Abstract
Many central metabolic processes require iron as a cofactor and take place in specific subcellular compartments such as the mitochondrion or the chloroplast. Proper iron allocation in the different organelles is thus critical to maintain cell function and integrity. To study the dynamics of iron distribution in plant cells, we have sought to identify the different intracellular iron pools by combining three complementary imaging approaches, histochemistry, micro particle-induced x-ray emission, and synchrotron radiation micro X-ray fluorescence. Pea (Pisum sativum) embryo was used as a model in this study because of its large cell size and high iron content. Histochemical staining with ferrocyanide and diaminobenzidine (Perls/diaminobenzidine) strongly labeled a unique structure in each cell, which co-labeled with the DNA fluorescent stain DAPI, thus corresponding to the nucleus. The unexpected presence of iron in the nucleus was confirmed by elemental imaging using micro particle-induced x-ray emission. X-ray fluorescence on cryo-sectioned embryos further established that, quantitatively, the iron concentration found in the nucleus was higher than in the expected iron-rich organelles such as plastids or vacuoles. Moreover, within the nucleus, iron was particularly accumulated in a subcompartment that was identified as the nucleolus as it was shown to transiently disassemble during cell division. Taken together, our data uncover an as yet unidentified although abundant iron pool in the cell, which is located in the nuclei of healthy, actively dividing plant tissues. This result paves the way for the discovery of a novel cellular function for iron related to nucleus/nucleolus-associated processes.
Keywords: Embryo, Fluorescence, Imaging, Iron, Nucleolus, Nucleus
Introduction
Metal ions play multiple structural and catalytic functions in living cells. Iron is among the most important essential metals because it serves as a cofactor in many metabolic processes such as respiration, photosynthesis, cell division, and fatty acid and branched amino acid biosynthesis. Most of these reactions take place in intracellular organelles that represent as many sinks for iron. The subcellular distribution of iron and its dynamics between compartments are scarcely documented and rely mostly on fragmented biochemical estimations. For instance, with a complete electron transfer chain containing 22 iron atoms, chloroplasts are considered as obvious sites of high iron accumulation in plant cells (1). In animal cells, mitochondria and lysosomes, in particular those involved in autophagic degradation of iron-rich macromolecules, are expected to contain rather high concentrations of iron as well (2). Because iron can react with oxygen and generate oxidative stress, living cells prevent both iron-promoted toxicity and iron shortage by maintaining a strict balance between transport, storage, and recycling of iron in each compartment. The deregulation of iron compartmentalization is very often associated, in animal systems, with neurodegenerative pathologies. For example, mutations of the mitochondrial protein frataxin result in increased iron accumulation as well as altered iron-sulfur cluster assembly in mitochondria, causing a human disease called Friedreich ataxia (3, 4). Likewise, the etiology of Alzheimer and Parkinson diseases is associated with the accumulation of iron deposits in specific regions of the brain (5). In the model plant species Arabidopsis thaliana, the alteration of iron distribution in two important organelles, the chloroplast and the vacuole, severely impacts plant growth. Indeed, the disruption of the FRO7 gene, encoding a ferric reductase located at the chloroplast surface, results in decreased chloroplastic iron accumulation, severe leaf chlorosis, and impaired growth on iron-limited soils (6). In addition, the vacuoles were identified as the main storage site of iron in mature Arabidopsis embryos (7). During embryo development, iron loading into these vacuoles is mediated by VIT1, an iron/manganese tonoplastic transporter (8), whereas the remobilization of this vacuolar iron pool during early germination requires the efflux activity of the two cation metal transporters NRAMP3 and NRAMP4 (7, 9). Abolition of vacuolar influx in a vit1 mutant or efflux in an nramp3 nramp4 double mutant similarly impairs post-germinative growth under iron-limited conditions (8, 9). Crucial in both studies was having recourse to elemental imaging techniques (synchrotron radiation x-ray fluorescence for the study of VIT1 and energy-dispersive x-ray microscopy for NRAMP3 and NRAMP4) to reveal the subtle changes in iron distribution occurring in seeds of the mutants, not detectable otherwise. These two studies perfectly illustrated the power of combining molecular genetics with sophisticated elemental imaging approaches to unravel the function of several transporters in metal homeostasis.
