In PNAS, Kapoor et al. (1) discover a Flaviviridae agent most closely related to hepatitis C virus (HCV); the identified RNA genome was isolated from domestic dogs with respiratory illness. This might shed new light on the origin and evolution of HCV and could lead to surrogate model systems for this important human pathogen. Over 100 million people are affected by hepatitis C worldwide, but research has been hampered by a lack of widely available animal models and representative culture systems. Thus, discovery of an HCV homolog in dogs gives hope for finding new ways to study this deadly human virus.
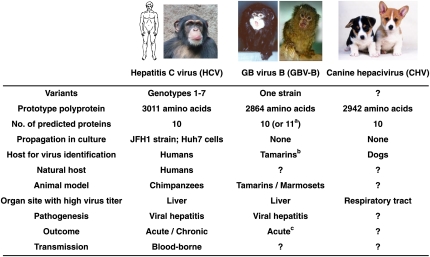
The host range of HCV is restricted, and natural infection was detected only in humans; higher primates are susceptible to experimental infection (Fig. 1). However, an HCV homolog has not been detected in higher primates to date. The only other known homolog is GB virus B (GBV-B), which causes hepatitis in experimentally infected New World monkeys, but its true origin remains unknown (2) (Fig. 1). Thus, the finding of a homolog in dogs raises the interesting proposition that HCV might have evolved as a result of the close contact between dogs and humans during the past several thousand years. Kapoor et al. (1) estimate a divergence time of the dog and human viruses from a common ancestor within the past 500–1,000 y, but it is widely recognized that such calculations should be interpreted with great caution. As pointed out by Kapoor et al. (1), a relatively recent zoonotic origin of HCV would fit well with the high pathogenic potential of HCV in humans. However, it will be of interest to determine whether there are further HCV homologs in other mammals. They could be a widespread group of host-specific viruses causing little or no disease, as observed for a more distantly related cluster of Flaviviridae (suggested genus Pegivirus), GBV-A and GBV-C viruses, in primates and humans (2–5).
Fig. 1.
Prototype HCV species and suggested species within the genus Hepacivirus of the Flaviviridae family of viruses and their natural and experimental hosts. aThe p13 protein of GBV-B might be processed into a p7 protein (equivalent to HCV p7) and a p6 protein. The p6 protein is not required for in vivo infectivity (8). bThe tamarin from which GBV-B was isolated had been inoculated with the GB-agent (2). cPersistent infection has been reported with recombinant GBV-B (8).
The International Committee of Taxonomy of Viruses deals with classification of viruses in a formalized way after researchers submit suggestions. Kapoor et al. (1) call the HCV homolog isolated from dogs canine hepacivirus (CHV). Based on analysis of the evolutionary conserved helicase and RNA polymerase regions of members of the different genera of Flaviviridae, it is apparent that the genome sequence of CHV clusters with HCV and GBV-B. HCV is the sole recognized species member of the genus Hepacivirus; GBV-B is a suggested species. Based on a detailed phylogenetic analysis, Kapoor et al. (1) suggest that CHV should be classified as a unique Hepacivirus species. However, HCV and GBV-B are hepatotropic, and it is not clear whether that is the case for CHV (Fig. 1). Also, although CHV was detected in dogs from different outbreaks of respiratory disease, it should be confirmed that this is a canine virus. Given the heterogeneity of HCV, it was surprising that CHV sequences recovered from respiratory or liver samples from dogs in different outbreaks were practically identical. These researchers screened a number of healthy dogs and did not detect CHV. Additional studies will be required to confirm the canine origin of this virus. It would be relevant to develop a specific serological test to screen dogs and other animals to determine how widespread infections with CHV are.
The study by Kapoor et al. (1) is limited to a detailed analysis of the near-complete genome of CHV. Given the close relatedness to HCV, such data are of particular interest during a time when new sequence-independent amplification and sequencing technologies, also applied here, will result in detection of numerous new viruses, which may or may not have any disease relevance. The challenge will be to determine whether such unique viruses, including CHV, cause disease in the host where they were discovered or in other susceptible animals. Questions about virulence and pathogenesis and, as mentioned above, the epidemiology of this virus are important to address. The CHV genome was found at a high titer in respiratory samples from sick dogs, and the virus was not detected in healthy dogs (Fig. 1). This would suggest that CHV played a role in the respiratory illness observed in these particular dogs. It will, however, be important to perform animal transmission studies using serial dilutions of high-titer virus stocks to demonstrate that CHV is a transmissible agent that causes respiratory illness. This would begin to address Koch's postulate for a disease-causing agent. Other disease outbreaks in dogs around the world should be examined. At present, there are no definitive data on target organs or disease association of this virus. How is it transmitted to other dogs? Does it infect other mammals? It would be of interest to determine whether different variants of CHV exist as observed for HCV with diverse major genotypes.
