb1-1). Pol II and Top2 ChIP DNA from the rpb1-1 mutant were compared with that from the RPB1 wild type at 37 °C (29), using Agilent microarrays as above. The relative change in binding data across 6215 ORFs for which we had sufficient probes and their associated promoters were then averaged. As expected there is a significant decrease in Pol II enrichment across the ORF at the nonpermissive temperature in the rpb1-1 strain. The greatest decrease in Pol II binding occurred in the promoter region adjacent to the translation start site (Fig. 3). In contrast, there is a slight increase in Top2 enrichment over the promoter (Fig. 3). There is also a small decrease in Top2 binding at the end of the ORF (Fig. 3). This suggests that Top2 is recruited to the promoter and may then migrate into the ORF either via a direct interaction with the transcriptional machinery or following the production of transcription-induced supercoils. In the absence of transcription, Top2 could then be unable to migrate and thus accumulates at the promoter. We conclude that Pol II is not required for recruitment of Top2 to the promoter.
Fig. 3.
Top2 is not recruited to promoters by RNA Pol II. Pol II and Top2 binding were compared following inhibition of Pol II (strain Z460). Cells were grown to log phase at 25 °C and then shifted to 37 °C for 45 min prior to harvesting chromatin. ORFs were separated into 10 equal sized bins and their associated promoters were divided into five 100-bp bins. Data for 6215 ORFs were averaged. Error bars represent the 95% confidence interval.
Top2 Is Required Redundantly with Top1 for Efficient Pol II Recruitment.
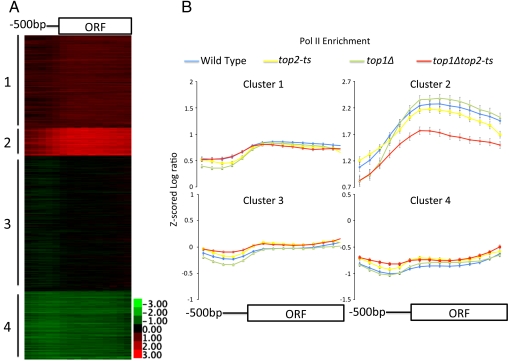
We then wished to ask whether Top2 is required for Pol II recruitment. In order to address this, we measured Pol II occupancy normalized to input DNA across the yeast genome using Agilent tiling arrays in wild-type and topoisomerase mutant cells. In order to ensure that we were, in fact, losing topoisomerase activity in the temperature-sensitive mutant we also measured Top2 binding in the mutants. Z scoring involves the transformation of microarray data so that the genome average is set to zero and the standard deviation is set to one facilitating interarray comparison. Normalized log ratio data from the Pol II arrays were Z scored and binding data across 6215 ORFs for which we had sufficient probes were extracted. Each ORF was divided into 10 equally sized bins, and its associated 5′ upstream region was divided into five 100-bp bins. As expected, growth at the top2-ts nonpermissive temperature caused a dramatic loss of Top2 from the genome (Fig. S4). Because of the strong correlation observed between transcriptional activity and Top2 binding, we wished to examine genes in which there was a high level of Pol II binding in order to determine whether Top2 loss affects transcription. Therefore we divided the Pol II binding data in wild type into four groups by K-means clustering. As shown in Fig. 4A, the genes most highly bound by RNA Pol II cluster together in group 2 (533 genes). The binding data in topoisomerase mutant cells was aligned to that in wild type, and the average binding within each cluster was computed (Fig. 4B). Consistent with previously published results (17), there is little if any effect of TOP2 mutation alone on Pol II binding (compare blue and yellow lines in Fig. 4B).
Fig. 4.
Top2 acts redundantly with Top1 to enhance Pol II recruitment. Log ratio data from ChIP of Pol II in wild type (W303-1a) normalized to input, was Z scored and the average binding across 6215 promoters (divided into five 100-bp bins) and ORFs (divided into 10 equal size bins) was determined and separated into four groups using K-means clustering (A). Number of genes in each cluster: cluster 1 (n = 1,774), cluster 2 (n = 533), cluster 3 (n = 2,601), and cluster 4 (n = 1,303). Average Z scored Pol II binding data for each cluster identified in A was determined in wild-type (W303-1a, blue line), top1Δ (AMR51, green line), top2-ts (RS191, yellow line), and top1Δtop2-ts (RS192, red line) strains (B). Cells were grown to log phase at 25 °C and then shifted to 37 °C for 60 min prior to harvesting chromatin. Error bars represent the 95% confidence interval.
