Abstract
Bacteria optimize the use of their motility appendages to move efficiently on a wide range of surfaces prior to forming multicellular bacterial biofilms. The “twitching” motility mode employed by many bacterial species for surface exploration uses type-IV pili (TFP) as linear actuators to enable directional crawling. In addition to linear motion, however, motility requires turns and changes of direction. Moreover, the motility mechanism must be adaptable to the continually changing surface conditions encountered during biofilm formation. Here, we develop a novel two-point tracking algorithm to dissect twitching motility in this context. We show that TFP-mediated crawling in Pseudomonas aeruginosa consistently alternates between two distinct actions: a translation of constant velocity and a combined translation-rotation that is approximately 20× faster in instantaneous velocity. Orientational distributions of these actions suggest that the former is due to pulling by multiple TFP, whereas the latter is due to release by single TFP. The release action leads to a fast “slingshot” motion that can turn the cell body efficiently by oversteering. Furthermore, the large velocity of the slingshot motion enables bacteria to move efficiently through environments that contain shear-thinning viscoelastic fluids, such as the extracellular polymeric substances (EPS) that bacteria secrete on surfaces during biofilm formation.
Keywords: extracellular polysaccharides, cystic fibrosis, biometric identification, PAO1
Bacterial biofilms are multicellular communities that adhere to almost any surface and are fundamental to the ecology and biology of bacteria (1). To assemble into microcolonies and form biofilms, bacteria must adapt their motility mechanisms for surface locomotion (2, 3). For example, both flagella (4) and excreted surfactants (5) enable collective surface motility modes (6, 7) that allow bacteria to colonize surfaces. Many bacterial species, including the opportunistic pathogen Pseudomonas aeruginosa, which contributes to fatal airway infections in cystic fibrosis (8, 9), the causative agent for gonorrhea Neisseria gonorrhoeae, and the predatory soil bacterium Myxococcus xanthus (10), use type-IV pili (TFP) to move on surfaces (11–15). TFP are associated with the “twitching” collective motility mode (16), in which cells exhibit apparently random irregular motions. A single type-IV pilus undergoes cycles of repeated extension-adhesion and retraction-release (17, 18) that are driven by an ATP motor (19, 20). Single TFP can generate forces of up to approximately 100 pN (21, 22), and multiple pili can cooperatively generate forces of up to approximately 1 nN (23), to enable motion on surfaces. To traverse distances that are significantly longer than the extension distance of a single pilus (typically several microns) (17), bacteria deploy multiple pili using a “tug-of-war” mechanism (24). These studies show that TFP act as linear actuators (17) to enable directional motion.
What is not known is how the collective deployment of TFP results in the irregular motions characteristic of twitching motility. These irregular motions must result from changes in the direction of the motion of the bacteria on the surface. In M. xanthus, pili appear sequentially on different poles of the bacteria and thereby enable the bacterium to reverse direction over time scales of minutes (25). However, this mechanism is not seen in other species that possess pili, such as P. aeruginosa. How TFP linear actuators allow P. aeruginosa and other species to change directions or execute “turns” on surfaces, as required to efficiently explore surfaces, is not known.
Here, we combine microscopy with tracking methods that yield trajectories with high spatial and temporal resolution to elucidate TFP-driven crawling in single cells of P. aeruginosa. To capture the full complexity of the motions that characterize twitching motility, we developed a two-point tracking algorithm to record independent trajectories for the leading (pLead) and trailing (pTrail) poles of each cell. Despite the seeming irregularity of motion, we find that the trajectories of each pole can consistently (over a total of 12,000 actions) be decomposed into an alternating sequence of two qualitatively distinct types of movements: a linear translation of constant velocity and variable duration in time (0.3–10 sec), followed by a combined translation-rotation that is on average approximately 20× faster in instantaneous velocity with a short duration (approximately 100 ms). Surprisingly, we find that the fast motions contribute as much to total displacement as the slow linear translations that are driven by pulling of TFP, although the latter is the standard model for TFP motility (17) and occur for approximately 95% of the total time of motion. We propose a model in which slow translation is due to multiple TFP pulling to exert force via retraction, whereas fast rotation is due to release of a single type-IV pilus in the presence of other TFP under tension so that the bacterium rapidly “slingshots” to a new equilibrium position and orientation. Indeed, the orientations of “pull” actions are broadly distributed, reflecting the vector sum of forces from multiple TFP, whereas the orientations of “release” actions are narrowly distributed (< 10 degrees), which we attribute to the angular distribution of motility-active TFP in individual cells. Similarly, pull speeds vary by a factor of approximately 20× with variable durations, consistent with the deployment of varying numbers of TFP pulling in different directions, whereas release speeds vary by a factor of approximately 10× but with consistently short durations. These results suggest a twitching motility model in which the release of TFP from a surface has a single characteristic time scale and the force generated by a single type-IV pilus is broadly distributed, consistent with direct measurements (10–100 pN) (22, 23). Based on our model, we postulate that the TFP release cycle allows bacteria to turn efficiently by “oversteering,” in which the rear pole of the cell loses traction with the surface before linear translation resumes. Moreover, this mechanism may also enable bacteria to reduce the local viscosity in shear-thinning media, such as the extracellular polymeric substances (EPS) that compose the biofilm matrix (26).