We have undertaken the analysis of iron distribution in plant cells using several imaging approaches. In particular, we have utilized the Perls/DAB3 histochemical staining method, which we had previously adapted to plant cells and which is characterized by a sensitivity and resolution high enough to allow intracellular iron detection (7).
Here we report on the unexpected finding that the nucleus is also an iron-rich organelle in plant cells. It is likely that this compartment has been ignored, or at least overlooked, in the past because high spatial resolution iron distribution has so far been addressed in very few reported studies.
EXPERIMENTAL PROCEDURES
Histochemical Staining of Iron with the Perls/DAB Procedure
Pea embryos were dissected from developing seeds of pea plants (Pisum sativum) grown in pots in a greenhouse and irrigated with water. Embryos from pea and leaf fragments from A. thaliana and tomato (Lycopersicon esculentum) were vacuum-infiltrated with the fixation solution containing 2% (w/v) paraformaldehyde, 1% (v/v) glutaraldehyde, 1% (w/v) caffeine in 100 mm phosphate buffer (pH 7) for 30 min and incubated for 15 h in the same solution. The fixed samples were washed with 0.1 m phosphate buffer (pH 7.4) three times and dehydrated in successive baths of 50, 70, 90, 95, and 100 ethanol, butanol/ethanol 1:1 (v/v), and 100% butanol. Then, the tissues were embedded in the Technovit 7100 resin (Heraeus Kulzer) according to the manufacturer's instructions, and thin sections (5 mm) were made. The sections were deposited on glass slides that were incubated for 45 min in Perls stain solution, except for negative controls. After washing with distillated water, the glass slides were incubated in a methanol solution containing 0.01 m NaN3 and 0.3% (v/v) H2O2 for 1 h and then washed with 0.1 m phosphate buffer (pH 7.4). For the intensification reaction, samples were then incubated between 10 and 30 min in a 0.1 m phosphate buffer (pH 7.4) solution containing 0.025% (w/v) DAB (Sigma), 0.005% (v/v) H2O2, and 0.005% (w/v) CoCl2 (intensification solution). Rinsing with distilled water stopped the reaction.
Double Staining with Perls/DAB and DAPI
The thin sections were first stained with Perls/DAB, as described above, and then stained with DAPI. The glass slides were incubated in a 2 μg·ml−1 (w/v) solution of DAPI for 5 min in the dark and rinsed with distilled water. Observations were realized using a Leica DM6000 microscope equipped with an A4 filter (excitation, 340–380 nm; emission, 450–490 nm).
Cryofixation and Cryosections of Pea Embryos
Embryos, dissected from seeds, were immersed in optimum cutting formulation (OCT, Sakura Finetek, Leiden, The Netherlands) and rapidly frozen in liquid nitrogen. The solid blocks obtained were then sectioned (30–40 μm thickness) using a cryomicrotome (the sample holder was kept at −50 °C, whereas the sample atmosphere was set at −18 °C). The resulting slides were deposited on Ultralene films (Spex Certiprep) and kept in liquid nitrogen until introduced in the analysis chamber of the Light for Ultimate Characterization by Imaging and Absorption (LUCIA) synchrotron beamline.
RESULTS
The pea (P. sativum) embryo was chosen as plant material in this work for several reasons: (i) its high iron content (200–400 ppm) that is above the detection threshold of elemental imaging techniques (micro particle-induced x-ray emission (μPIXE) and micro X-ray fluorescence (μXRF)); (ii) the large size of its cells and organelles that facilitates imaging analyses; and (iii) the fact that, together with other legume seeds, it represents a widespread staple food for humans, which makes it an excellent target for biotechnological engineering of seed nutritional quality.