Because Top2 is so clearly enriched at highly transcribed genes (Fig. S5), we next asked whether Top1 might play a redundant role with Top2 during recruitment of Pol II. First we examined Pol II binding in a top1Δ strain, and as shown in Fig. 4B, it has no significant effect on Pol II recruitment in any of the clusters (compare blue and green lines in Fig. 4B). Moreover, deletion of TOP1 did not alter the distribution of Top2 throughout the genome (Fig. S4). Our findings are consistent with previous reports that top1Δ affects only a small subset of telomere-proximal genes (32). We then examined the genome-wide binding of Pol II in a top1Δtop2-ts double mutant. As shown in Fig. 4B (compare red and blue lines), there is a significant decrease in Pol II across the ORFs of genes found in cluster 2. Loss of topoisomerase activity has no obvious effect on genes in cluster 1, 3, or 4 (Fig. 4B), which are more weakly transcribed. Therefore, we conclude that Top1 and Top2 are redundantly required for efficient recruitment of RNA Pol II to highly transcribed genes.
Previous in vitro experiments (16) as well as a recent report in S. pombe demonstrated an important role for topoisomerase activity for the transcription of long genes (11). We wished to know whether this was the case in S. cerevisiae as well. Therefore, we sorted genes based upon the length of their coding regions, and we plotted the average Pol II binding from -1,000 to +3,000 bp across the promoter and ORF. As shown in Fig. S7, there is no significant change in Pol II binding in strains lacking topoisomerase activity, as compared to wild type, when the data are clustered according to gene length.
Nucleosome Remodeling Occurs Upstream of Top2 Binding at PHO84.
An early step in gene activation is the recruitment of ATP-dependant nucleosome remodeling complexes that displace nucleosomes and promote transcriptional activation (33–35). If these remodeling complexes cause nucleosome loss, our results predict that this would enable Top2 binding at the promoter as an intermediate step in transcriptional activation. SWI/SNF is a nucleosome remodeling complex that evicts or slides nucleosomes (33, 36). Previous work has shown that SWI/SNF mediated nucleosome remodeling is an important regulator of approximately 1% of the genome and that the acid phosphatase genes, including PHO84, are among the genes most affected in a swi/snf mutant (37).
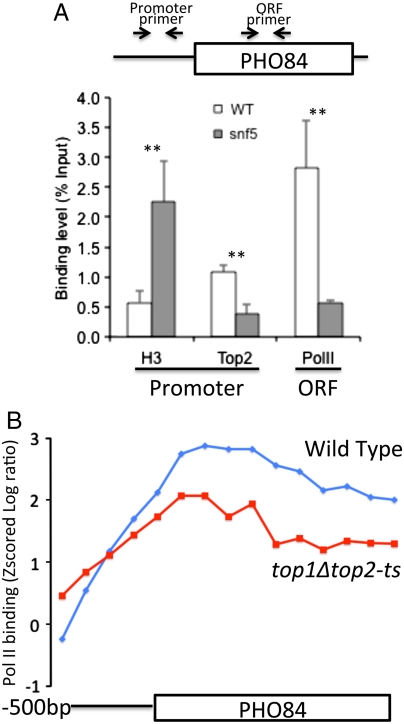
To assess the relationship between nucleosome remodeling, Top2 binding and transcription at PHO84 we determined the levels of histone H3, Top2, and Pol II in a yeast mutant lacking Snf5, an essential component of the SWI/SNF complex (Fig. 5A). We confirmed that Snf5 is involved in nucleosome displacement because histone H3 levels at the PHO84 promoter are significantly higher in the snf5 mutant than in wild-type yeast. Our ChIP analysis also shows that Top2 and Pol II binding to the PHO84 promoter decreases significantly (about 3-fold and 6-fold, respectively) in the snf5 mutant (Fig. 5A). We conclude that SWI/SNF mediated chromatin remodeling leads to nucleosome loss and allows for Top2 binding and transcription of PHO84.
Fig. 5.