Results
Two-Point Tracking Reveals Alternating Pulses and Square Waves in Trajectories.
Traditional high-throughput algorithms applied to locate spherical colloidal particles (27) identify the positions of the particle centroids. However, P. aeruginosa are rod-shaped and therefore must be characterized by both the positions of the centroids and the orientations of the cells. In addition, the TFP of P. aeruginosa are distributed anisotropically along the body and are located primarily at the poles at the bacterium (17, 28, 29). Relating the motion of the body of the bacterium to the deployment of pili located at the poles thus requires new methods to separately analyze the motion of each pole. We therefore modified standard particle-tracking algorithms to separately track the two poles of rod-like bacteria in a database of microscopy movies (Fig. S1). We fitted an ellipse to each image of a bacterium (30) and identified the two poles with the two foci of the ellipse. Although bacteria are closer to spherocylinders than ellipses, we empirically find this to be a computationally efficient and reliable way to locate the poles. Because the separation between the two poles of the bacterium was larger than the displacement of the centroid of the bacterium at our fast imaging rate (10 frames/ sec), we could use standard tracking algorithms (27) to separately track the two poles over time. (For schematic diagram, see Fig. S1.)
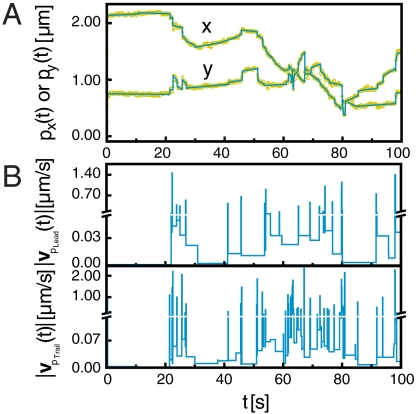
The positions of the poles as a function of time exhibit significant noise that reflects the errors in the ellipse fitting, as shown for the leading pole pLead over a time interval of 100 sec in Fig. 1(A) (points). We estimate this noise to be approximately 0.03 μm (0.5 pixels), which sets a lower threshold of approximately 0.3 μm/s on the velocities that can be resolved given the imaging rate of 10 frames/ sec. To accurately extract velocities from the trajectory that fall below this noise threshold, we first subdivide the trajectory into segments using a noise threshold of 2 pixels (0.12 μm). If the position varies less than this threshold, we calculate the velocity using linear regression across the entire segment (solid line in Fig. 1A). For extremely fast motions, the position varies by more than the noise threshold, and we directly calculate the velocity of the pole from its net displacement. This strategy allows us to accurately measure velocities ranging from 0.001–20 μm/s over durations ranging from 0.1–10 s (Movie S1). Comparison of the trajectory obtained via regression (solid line) with the positions measured from our experiment (symbols) shows excellent agreement; we estimate the relative error in the calculation of the velocity amplitude from the slope of the trajectory segment to be less than 5%.
Fig. 1.
Analysis of the two-point velocity profiles of the ΔfliM mutant. (A) x and y position of one focus (px(t) and py(t)) as a function of time. Yellow symbols show raw data and cyan lines indicate the denoising regression (threshold = 2 pixels). (B) Velocity amplitudes (|vpLead(t)| and |vpTrail(t)|) of foci pLead and pTrail as a function of time. Velocity amplitudes are calculated from the regression data.
The resultant plots of the velocity amplitude obtained from the regression analysis versus time exhibit high-speed pulses that regularly alternate with low-speed square waves, as shown for the leading (pLead) and trailing (pTrail) poles of a single bacterium in Fig. 1B. Over one order of magnitude separates the characteristic velocities of pulses and square waves, indicating that twitching is an inherently multiscale phenomenon. Furthermore, the durations in time of the high-speed pulses are shorter and more narrowly distributed compared to those of the low-speed square waves. The velocity amplitude profiles of pLead and pTrail exhibit significant differences, with more high-speed velocity pulses found in the velocity profile of the trailing pole than that of the leading pole. Together, these observations suggest that distinct physical mechanisms that depend on both the orientation and the shape anisotropy of the bacterium may be responsible for the pulses and square waves.