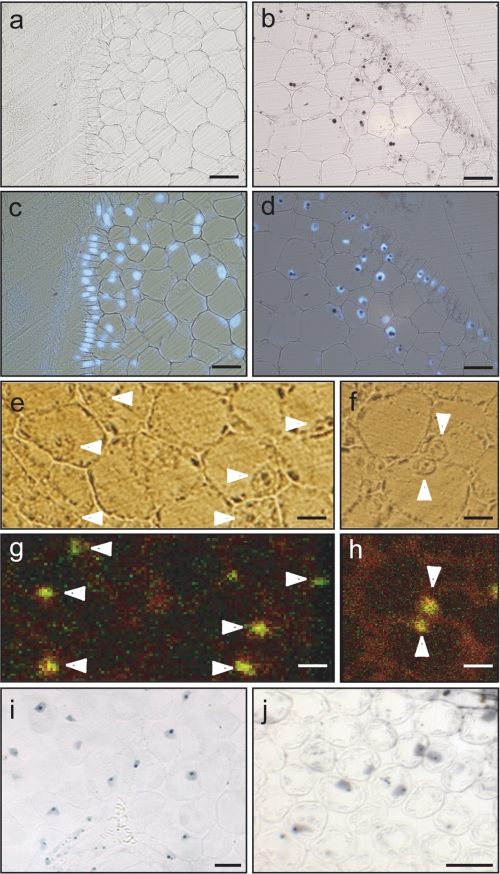
In a first approach, pea embryos were dissected from developing seeds and stained with Perls/DAB. Pea embryonic cells exhibited a strongly stained structure (Fig. 1b), invisible in control sections without Perls (Fig. 1a), which resembled a nucleus (Fig. 1a). The nuclear identity of this compartment was confirmed by counterstaining the histological sections with DAPI (Fig. 1, c and d). Although we had previously established that the Perls/DAB technique is specific for iron, we sought to confirm this result with an elemental imaging approach (7). Unstained histological sections were thus analyzed by μPIXE using a proton beam of 1-μm diameter at the AIFIRA facility (10). As expected, whenever a nucleus could be identified by light microscopy (Fig. 1, e and f), the μPIXE imaging analysis revealed the presence of high concentrations of iron (Fig. 1, g and h), confirming the existence of a pool of iron in the nucleus. This observation is not a particular feature of pea embryos because in leaf cells from A. thaliana and tomato, the nuclei were also strongly stained with Perls/DAB (Fig. 1, i and j, respectively).
FIGURE 1.
Plant cells accumulate iron in the nucleus. a–j, histochemical analysis of pea embryonic cells (a–d), A. thaliana (i), and tomato leaf mesophyll cells (j). e–h, elemental imaging was performed on sections of pea embryos embedded in resin. a, negative control of the Perls/DAB staining where the first reaction with potassium ferrocyanide was omitted. b, Perls/DAB staining of iron. c, DAPI-stained nuclei revealed by epifluorescence (from control section shown in panel a). d, merge of Perls/DAB and DAPI reactions. e and f, bright field microscopy of unstained cells. g and h, μPIXE analysis of the same sections with iron imaging in green and phosphorus imaging in red. Samples were scanned with a 3.0-MeV proton beam focused to 1-μm size and measuring the X-rays emitted at 6.4 keV for iron and at 2.0 KeV for phosphorus. White arrows point out the cell nucleus. Bar = 40 μm for panels a–d and i–j, bar = 20 μm for panels e–h.