Loss of SWI/SNF function leads to defects in histone eviction and decreased recruitment of Top2 and Pol II at PHO84. H3, Top2, and Pol II enrichment were determined using ChIP at the promoter or coding region of the PHO84 gene (A). Consistent with previous reports (37), H3 occupancy at the promoter increased and Pol II binding within the coding region decreased significantly following loss of SWI/SNF in the snf5 deletion strain as compared to wild type (BY4741). Top2 binding also showed a significant decrease following snf5 deletion. The average of three experiments is shown. Error bars represent the standard deviation. Double asterisks indicate p < 0.01 in a one-sided t test. (B) The binding of Pol II across the promoter (divided into five 100-bp bins) and ORF (divided into 10 equal size bins) of the PHO84 gene in wild type (W303-1a, blue line) and top1Δtop2-ts (RS192, red line) is shown as determined from ChIP-chip data.
To determine whether the recruitment of Top2 is important for transcription of PHO84, we examined our Pol II array data in the top1Δtop2-ts mutant. Shown in Fig. 5B is the average Pol II binding across the PHO84 ORF. Pol II binding is decreased in the top1Δtop2-ts mutant strain in comparison to wild type as determined by our microarray analysis. We conclude that SWI/SNF recruitment during gene activation results not only in nucleosome eviction but also in subsequent Top2 binding at the PHO84 promoter and improved recruitment of RNA Pol II.
Discussion
In this paper, we show that there is an inverse relationship between nucleosome presence and Top2 binding at promoters. We find that natural or genetically induced nucleosome loss enables topoisomerase II binding. Conversely, Top2 occupancy has little effect on nucleosome presence. Although Top2 presence correlates strongly with increased gene transcription, we find that RNA polymerase II presence is not required for Top2 binding. However, topoisomerases Top1 and Top2 function redundantly to facilitate recruitment of the transcriptional machinery to highly active genes, and it suggests that Top1 and Top2 may function to relieve the torsional stress introduced by multiple rounds of transcription. Finally we find that Swi/Snf disruption of nucleosomes at the PHO84 promoter is required for Top2 binding at this site arguing that nucleosome remodeling enables Top2 binding and subsequent RNA polymerase recruitment.
Nucleosome Eviction Enables Top2 Recruitment.
Consistent with previous reports we have shown that Top2 is found at gene promoters in a pattern correlated with transcription (17), but it was unclear how Top2 is recruited to these genomic loci. Nucleosome loss can occur in vivo in yeast through a number of different mechanisms. These include the presence of natural NFRs at promoters (38), nucleosome loss upon gene activation such as that which occurs at PHO5 (25, 26, 39), or genetically stimulated nucleosome depletion by shutting off histone H4 synthesis on a GAL1-H4 construct (23). We demonstrate that in all cases histone depletion is sufficient to trigger Top2 recruitment, suggesting that Top2 binding is a default state that occurs whenever nucleosomes are evicted from the promoter (Fig. 2A). Top2 is found in complexes with RNA Pol II and could also be recruited through direct interaction with the transcription complex (15). Alternatively, it could be recruited to supercoiled DNA produced by active transcription as suggested by the twin-supercoiled-domain model (5). However, we show that Top2 is not recruited by transcription as Top2 binding does not decrease when RNA Pol II is inhibited (Fig. 3). Chromatin remodeling and nucleosome eviction by complexes such as Swi/Snf also occur prior to Top2 recruitment (Fig. 5A). The simplest explanation for our data is that the formation of naked DNA following nucleosome eviction by chromatin remodelers prior to recruitment of RNA Pol II is sufficient to provide a binding site for Top2. However, we cannot exclude the possibility that an additional factor is needed to target Top2 to nucleosome-free promoter regions.
Top2 Is Not Required for Nucleosome Assembly or Disassembly in Budding Yeast.
Previous studies have produced contradictory conclusions regarding the requirement for topoisomerase activity during nucleosome assembly and eviction. They have demonstrated a requirement for topoisomerases during nucleosome remodeling in budding yeast in vitro (8, 12) and in S. pombe in vivo (11), but not in Xenopus oocytes (9, 40). We find that in S. cerevisiae there is no effect of loss of topoisomerase activity on nucleosome density in vivo (Fig. 2B). These contradictory findings may be explained by the fact that our experiments were done in living yeast, as well as by differences between the species studied. For instance, the binding pattern of Top2 varies significantly between budding yeast and S. pombe where Top1 is more highly correlated with transcription (11). Alternatively, the increase in nucleosome occupancy observed by Durand-Dubief et al. could be secondary to decreased transcription in the topoisomerase mutant strain (11). We do see a very mild increase in nucleosome occupancy at gene promoters following topoisomerase inactivation (Fig. S8); however, we believe that this is secondary to alterations in transcription as there is no correlation between the degree of Top2 loss and the change in H3 occupancy, a finding observed both at ORFs (Fig. 2B) and at promoters (Fig. S9). Therefore, we conclude that Top2 is not required for nucleosome disassembly in vivo in S. cerevisiae.