For the study of human pathogens, it is desirable to have a small animal model, preferably a mouse model. For HCV, only the chimpanzee model permits studies of infectivity and pathogenesis (6) (Fig. 1). The human liver chimeric mouse model permits HCV infectivity studies and has been useful in studies of passive immunoprophylaxis, but it does not permit studies of the adaptive immune responses of critical importance for vaccine development (6). Recently, a model with immunocompetent genetically humanized mice that permitted entry and neutralization studies was reported (7). However, there is a need for further developments in this field. One possibility is to use surrogate models like simian immunodeficiency virus in Old World monkeys that have served as a model for studies of HIV in humans. GBV-B has been explored as a surrogate model for HCV because this homolog is associated with acute (and, in a few cases, chronic) hepatitis in experimental infections in tamarins (8) (Fig. 1). It remains to be determined whether CHV in dogs or potentially other animal species could serve as a surrogate model for HCV. It is unclear whether this virus replicates in the liver and whether it induces hepatitis. Kapoor et al. (1) report that they detected CHV in liver samples from dogs at low titers; furthermore, in situ hybridization was used to detect the virus in the cytoplasm of hepatocytes. For HCV and GBV-B, the ability to cause viral hepatitis, without the influence of coinfecting agents, was formally proven by inoculating chimpanzees and tamarins, respectively, with RNA transcripts from synthetic DNA recombinants (9–11). It was not determined whether CHV causes persistent infections; the presence of a high percentage of genome-scale ordered RNA structures in its genome could signify a potential for persistence in the natural host. Thus, it will be important to screen large cohorts of dogs to determine whether persistence exists in this animal species.
For virologists, analysis of the complete sequence of a novel virus related to HCV is of great interest. The analysis convincingly confirmed a closer relatedness of CHV to HCV compared with that of GBV-B throughout the genome. Knowledge about protein motifs conserved between HCV and CHV could yield important information of relevance for future functional studies, and Kapoor et al. (1) make a number of observations regarding the predicted CHV polyprotein (Fig. 1). The 5′ terminal CHV sequence was determined by the RACE procedure. Although this region had features similar to those of the HCV 5′ UTR, it is notable that the structures at the 5′ end of the CHV genome were different from those of HCV. It remains unknown what the exact role of the HCV 5′ terminal stem-loop structure is, although this structure is critical for the major HCV genotypes in vitro (12). Even though Kapoor et al. (1) use a number of different approaches to determine the 3′ end of CHV, the nature of this sequence needs to be studied further. HCV and GBV-B have a poly-U tract within the 3′ UTR (9, 13). This was apparently not the case for CHV; instead, CHV appears to have a poly-A stretch close to the 3′ end. Thus, although HCV and CHV are more closely related based on phylogenetic analysis of coding sequences, there are genetic elements that appear to be more similar in HCV and GBV-B.
Initial attempts to culture CHV were not successful. Given the great difficulties encountered in culturing HCV (and GBV-B) (Fig. 1), culture systems for this previously undescribed related virus would be relevant. Only a single strain of HCV, the genotype 2a strain JFH1 from a Japanese patient, has been found to grow robustly in culture, in human hepatoma cell lines (14). However, viable JFH1-based recombinants with gene elements specific to genotypes 1–7 were developed (15, 16). Thus, it might be possible to generate HCV/CHV chimeras that could be cultivated, which would be of interest in functional studies and potentially could have relevance in HCV vaccine development, as found for different species of the genus Flavivirus.
Recently, the importance of microRNA (miR)-122 for HCV replication has been the subject of numerous investigations (17), and an antisense locked nucleic acid directed against this miR appeared to have therapeutical effect on HCV in vivo and in vitro (12, 18). The effect of miR-122 on HCV is mediated through specific binding to seeding sites in the HCV 5′ UTR (19). Kapoor et al. (1) do not identify equivalent sites in CHV. Thus, this virus might not depend on a specific miR for its function. It is of note that recombinant HCV lacking one of the seeding sites in the 5′ UTR was recently generated and that this virus had efficient growth in vitro (12).