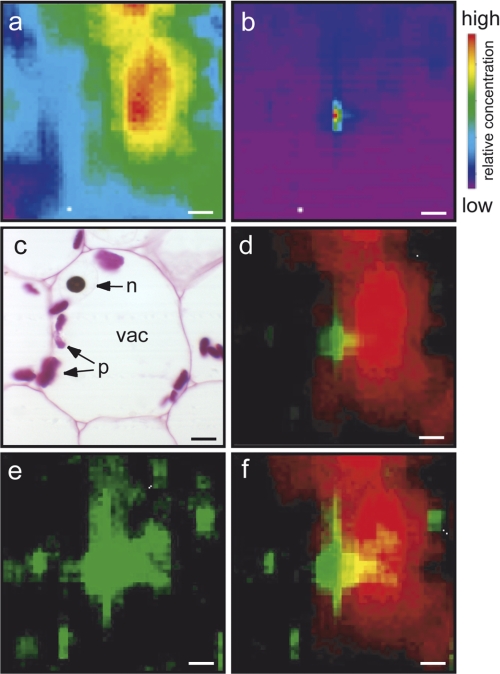
To rule out the possibility that this iron distribution was somehow caused by sample processing for histological analyses, we then performed synchrotron radiation μXRF on cryofixed and cryosectioned pea embryos. The cryosections were kept in liquid nitrogen until the introduction in the beamline chamber, and the following analyses were performed in cryogenic conditions (130 K) to maintain the cellular and chemical integrity throughout the process of elemental mapping. The average size of a pea embryonic cell, its vacuole, and its nucleus are, respectively, 60–70, 40–50, and 15 μm. Given that the lateral resolution of the synchrotron beam utilized is 2.5 × 3.5 μm, we could analyze zones of 100 × 100 μm and obtain elemental mapping within compartments of a whole cell. The mapping of potassium was used as a canvas to locate the vacuolar compartment, where most of the potassium is stored, and thus depict cell-to-cell limits and focus on a single cell (Fig. 2a). Iron detection showed a strong 20 × 10-μm signal (Fig. 2b), a size that is compatible with that of the nucleus, clearly visible on a histological section of a comparable cell (Fig. 2c). Merging both iron and potassium maps clearly showed that the iron-rich zones are adjacent to the potassium-rich vacuolar structures (Fig. 2d), which again is consistent with iron being located in the nucleus. By increasing the fluorescence scale so as to saturate the nuclear signal, several other pools of iron appeared in the cell periphery and in the vacuole (Fig. 2, e and f). Taken together, these results clearly establish that, besides the usual organelles, the nucleus represents a novel site of massive iron accumulation in the cell.
FIGURE 2.
Elemental μXRF mapping on cryosections of pea embryos. a, mapping of potassium. b, mapping of iron. c, pea cell from resin-embedded embryo, stained with Perls/DAB and Schiff to reveal iron and carbohydrates, respectively. n, nucleus, vac, vacuole, p, plastids. d, merge of potassium (red) and iron (green) signals. e, iron fluorescence intensity with modified fluorescence levels to reveal minor pools of iron. f, merge of potassium fluorescence (red) with the readjusted iron fluorescence from e. The synchrotron beam (size: 2.5 × 3.5 μm) was set at 7200 eV. Bar = 10 μm.
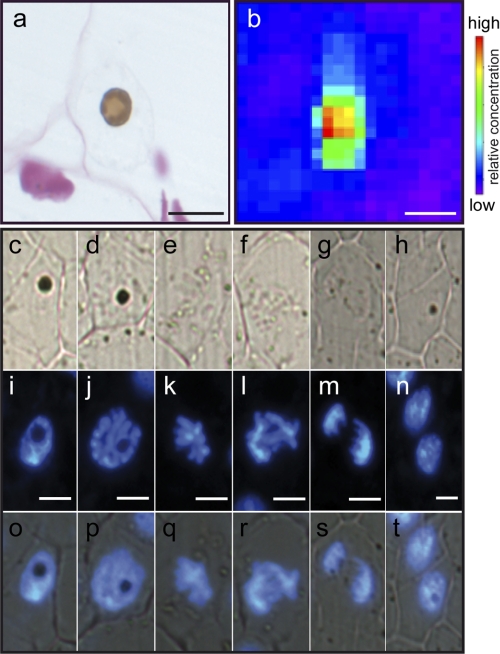
A detailed observation of the Perls/DAB-stained iron pool within the nucleus indicated that it is restricted to a subnuclear domain (Fig. 3a, see also Fig. 1d) that is reminiscent in size and shape to the nucleolus. Because one of the features of the nucleolus is its disorganization during mitosis, we followed the fate of the Perls/DAB-stained nuclear structure during the different stages of cell division, easily identified in pea embryo histological sections stained with DAPI (Fig. 3, i–n). Clearly, the iron-rich structures, still visible during interphase (Fig. 3, c, i, and o) and prophase (Fig. 3, d, j, and p), disappeared from metaphase to telophase (Fig. 3, e, f, g, k, l, m, q, r, s), reappearing in the daughter nuclei at the interphase stage (Fig. 3, h, n, and t). This observation unambiguously identifies the nucleolus as the iron-rich structure in pea cell nuclei. Consistently, fine mapping of the nucleus by μXRF showed that on a section representing a 20 × 10-μm-large nucleus, iron concentration is highest in a 5 × 5-μm sector, a size that is compatible with the measured size of the nucleolus in pea embryo cells (Fig. 3, a and b).