Top2 and Top1 Act Redundantly to Enhance Pol II Recruitment at Highly Transcribed Genes in Vivo.
As both Top1 and Top2 can relieve positive and negative supercoils, it is believed that they can substitute for one another in many contexts (1, 27). It has been proposed that transcription in prokaryotes requires topoisomerase activity due to the twin-supercoiled-domain model (5, 6). However, previous reports in budding yeast have demonstrated little or no defect in transcription in topoisomerase mutants (2, 17). We have now shown that Top2 inhibition in budding yeast leads to decreased transcription as measured by Pol II occupancy. However, this occurs only at highly transcribed genes and in a top1Δ background, suggesting that Top2 acts to promote Pol II recruitment redundantly with Top1, and therefore that it is the DNA relaxase activity that is important in transcription. These results are consistent with a recently published report in S. pombe showing that Top1 and Top2 are redundantly important for transcription (11). However, it appears that Top1 is the dominant topoisomerase during transcription in S. pombe.
How might topoisomerase activity be important for transcription? We find that, consistent with previous reports (11, 14), topoisomerase function is most important at highly active genes, suggesting that the topological problems introduced by transcription are most severe when a gene has multiple copies of polymerase actively transcribing it. Relaxing supercoiled DNA could be required either at the promoter prior to polymerase recruitment or after recruitment, during transcription elongation. One feature of the twin-supercoiled-domain model is that it predicts that local negative supercoiling will be the highest at the promoter (6), a feature that may be exacerbated by the concurrent removal of nucleosomes (41). Studies in mammalian cells have demonstrated a requirement for Top2 catalytic activity in order to recruit Pol II to the promoters of some nuclear receptor targets (42). One possibility then is that topoisomerase catalytic activity at the promoter is important for resolving negative supercoils produced either by nucleosome disassembly or by previous rounds of transcription, which could act as a structural impediment to further Pol II recruitment.
In contrast to studies in S. pombe, we did not see any association between gene length and a requirement for topoisomerase activity, something that might be expected if it were important for elongation (11). The S. pombe genome is significantly more complex as its genes contain many more introns and have longer intergenic segments than do genes in S. cerevisiae (43). Therefore, the greater length of transcripts in S. pombe may increase the need for topoisomerase activity during elongation. In conclusion our genome-wide and gene specific studies provide a likely mechanism by which Top2 is recruited to promoters to enhance activity at highly transcribed genes in vivo. How Top2 promotes transcription in eukaryotes remains an important area for future study.
Experimental Procedures
Yeast Strains and Media.
Lists of yeast strains and primer sequences used in this study are provided in Tables S1 and S2. Cells were grown in YEP (yeast extract peptone) dextrose media unless otherwise mentioned.
Chromatin Immunoprecipitation and Genome-Wide Mapping.
ChIP assays were performed as previously described (44). Briefly, cross-linked whole cell extract was incubated overnight with antibodies directed against histone H3 C terminus (Abcam, 1791), Top2 (TopoGEN Inc., 2014), or Pol II C terminus (Covance, 8WG16) together with Protein A-agarose beads. The Top2 and H3 ChIP-chip assays with Affymetrix microarrays were performed in a wild-type strain as previously described (45). Subsequent ChIP-chip assays were done using two-color Agilent 244-k tiling arrays (G4491A). Amplification and labeling was done as previously described (46). Hybridization and washing were done according to the manufacturer’s instructions. Following array scanning, features were extracted using Agilent Feature Extraction and normalized using Agilent Chip Analytics with the default settings. To Z score data, the genome average was subtracted from the log ratio and then divided by the genome standard deviation for each probe. Average probe signal was extracted within bins as described in the figure legends. Genes were selected such that at least 50% of bins contained valid signal. K-means clustering was performed using Cluster 3.0 (47) (1,000 runs) and visualized in Java Treeview (48). For nucleosome depletion, strain UKY403, containing pGal-H4, was grown in YEP dextrose for 6 h following shift from YEP galactose (YEPG) culture. Control cells were obtained from identical cultures grown in YEPG. To inhibit Top2 activity the top2-ts mutants were compared with wild type after shift from 25 °C to 37 °C for 30 or 60 min. The rpb1-1 mutant and isogenic wild-type strains were treated as previously described (29). For PHO5 induction, wild-type yeast cells were grown to log phase in SDcomplete and then shifted to SD-phosphate media for the indicated times.