Extant results on TFP-driven motility suggest two potential models to explain the origin of the alternating high-speed and low-speed motions that we observed in the trajectories. First, multiple TFP pulling in coordination may allow the bacterium to move at higher speeds. However, motility experiments on Neisseria gonorrheae mutants showed that the velocity was independent of the average number of TFP (16, 22); moreover, the force exerted by multiple TFP pulling in coordination scaled only linearly with the number of TFP (23). Second, the pilus motor that drives the retraction may exhibit multiple states, each with its own characteristic velocity. This scenario was found in N. gonorrheae, in which TFP retracted at two distinct velocities separated by a factor of five only in the regime of low loading forces (31), but is unlikely to explain the 20-fold difference between our low- and high-speed velocities. Moreover, neither model suggests any natural explanation for the striking contrast in the distribution of durations of the two movement types, the monodisperse duration of the high-speed velocity pulses contrasted with the polydisperse duration of the low-speed velocity plateaus. We therefore conclude that neither model can explain three key features of our trajectories: (i) the large separation in the characteristic scales of velocity and duration, (ii) the regular alternation between low and high speeds, and (iii) the drastic difference in the distributions of movement duration between low and high speeds. These features suggest that a new model is needed to understand how bacteria deploy multiple TFP.
Bacteria Move Faster During Turns than During Pure Linear Motion.
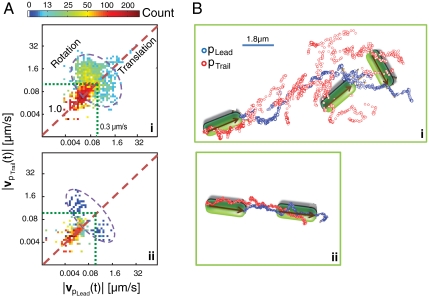
To obtain new insight into how bacteria deploy TFP, we first examine the rotational motion of a crawling bacterium. To distinguish rotational motion from translational motion in the database of trajectories, we analyze the correlation between the motions of the two poles. For translational motion along the cell body axis and for “parallel transport” of the cell in which a constant cell orientation is maintained, the magnitudes of the instantaneous velocities of the two poles are nearly equal (i.e., |vpLead(t)| = |vpTrail(t)|), whereas for rotational motion the magnitudes of the velocities are not equal (i.e., |vpLead(t)| ≠ |vpTrail(t)|). Individual cells deploy translations and rotations differently, as shown by the two-dimensional histograms over the instantaneous velocity amplitudes |vpLead(t)| and |vpTrail(t)| for two different cells in Fig. 2A, i–ii, and therefore exhibit morphologically distinct trajectories as shown in Fig. 2B. For example, the trajectory of the trailing pole pTrail for bacterium (i) is only weakly correlated with the trajectory of the leading pole pLead, consistent with a statistical preference for rotational motion, whereas the trajectories of pLead and pTrail of bacterium (ii) are highly correlated, consistent with a statistical preference for linear translational motion. Although the motions of these bacteria are different, it can be clearly seen that high-speed motions are dominated by rotations, whereas low-speed motions are dominated by translation. This behavior is a generic feature of TFP-driven motility in P. aeruginosa.
Fig. 2.
Identical ΔfliM bacteria exhibit individuation of motion preferences. (A) 2D histogram of |vpLead(t)| versus |vpTrail(t)|, from datasets of two different ΔfliM mutant bacteria. The dashed line (slope = 1.0) is a guide to the eye to indicate translational motion (|vpLead(t)| = |vpTrail(t)|); the dashed circle is a guide to the eye to indicate rotational motion (|vpLead(t)| ≠ |vpTrail(t)|); the dotted green lines indicate the velocity threshold (|vpLead,pTrail| = 0.3 μm/s) separating release actions and pull actions. Genetically identical bacteria exhibit distinct individual motion preferences: (i) rotation; (ii) translation. (B) Portion of trajectories from (top) bacterium (i), with schematic illustrating rotational motion, and (bottom) bacterium (ii), with schematic illustrating translational motion. In (B) the blue and red circles indicate positions of pLead and pTrail, respectively.
Motion Can Be Decomposed into Two Distinct and Alternating Actions.