FIGURE 3.
Iron-specific accumulation in the nucleolus. a, close-up view of a nucleus from a Perls/DAB- and Schiff-stained embryonic cell. b, fine μXRF iron mapping of a nucleus. c–t, Perls/DAB and DAPI co-staining of embryonic cells at the different stages of mitosis. c–h, bright field microscopy; i–n, UV microscopy of the same sections to reveal DAPI epifluorescence; o–t, merge of both signals. Bar = 10 μm.
DISCUSSION
The analysis of subcellular iron localization by histochemistry (Perls/DAB) and elemental microanalyses (μPIXE and μXRF) has established without doubt that the nucleus contains detectable amounts of iron. This finding was not restricted to pea because strong nuclear iron staining was also observed in leaves of other species such as A. thaliana and tomato (Fig. 1, i–j). Furthermore, we have shown that within the nucleus, the iron concentration in the nucleolus is the highest encountered among the intracellular compartments. This result is novel and completely unexpected. Indeed, the accumulation of a highly reactive and potentially toxic metal in the very close vicinity of DNA and RNA is rather counterintuitive. In mammals, the nuclei of healthy cells barely contain detectable amounts of iron (11–14), iron being located mainly in the cytoplasm, whereas strong iron accumulations in nuclei have only been reported in neurodegenerative tissues, such as in cerebral cortex neurons with neuroferritinopathy (15, 16) or in hippocampal glial cells from Alzheimer disease (17, 18). Therefore our findings challenge the widely admitted idea that in animals, the presence of iron in nuclei is a sign of disease.
Our finding raises the important question of the role that nucleolar iron plays in plant cells. If the nucleolus requires such high amounts of iron, it is conceivable that iron atoms might be associated to a very abundant class of molecules. Because the main function of the nucleolus is to synthesize ribosomal RNA, it is tempting to speculate that iron is somehow involved in the metabolism of rRNA. Moreover, because it is difficult to conceive that such a high amount of iron corresponds to the cofactor of a specific enzyme, a possibility exists that iron in the nucleolus might be directly bound to rRNA. Consistent with this hypothesis, rRNA has been shown to bind high amounts of iron, both in vitro (17, 19) and in vivo (17). It is thus possible that in plants, iron is required during ribosomal RNA biosynthesis in the nucleolus, either by promoting or stabilizing the secondary structure or by catalyzing maturation of the different rRNA subunits.
Taken together, our data have revealed a new intracellular site where high amounts of iron atoms are needed, for a function that remains to be identified. Consequently, this finding should lead to reconsidering the overall scheme of iron requirements and distribution at the intracellular level in plant cells, including the nucleolus as a new player from now on. This work has clearly illustrated that healthy cells can accumulate and tolerate high amounts of iron in their nucleus without being a sign of a particular dysfunction. This peculiar feature of plant nuclei could reflect the fact that iron plays a plant-specific function. Plant cells must have evolved highly efficient mechanisms, still unidentified, to protect the nucleus from the potential deleterious effects of iron.
Acknowledgments
We thank the SOLEIL Synchrotron for provision of beamtime (Project 20100264) as well as Nicolas Trcera, Pierre Lagarde, and Anne-Marie Flank from the LUCIA beamline for unconditional help and support.
This work was supported by the Centre National de la Recherche Scientifique, l'Institut National de la Recherche Agronomique, and by Grant 23643 (Circulation of Iron within the Plant and Delivery to the Seed (CIDS)).