Supplementary Material
Acknowledgments.
We thank members of the Grunstein lab for helpful discussion and critical reading of the manuscript and, in particular, Wei Xie and Graham Davies for computational assistance. We are also grateful to Dr. Sternglanz for topoisomerase mutants, and the UCLA microarray core facility for array services. This work was supported by grants from the National Institutes of Health Medical Scientist Training Program and a Jonsson Comprehensive Cancer Center Foundation fellowship (to A.S.S.), a grant from the Nakajima Foundation (to T.K.), and National Institutes of Health Grant GM23674 (to M.G.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE22626).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1106834108/-/DCSupplemental.
References
- 1.Wang JC. Cellular roles of DNA topoisomerases: A molecular perspective. Nat Rev Mol Cell Biol. 2002;3:430–440. doi: 10.1038/nrm831. [DOI] [PubMed] [Google Scholar]
- 2.Brill SJ, DiNardo S, Voelkel-Meiman K, Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. Nature. 1987;326:414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- 3.DiNardo S, Voelkel K, Sternglanz R. DNA topoisomerase II mutant of Saccharomyces cerevisiae: Topoisomerase II is required for segregation of daughter molecules at the termination of DNA replication. Proc Natl Acad Sci USA. 1984;81:2616–2620. doi: 10.1073/pnas.81.9.2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uemura T, et al. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50:917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 5.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 7.Giaever GN, Wang JC. Supercoiling of intracellular DNA can occur in eukaryotic cells. Cell. 1988;55:849–856. doi: 10.1016/0092-8674(88)90140-7. [DOI] [PubMed] [Google Scholar]
- 8.Garinther WI, Schultz MC. Topoisomerase function during replication-independent chromatin assembly in yeast. Mol Cell Biol. 1997;17:3520–3526. doi: 10.1128/mcb.17.7.3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Germe T, Hyrien O. Topoisomerase II-DNA complexes trapped by ICRF-193 perturb chromatin structure. EMBO Rep. 2005;6:729–735. doi: 10.1038/sj.embor.7400465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pfaffle P, Jackson V. Studies on rates of nucleosome formation with DNA under stress. J Biol Chem. 1990;265:16821–16829. [PubMed] [Google Scholar]
- 11.Durand-Dubief M, Persson J, Norman U, Hartsuiker E, Ekwall K. Topoisomerase I regulates open chromatin and controls gene expression in vivo. EMBO J. 2010;29:2126–2134. doi: 10.1038/emboj.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gavin I, Horn PJ, Peterson CL. SWI/SNF chromatin remodeling requires changes in DNA topology. Mol Cell. 2001;7:97–104. doi: 10.1016/s1097-2765(01)00158-7. [DOI] [PubMed] [Google Scholar]
- 13.Salceda J, Fernandez X, Roca J. Topoisomerase II not topoisomerase I, is the proficient relaxase of nucleosomal DNA. EMBO J. 2006;25:2575–2583. doi: 10.1038/sj.emboj.7601142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schultz MC, Brill SJ, Ju Q, Sternglanz R, Reeder RH. Topoisomerases and yeast rRNA transcription: Negative supercoiling stimulates initiation and topoisomerase activity is required for elongation. Genes Dev. 1992;6:1332–1341. doi: 10.1101/gad.6.7.1332. [DOI] [PubMed] [Google Scholar]
- 15.Mondal N, Parvin JD. DNA topoisomerase IIalpha is required for RNA polymerase II transcription on chromatin templates. Nature. 2001;413:435–438. doi: 10.1038/35096590. [DOI] [PubMed] [Google Scholar]
- 16.Mondal N, et al. Elongation by RNA polymerase II on chromatin templates requires topoisomerase activity. Nucleic Acids Res. 2003;31:5016–5024. doi: 10.1093/nar/gkg705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bermejo R, et al. Genome-organizing factors Top2 and Hmo1 prevent chromosome fragility at sites of S phase transcription. Cell. 2009;138:870–884. doi: 10.1016/j.cell.2009.06.022. [DOI] [PubMed] [Google Scholar]
- 18.Aparicio O, et al. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo. Curr Protoc Mol Biol. 2005 doi: 10.1002/0471142727.mb2103s69. Chapter 21:Unit 21.23. [DOI] [PubMed] [Google Scholar]