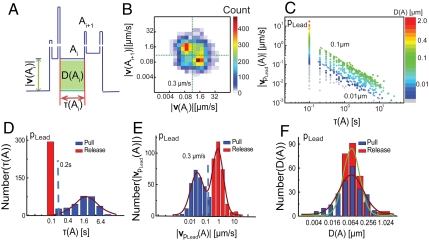
To elucidate the motility events that constitute twitching, we decompose the complex motion of the leading pole of the bacterium into a sequence of unique and discrete actions. Each trajectory can be described as a series of N distinct actions Ai characterized by a duration time τ(Ai) and a constant velocity amplitude |v(Ai)| with displacement D(Ai), as shown schematically in Fig. 3A. We hypothesize that these discrete actions must ultimately derive from the deployment of TFP. We first investigate the correlation of sequential actions (actions Ai and Ai+1) over a specific trajectory. The 2D histogram over the velocity amplitudes |vpLead(Ai)| and |vpLead(Ai+1)| exhibits two distinct maxima (Fig. 3B), indicating that slow (fast) actions typically succeed fast (slow) actions. Similarly, the 2D histogram over the duration times |τ(Ai)| and |τ(Ai+1)| reveals that short (long) actions are typically followed by long (short) actions (Fig. S2). Furthermore, by analyzing all the discrete actions in a trajectory (Fig. 3C), we find that an overwhelming fraction of high velocity actions have short durations (< 200 ms). These results indicate that the motility cycle of TFP-driven twitching for the database of trajectories consists of alternating low velocity plateaus of long duration and high velocity pulses of short duration.
Fig. 3.
Analysis of the velocity of leading pole pLead reveals distinct pulls and releases. (A) Schematic definition of an action in the velocity profile: For the ith action Ai, |vpLead(Ai)| is the velocity amplitude, τ(Ai) is the duration time, and D(Ai) is the total displacement. (B) 2D histogram of the velocity amplitude of connected actions (Ai and Ai+1) for leading pole pLead. (C) Velocity amplitude |vpLead(A)| as a function of duration τ(A) for the leading pole pLead in the trajectory of a single ΔfliM bacterium (N = 12,000 points); the color scale indicates the total displacement D(A). The two dashed lines (slope = -1.0) are guides to the eye indicating total displacements between 0.01 μm and 0.1 μm. (D) Histogram of the duration τ(A). The dashed line indicates the time threshold (τc = 0.2 s) separating release actions (red bars) from pull actions (blue bars). (E) Histogram of the velocity amplitude |vpLead(A)|. The dashed line indicates the velocity threshold (|vpLead,c| = 0.3 μm/s) separating release actions (red bars) from pull actions (blue bars). (F) Histogram of the displacement D(A) of release actions (red bars) and of pull actions (blue bars). In D–F the line is the Gaussian fit to the distribution.
The distributions of the duration and velocity of actions are bimodal, with characteristic scales that are separated by over one order of magnitude (Fig. 3D and E). Slow actions (of velocity < 0.3 μm) exhibit long duration times (300 ms - 20 s), whereas fast actions (of velocity > 0.3 μm) exhibit short duration times (≤ 200 ms). The distribution of durations for slow actions is significantly broader [full-width half-max (FWHM) of approximately 5 s] than that of fast actions (FWHM of approximately 200 ms). The average duration of fast actions is approximately 20 times shorter than that of slow actions (Fig. 3D). However, the average velocity amplitude of fast actions is approximately 20 times greater than that of slow actions (Fig. 3E). This implies that the average displacements for these two processes are approximately equal (Fig. 3F). This surprising result suggests that the standard model of TFP-driven pulling that is used to explain twitching motility (17) must be extended to include a second physical process that also contributes significantly to total displacement.
Fast and Slow Actions Exhibit Distinct Orientation Preferences.
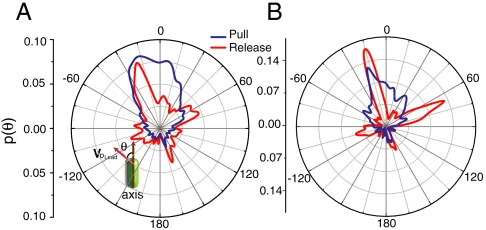
To elucidate the physical origin of these processes, we examined how fast and slow angles were oriented with respect to the body of the bacterium. We define the deviation angle θ as the angle between the velocity vector of the leading pole (pLead) and the body axis of the bacterium, as shown in the inset to Fig. 4A. The distribution of deviation angles p(θ) for slow actions (FWHM of approximately 90 degrees) is biased along a broad forward cone (Fig. 4B), as shown by data averaged over 10 different ΔfliM mutant cells. By contrast, the forward-directed maximum in p(θ) for fast actions is significantly narrower (FWHM of approximately 10 degrees), but fast actions can occur in multiple directions. Data for individual bacteria also exhibit similar distributions for slow and fast actions, as shown in Fig. 4B. The differences in the orientational distributions for slow and fast actions suggest that the underlying physical mechanisms have different spatial distributions.
Fig. 4.
Identical ΔfliM bacteria exhibit distinct orientation preferences. (Inset A) Schematic plot of the pLead action velocity vector; θ is the deviation angle between the pLead velocity vector and the body axis of the bacterium, with clockwise (counterclockwise) motion defined as positive (negative). (A) Normalized distribution of the deviation angle (θ), p(θ) calculated from 10 trajectories of different single ΔfliM bacteria, including approximately 180,000 images. (B) Distribution of deviation angle for a single bacterium, showing that single cells also exhibit orientational preference. In A and B distributions for release actions (red line) and pull actions (blue line) are decomposed from the entire trajectory.