The identification of a unique virus related to HCV by Kapoor et al. (1) is a major accomplishment, with exciting possibilities for future HCV research. It also raises the important perspective of determining whether viruses closely related to HCV might be more widespread in mammals than previously believed. The role of this previously undescribed virus in disease in dogs remains to be addressed. For veterinarians, it will be relevant to determine whether CHV causes severe respiratory illness in dogs. For basic science, the frequent discovery of viruses resulting from the advance of “deep sequencing” is very exciting and will lead to major advances in our understanding of the evolution and functionality of genetic elements. For practitioners treating human and animal diseases, the key question, however, will be whether these viruses cause disease and how to treat such diseases. Often, the identified agents will be innocent bystanders, as was the case for GBV-C, which was initially proposed as a unique human hepatitis virus (5, 20).
Footnotes
The author declares no conflict of interest.
See companion article on page 11608 of issue 28 in volume 108.
References
- 1.Kapoor A, et al. Characterization of a canine homolog of hepatitis C virus. Proc Natl Acad Sci USA. 2011;108:11608–11613. doi: 10.1073/pnas.1101794108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simons JN, et al. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc Natl Acad Sci USA. 1995;92:3401–3405. doi: 10.1073/pnas.92.8.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams NJ, et al. Detection in chimpanzees of a novel flavivirus related to GB virus-C/hepatitis G virus. J Gen Virol. 1998;79:1871–1877. doi: 10.1099/0022-1317-79-8-1871. [DOI] [PubMed] [Google Scholar]
- 4.Bukh J, Apgar CL. Five new or recently discovered (GBV-A) virus species are indigenous to New World monkeys and may constitute a separate genus of the Flaviviridae. Virology. 1997;229:429–436. doi: 10.1006/viro.1997.8461. [DOI] [PubMed] [Google Scholar]
- 5.Simons JN, et al. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 6.Bukh J, et al. Challenge pools of hepatitis C virus genotypes 1-6 prototype strains: Replication fitness and pathogenicity in chimpanzees and human liver-chimeric mouse models. J Infect Dis. 2010;201:1381–1389. doi: 10.1086/651579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dorner M, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takikawa S, et al. Functional analyses of GB virus B p13 protein: Development of a recombinant GB virus B hepatitis virus with a p7 protein. Proc Natl Acad Sci USA. 2006;103:3345–3350. doi: 10.1073/pnas.0511297103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bukh J, Apgar CL, Yanagi M. Toward a surrogate model for hepatitis C virus: An infectious molecular clone of the GB virus-B hepatitis agent. Virology. 1999;262:470–478. doi: 10.1006/viro.1999.9941. [DOI] [PubMed] [Google Scholar]
- 10.Kolykhalov AA, et al. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science. 1997;277:570–574. doi: 10.1126/science.277.5325.570. [DOI] [PubMed] [Google Scholar]
- 11.Yanagi M, Purcell RH, Emerson SU, Bukh J. Transcripts from a single full-length cDNA clone of hepatitis C virus are infectious when directly transfected into the liver of a chimpanzee. Proc Natl Acad Sci USA. 1997;94:8738–8743. doi: 10.1073/pnas.94.16.8738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li YP, Gottwein JM, Scheel TK, Jensen TB, Bukh J. MicroRNA-122 antagonism against hepatitis C virus genotypes 1-6 and reduced efficacy by host RNA insertion or mutations in the HCV 5′ UTR. Proc Natl Acad Sci USA. 2011;108:4991–4996. doi: 10.1073/pnas.1016606108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolykhalov AA, Feinstone SM, Rice CM. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J Virol. 1996;70:3363–3371. doi: 10.1128/jvi.70.6.3363-3371.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakita T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottwein JM, et al. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: Role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology. 2009;49:364–377. doi: 10.1002/hep.22673. [DOI] [PubMed] [Google Scholar]
- 16.Scheel TK, Gottwein JM, Mikkelsen LS, Jensen TB, Bukh J. Recombinant HCV variants with NS5A from genotypes 1-7 have different sensitivities to an NS5A inhibitor but not interferon-α. Gastroenterology. 2011;140:1032–1042. doi: 10.1053/j.gastro.2010.11.036. [DOI] [PubMed] [Google Scholar]
- 17.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 18.Lanford RE, et al. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science. 2010;327:198–201. doi: 10.1126/science.1178178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Machlin ES, Sarnow P, Sagan SM. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc Natl Acad Sci USA. 2011;108:3193–3198. doi: 10.1073/pnas.1012464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Linnen J, et al. Molecular cloning and disease association of hepatitis G virus: A transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]