- 19.Bermejo R, et al. Top1- and Top2-mediated topological transitions at replication forks ensure fork progression and stability and prevent DNA damage checkpoint activation. Genes Dev. 2007;21:1921–1936. doi: 10.1101/gad.432107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pokholok DK, et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 21.Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Yuan GC, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 23.Han M, Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 24.Almer A, Rudolph H, Hinnen A, Horz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boeger H, Griesenbeck J, Strattan JS, Kornberg RD. Nucleosomes unfold completely at a transcriptionally active promoter. Mol Cell. 2003;11:1587–1598. doi: 10.1016/s1097-2765(03)00231-4. [DOI] [PubMed] [Google Scholar]
- 26.Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11:1599–1607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 27.Champoux JJ. DNA topoisomerases: structure, function, and mechanism. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 28.Lee W, et al. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 29.Holstege FC, et al. Dissecting the regulatory circuitry of a eukaryotic genome. Cell. 1998;95:717–728. doi: 10.1016/s0092-8674(00)81641-4. [DOI] [PubMed] [Google Scholar]
- 30.Lee CK, Shibata Y, Rao B, Strahl BD, Lieb JD. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat Genet. 2004;36:900–905. doi: 10.1038/ng1400. [DOI] [PubMed] [Google Scholar]
- 31.Venters BJ, et al. A comprehensive genomic binding map of gene and chromatin regulatory proteins in Saccharomyces. Mol Cell. 2011;41:480–492. doi: 10.1016/j.molcel.2011.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lotito L, et al. Global transcription regulation by DNA topoisomerase I in exponentially growing Saccharomyces cerevisiae cells: Activation of telomere-proximal genes by TOP1 deletion. J Mol Biol. 2008;377:311–322. doi: 10.1016/j.jmb.2008.01.037. [DOI] [PubMed] [Google Scholar]
- 33.Hirschhorn JN, Brown SA, Clark CD, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 34.Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 35.Owen-Hughes T, Utley RT, Cote J, Peterson CL, Workman JL. Persistent site-specific remodeling of a nucleosome array by transient action of the SWI/SNF complex. Science. 1996;273:513–516. doi: 10.1126/science.273.5274.513. [DOI] [PubMed] [Google Scholar]
- 36.Kassabov SR, Zhang B, Persinger J, Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 37.Sudarsanam P, Iyer VR, Brown PO, Winston F. Whole-genome expression analysis of snf/swi mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Segal E, et al. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dion MF, et al. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 40.Hirano T, Mitchison TJ. Topoisomerase II does not play a scaffolding role in the organization of mitotic chromosomes assembled in Xenopus egg extracts. J Cell Biol. 1993;120:601–612. doi: 10.1083/jcb.120.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nitiss JL. DNA topoisomerase II and its growing repertoire of biological functions. Nat Rev Cancer. 2009;9:327–337. doi: 10.1038/nrc2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ju BG, et al. A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science. 2006;312:1798–1802. doi: 10.1126/science.1127196. [DOI] [PubMed] [Google Scholar]
- 43.Wood V, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- 44.Hecht A, Strahl-Bolsinger S, Grunstein M. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature. 1996;383:92–96. doi: 10.1038/383092a0. [DOI] [PubMed] [Google Scholar]
- 45.Sperling AS, Grunstein M. Histone H3 N-terminus regulates higher order structure of yeast heterochromatin. Proc Natl Acad Sci USA. 2009;106:13153–13159. doi: 10.1073/pnas.0906866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Robyr D, Grunstein M. Genomewide histone acetylation microarrays. Methods. 2003;31:83–89. doi: 10.1016/s1046-2023(03)00091-4. [DOI] [PubMed] [Google Scholar]
- 47.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 48.Saldanha AJ. Java Treeview—Extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.