Discussion
Proposed Model of Twitching Motility.
The highly temporally and spatially resolved measurements presented in Figs. 1–4 reveal two distinct physical processes during crawling: a slow action with low velocity and long duration that is correlated with linear translation along the forward direction, and a fast action with high velocity and short duration that is correlated with multidirectional rotation. The characteristic scales for the velocity and duration of these actions are separated by over an order of magnitude but contribute nearly equally to the total displacement of the bacterium. Most strikingly, these two actions regularly alternate: Velocity pulses are consistently followed by velocity plateaus and vice versa. This pattern is observed for the over 12,000 actions that we analyzed. Existing models for the deployment of TFP motility do not capture these features.
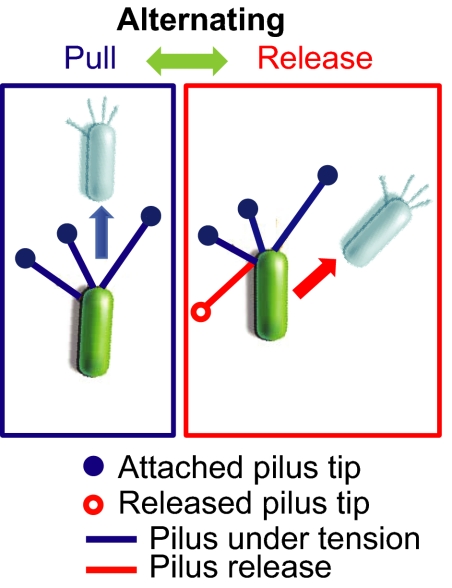
To explain the complex motion of the twitching bacterium revealed by our measurements, we instead propose a model for crawling that relates the velocity of motions to differences in the deployment of multiple pili, as shown in Fig. 5. In our model, the low-speed square waves or pulls result from the cooperative retraction of multiple pili that are preferentially located at the poles, leading to slow and steady motion along the body axis of the bacterium as previously described (17). The unique feature of our model is a mechanism for the high-speed releases, which we attribute to the release of a single type-IV pilus while the other TFP are under tension.
Fig. 5.
Model for twitching motility. Schematic illustrating the mechanisms for the alternating pull (left) and release (right) actions of a crawling bacterium.
This simple model allows us to explain the features of twitching motility highlighted by our analysis. Rotation of the trailing pole pTrail (Fig. 2) occurs when a tethered pilus is rapidly released. The width of the distribution of durations of pull actions is larger than that of release actions (Fig. 3D), consistent with the measured polydispersity in the lengths of TFP (17); we expect that the duration of TFP pulls should correlate with the length of TFP, whereas the duration of TFP releases should be largely independent of TFP length (see Fig. S2). The large velocities of release actions (Fig. 3E) result from the rapid equilibration of the cell position away from the direction of the released pilus to a new position defined by the vector sum of forces from all other TFP in tension. The difference in the distributions of deviation angle between pulls and releases (Fig. 4) results from the spatial asymmetry of tethered pili: If the released type-IV pilus is positioned off-center from the major axis of the bacterium, the resultant vector sum of the remaining forces due to tethered TFP may cause the bacterium to rapidly rotate to a new position. The strong forward anisotropy of p(θ) for pull actions therefore results from the retraction of multiple TFP at the leading pole. By contrast, the narrow distribution of release actions along the forward direction reflects the anticorrelation with the orientation of a single pilus, and the multidirectionality of release actions reflects the release of either leading or trailing TFP.
The rapid equilibration of the cell body during the release action affords the bacterium a natural and efficient mechanism to rotate its orientation through large angles. If the vector sum of force due to TFP under tension contains a component orthogonal to the long body axis, the bacterium can turn while attached to the surface via oversteering during release. Our model suggests that this slingshot-like motion of TFP release contributes as much to twitching motility as the pull motion from the TFP retraction cycle (Movie S2), which is the standard model for TFP motility (17).
Why Do Bacteria Use a Slingshot Release Mechanism for Propulsion?