- DAB
- diaminobenzidine
- μPIXE
- micro particle-induced x-ray emission
- μXRF
- micro X-ray fluorescence.
REFERENCES
- 1. Wollman F. A., Minai L., Nechushtai R. (1999) Biochim. Biophys. Acta 1411, 21–85 [DOI] [PubMed] [Google Scholar]
- 2. Kurz T., Terman A., Gustafsson B., Brunk U. T. (2008) Histochem. Cell Biol. 129, 389–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gordon N. (2000) Brain Dev. 22, 465–468 [DOI] [PubMed] [Google Scholar]
- 4. Mühlenhoff U., Richhardt N., Ristow M., Kispal G., Lill R. (2002) Hum. Mol. Genet. 11, 2025–2036 [DOI] [PubMed] [Google Scholar]
- 5. Zecca L., Youdim M. B., Riederer P., Connor J. R., Crichton R. R. (2004) Nat. Rev. Neurosci. 5, 863–873 [DOI] [PubMed] [Google Scholar]
- 6. Jeong J., Cohu C., Kerkeb L., Pilon M., Connolly E. L., Guerinot M. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 10619–10624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Roschzttardtz H., Conéjéro G., Curie C., Mari S. (2009) Plant Physiol. 151, 1329–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim S. A., Punshon T., Lanzirotti A., Li L., Alonso J. M., Ecker J. R., Kaplan J., Guerinot M. L. (2006) Science 314, 1295–1298 [DOI] [PubMed] [Google Scholar]
- 9. Lanquar V., Lelièvre F., Bolte S., Hamès C., Alcon C., Neumann D., Vansuyt G., Curie C., Schröder A., Krämer U., Barbier-Brygoo H., Thomine S. (2005) EMBO J. 24, 4041–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carmona A., Devès G., Ortega R. (2008) Anal. Bioanal. Chem. 390, 1585–1594 [DOI] [PubMed] [Google Scholar]
- 11. Carmona A., Cloetens P., Deves G., Bohic S., Ortega R. (2008) J. Anal. At. Spectrom. 23, 1083–1088 [Google Scholar]
- 12. Matsuyama S., Shimura M., Fujii M., Maeshima K., Yumoto H., Mimura H., Sano Y., Yabashi M., Nishino Y., Tamasaku K., Ishizaka Y., Ishikawa T., Yamauchi K. (2010) X-Ray Spectrom. 39, 260–266 [Google Scholar]
- 13. Ortega R., Cloetens P., Devès G., Carmona A., Bohic S. (2007) PLoS ONE 2, e925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Witting P. K., Harris H. H., Rayner B. S., Aitken J. B., Dillon C. T., Stocker R., Lai B., Cai Z., Lay P. A. (2006) Biochemistry 45, 12500–12509 [DOI] [PubMed] [Google Scholar]
- 15. Schröder J. M. (2005) Acta Neuropathol. 109, 109–114 [DOI] [PubMed] [Google Scholar]
- 16. Vidal R., Miravalle L., Gao X., Barbeito A. G., Baraibar M. A., Hekmatyar S. K., Widel M., Bansal N., Delisle M. B., Ghetti B. (2008) J. Neurosci. 28, 60–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Honda K., Smith M. A., Zhu X., Baus D., Merrick W. C., Tartakoff A. M., Hattier T., Harris P. L., Siedlak S. L., Fujioka H., Liu Q., Moreira P. I., Miller F. P., Nunomura A., Shimohama S., Perry G. (2005) J. Biol. Chem. 280, 20978–20986 [DOI] [PubMed] [Google Scholar]
- 18. Quintana C., Bellefqih S., Laval J. Y., Guerquin-Kern J. L., Wu T. D., Avila J., Ferrer I., Arranz R., Patiño C. (2006) J. Struct. Biol. 153, 42–54 [DOI] [PubMed] [Google Scholar]
- 19. Berens C., Streicher B., Schroeder R., Hillen W. (1998) Chem. Biol. 5, 163–175 [DOI] [PubMed] [Google Scholar]