One possible explanation for the importance of the slingshot mechanism arises from the mechanical properties of the extracellular polymeric substances (EPS) that compose bacterial biofilms (26). Upon initial attachment to the surface, bacteria exhibit increased gene expression (32) for the production of EPS (33), which has been suggested as a precondition for irreversible attachment (34–36). For Pseudomonas, different components of the EPS appear to play different roles, with psl being more important for cell adhesion to surfaces (37), and pel being more important for cell–cell contact (38). In solution, the EPS of P. aeruginosa exhibits pseudoplastic shear-thinning rheology with viscosities η ≈ 0.1 and 0.05 Pa-s at shear rates of s and 1 s-1, respectively (39). These shear rates are representative of those experienced by a bacterium of typical length 1 μm moving under pull and release actions, respectively (Fig. 3F). Bacteria move in the low Reynolds number regime (Re ≈ 10-5) (40) in which inertial forces are negligible, and the viscous drag on the bacterium is therefore proportional to fluid viscosity (41). The rapid release mechanism may therefore allow bacteria to exploit the pseudoplastic rheology of the EPS and reduce viscous drag by approximately 200% based on our simple estimate. Moreover, this argument does not depend on the specific chemistry of the polymeric substances but only requires that the viscosity decrease with increasing shear rate. For example, the extracellular DNA (eDNA) also required for biofilm formation (42) is also capable of shear-thinning; the viscosity of a semidilute solution of DNA decreases by approximately 500% as the shear rate is increased from  to 1 s-1 (43). The slingshot mechanism suggested by our model would allow bacteria to exploit the biomechanics of pili to modify the local viscosity and thus facilitate surface movement through the EPS. This proposed physical strategy would be complementary to that of bacteria such as Helicobacter pylori, which use chemical modifications to reduce the local viscosity (44).
to 1 s-1 (43). The slingshot mechanism suggested by our model would allow bacteria to exploit the biomechanics of pili to modify the local viscosity and thus facilitate surface movement through the EPS. This proposed physical strategy would be complementary to that of bacteria such as Helicobacter pylori, which use chemical modifications to reduce the local viscosity (44).
In summary, we have investigated the complex sequence of motions that constitute twitching motility in P. aeruginosa using a two-point tracking algorithm. We find that linear translational pulls of constant velocity alternate with combined translational-rotational releases that are approximately 20× faster. Surprisingly, the contribution of the slingshot-like TFP release motion to the total distance traveled by the bacterium is comparable to that of TFP retraction, despite occurring for only approximately 5% of the total duration of the trajectory. After release of a single type-IV pilus, the cell body reorients along the vector sum of forces from multiple TFP under tension, thereby allowing it to turn while still surface-attached. The oversteering mechanism elucidated here, which is driven by the rapid release actions, enables changes of direction that are approximately 100× faster than those exhibited by M. xanthus (25). Moreover, the rapid slingshot release enables efficient locomotion in the shear-thinning macromolecular fluids encountered during biofilm formation. These methods show how P. aeruginosa deploy TFP to move and change direction on surfaces. Because twitching motions depend on the physical distributions of TFP on individual cells, analysis of motility patterns may enable new methods for biometric “fingerprinting” of individual cells for single cell diagnostics (see Fig. S2). Additionally, these methods can be easily extended to elucidate the complex motility patterns of other bacterial species using a variety of motility appendages.
Methods
Bacteria Strains and Flow Cell Experiment.
A ∆fliM isogenic mutant of Pseudomonas aeruginosa strain ATCC 15692 (45) was used to elucidate the origins of TFP-driven twitching motility. These rod-like bacteria move strictly using TFP, which exhibit polydisperse lengths of up to approximately 10 μm and retract over time scales of 1–10 sec (17, 46). Motility of attached P. aeruginosa cells was monitored in sterilized flow-cells containing FAB medium (47) with 0.6 mM glutamate flowing at 3.75 mL h-1 at constant temperature 30 ± 0.1 °C. An inoculum was prepared by growing bacteria in test tubes containing FAB medium with 30 mM carbon with shaking at 37 °C to OD600 ≈ 0.3. The cultures were diluted by adding 50 μL of the bacterial suspension into 950 μL of sterilized FAB (1∶20).
Microscopy and Tracking Algorithm.
Brightfield movies containing approximately 18,000 images were collected at 10 frames per second using an Olympus microscope equipped with a 100× oil objective. Typical movies contained < 10 rod-like cells of P. aeruginosa. To separately track the two poles of rod-like bacteria in a database of microscopy movies, we first estimated the positions of the poles of a given bacterium by fitting an ellipse to the cell body. We defined the two foci of the ellipse as the two poles. We then separately tracked the positions of the leading (pLead) and trailing (pTrail) poles as a function of time using standard algorithms (27). For our analysis, focus pLead is defined as the leading pole of the bacterium along the statistically preferred direction of motion.
To accurately calculate the velocity of bacteria below the noise limit of approximately 0.3 μm/s set by the errors from the ellipse fitting, we implemented a linear regression algorithm. We subdivided each trajectory into segments using a noise threshold of 2 pixels (0.12 μm). If the position in the segment varied less than this threshold, we calculated the velocity using linear regression across the entire segment (solid line in Fig. 1A). For the extremely rapid motions of duration two frames, we directly calculated the velocity of the pole as the total displacement divided by the frame time (0.1 sec).
Supplementary Material
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. B.C. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105073108/-/DCSupplemental.
References
- 1.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 2.O’Toole GA, Kolter R. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol. 1998;30:295–304. doi: 10.1046/j.1365-2958.1998.01062.x. [DOI] [PubMed] [Google Scholar]
- 3.Klausen M, Aaes-Jørgensen A, Molin S, Tolker-Nielsen T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol Microbiol. 2003;50:61–68. doi: 10.1046/j.1365-2958.2003.03677.x. [DOI] [PubMed] [Google Scholar]
- 4.Berg HC. The rotary motor of bacterial flagella. Annu Rev Biochem. 2003;72:19–54. doi: 10.1146/annurev.biochem.72.121801.161737. [DOI] [PubMed] [Google Scholar]
- 5.Angelini TE, Roper M, Kolter R, Weitz DA, Brenner MP. Bacillus subtilis spreads by surfing on waves of surfactant. Proc Natl Acad Sci USA. 2009;106:18109–18113. doi: 10.1073/pnas.0905890106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Verstraeten N, et al. Living on a surface: Swarming and biofilm formation. Trends Microbiol. 2008;16:496–506. doi: 10.1016/j.tim.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 7.McBride MJ. Bacterial gliding motility: Multiple mechanisms for cell movement over surfaces. Annu Rev Microbiol. 2001;55:49–75. doi: 10.1146/annurev.micro.55.1.49. [DOI] [PubMed] [Google Scholar]
- 8.Nguyen D, Singh PK. Evolving stealth: Genetic adaptation of Pseudomonas aeruginosa during cystic fibrosis infections. Proc Natl Acad Sci USA. 2006;103:8305–8306. doi: 10.1073/pnas.0602526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yoon SS, et al. Pseudomonas aeruginosa anaerobic respiration in biofilms: Relationships to cystic fibrosis pathogenesis. Dev Cell. 2002;3:593–603. doi: 10.1016/s1534-5807(02)00295-2. [DOI] [PubMed] [Google Scholar]
- 10.Shi W, Sun H. Type IV pilus-dependent motility and its possible role in bacterial pathogenesis. Infect Immun. 2002;70:1–4. doi: 10.1128/IAI.70.1.1-4.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattick JS. Type IV pili and twitching motility. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 12.Barken KB, et al. Roles of type IV pili, flagellum-mediated motility and extracellular DNA in the formation of mature multicellular structures in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2008;10:2331–2343. doi: 10.1111/j.1462-2920.2008.01658.x. [DOI] [PubMed] [Google Scholar]
- 13.Harshey RM. Bacterial motility on a surface: Many ways to a common goal. Annu Rev Microbiol. 2003;57:249–273. doi: 10.1146/annurev.micro.57.030502.091014. [DOI] [PubMed] [Google Scholar]
- 14.Heydorn A, et al. Statistical analysis of Pseudomonas aeruginosa biofilm development: Impact of mutations in genes involved in twitching motility, cell-to-cell signaling, and stationary-phase sigma factor expression. Appl Environ Microbiol. 2002;68:2008–2017. doi: 10.1128/AEM.68.4.2008-2017.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibiansky ML, et al. Bacteria use type IV pili to walk upright and detach from surfaces. Science. 2010;330:197. doi: 10.1126/science.1194238. [DOI] [PubMed] [Google Scholar]
- 16.Merz AJ, So M, Sheetz MP. Pilus retraction powers bacterial twitching motility. Nature. 2000;407:98–102. doi: 10.1038/35024105. [DOI] [PubMed] [Google Scholar]
- 17.Skerker JM, Berg HC. Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci USA. 2001;98:6901–6904. doi: 10.1073/pnas.121171698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaiser D. Bacterial swarming: A re-examination of cell-movement patterns. Curr Biol. 2007;17:R561–R570. doi: 10.1016/j.cub.2007.04.050. [DOI] [PubMed] [Google Scholar]
- 19.Nudleman E, Kaiser D. Pulling together with type IV pili. J Mol Microbiol Biotechnol. 2004;7:52–62. doi: 10.1159/000077869. [DOI] [PubMed] [Google Scholar]
- 20.Satyshur KA, et al. Crystal structures of the pilus retraction motor PilT suggest large domain movements and subunit cooperation drive motility. Structure. 2007;15:363–376. doi: 10.1016/j.str.2007.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merz AJ, Forest KT. Bacterial surface motility: Slime trails, grappling hooks and nozzles. Curr Biol. 2002;12:R297–R303. doi: 10.1016/s0960-9822(02)00806-0. [DOI] [PubMed] [Google Scholar]
- 22.Maier B, Potter L, So M, Seifert HS, Sheetz MP. Single pilus motor forces exceed 100 pN. Proc Natl Acad Sci USA. 2002;99:16012–16017. doi: 10.1073/pnas.242523299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biais N, Ladoux B, Higashi D, So M, Sheetz MP. Cooperative retraction of bundled type IV pili enables nanonewton force generation. PLoS Biol. 2008;6:e87. doi: 10.1371/journal.pbio.0060087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holz C, et al. Multiple pilus motors cooperate for persistent bacterial movement in two dimensions. Phys Rev Lett. 2010;104:178104. doi: 10.1103/PhysRevLett.104.178104. [DOI] [PubMed] [Google Scholar]
- 25.Sun H, Zusman D, Shi W. Type IV pilus of Myxococcus xanthus is a motility apparatus controlled by the frz chemosensory system. Curr Biol. 2000;10:1143–1146. doi: 10.1016/s0960-9822(00)00705-3. [DOI] [PubMed] [Google Scholar]
- 26.Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 27.Crocker JC, Grier DG. Methods of digital video microscopy for colloidal studies. J Colloid Interface Sci. 1996;179:298–310. [Google Scholar]
- 28.Weiss RL. The structure and occurrence of pili (fimbriae) on Pseudomonas aeruginosa. J Gen Microbiol. 1971;67:135–143. doi: 10.1099/00221287-67-2-135. [DOI] [PubMed] [Google Scholar]
- 29.Bradley DE. A study of pili on Pseudomonas aeruginosa. Genet Res. 1972;19:39–51. [Google Scholar]
- 30.Mohraz A, Solomon MJ. Direct visualization of colloidal rod assembly by confocal microscopy. Langmuir. 2005;21:5298–5306. doi: 10.1021/la046908a. [DOI] [PubMed] [Google Scholar]
- 31.Clausen M, Koomey M, Maier B. Dynamics of type IV pili is controlled by switching between multiple states. Biophys J. 2009;96:1169–1177. doi: 10.1016/j.bpj.2008.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies DG, Chakrabarty AM, Geesey GG. Exopolysaccharide production in biofilms: Substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl Environ Microbiol. 1993;59:1181–1186. doi: 10.1128/aem.59.4.1181-1186.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vandevivere P, Kirchman DL. Attachment stimulates exopolysaccharide synthesis by a bacterium. Appl Environ Microbiol. 1993;59:3280–3286. doi: 10.1128/aem.59.10.3280-3286.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsukawa M, Greenberg EP. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:4449–4456. doi: 10.1128/JB.186.14.4449-4456.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 36.Long G, Zhu P, Shen Y, Tong M. Influence of extracellular polymeric substances (EPS) on deposition kinetics of bacteria. Environ Sci Technol. 2009;43:2308–2314. doi: 10.1021/es802464v. [DOI] [PubMed] [Google Scholar]
- 37.Ma L, et al. Assembly and development of the Pseudomonas aeruginosa biofilm matrix. PLoS Pathog. 2009;5:e1000354. doi: 10.1371/journal.ppat.1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colvin KM, et al. The Pel polysaccharide can serve a structural and protective role in the biofilm matrix of Pseudomonas aeruginosa. PLoS Pathog. 2011;7:e1001264. doi: 10.1371/journal.ppat.1001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wloka M, Rehage H, Flemming H-C, Wingender J. Rheological properties of viscoelastic biofilm extracellular polymeric substances and comparison to the behavior of calcium alginate gels. Colloid Polym Sci. 2004;282:1067–1076. [Google Scholar]
- 40.Dusenbery DB. Living at Micro Scale. Cambridge, MA: Harvard Univ Press; 2009. [Google Scholar]
- 41.Berg H. Random Walks in Biology. Princeton, NJ: Princeton Univ Press; 1993. [Google Scholar]
- 42.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 43.Hemminger OL, Boukany PE, Wang S, Lee LJ. Flow pattern and molecular visualization of DNA solutions thorugh a 4∶1 planar micro-contraction. J Non-Newtonian Fluid Mech. 2010;165:1613–1624. [Google Scholar]
- 44.Celli JP, et al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci USA. 2009;106:14321–14326. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shrout JD, et al. The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional. Mol Microbiol. 2006;62:1264–1277. doi: 10.1111/j.1365-2958.2006.05421.x. [DOI] [PubMed] [Google Scholar]
- 46.Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004;2:363–378. doi: 10.1038/nrmicro885. [DOI] [PubMed] [Google Scholar]
- 47.Heydorn A, et al. Experimental reproducibility in flow-chamber biofilms. Microbiology. 2000;146:2409–2415. doi: 10.1099/00221287-146-10-2409. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.