Abstract
Arrestins bind active phosphorylated G protein-coupled receptors, blocking G protein activation and channeling the signaling to G protein-independent pathways. Free arrestin-3 and receptor-bound arrestin-3 scaffold the ASK1-MKK4-JNK3 module, promoting JNK3 phosphorylation, whereas highly homologous arrestin-2 does not. Here, we used arrestin-2/3 chimeras and mutants to identify key residues of arrestin-3 responsible for its ability to facilitate JNK3 activation. Our data demonstrate that both arrestin domains are involved in JNK3 activation, with the C-terminal domain being more important than the N-terminal domain. We found that Val-343 is the key contributor to this function, whereas Leu-278, Ser-280, His-350, Asp-351, His-352, and Ile-353 play supporting roles. We also show that the arrestin-3-specific difference in the arrangement of the β-strands in the C-terminal domain that underlies its lower selectivity for active phosphoreceptors does not play an appreciable role in its ability to enhance JNK3 activation. Importantly, the strength of the binding of ASK1 or JNK3, as revealed by the efficiency of co-immunoprecipitation, does not correlate with the ability of arrestin proteins to promote ASK1-dependent JNK3 phosphorylation. Thus, multiple residues on the non-receptor-binding side of arrestin-3 are crucial for JNK3 activation, and this function and the receptor-binding characteristics of arrestin can be manipulated independently by targeted mutagenesis.
Keywords: G Protein-coupled Receptors (GPCRs), Jun N-terminal kinase (JNK), MAP Kinases (MAPKs), Protein-Protein Interactions, Signal Transduction, Site-directed Mutagenesis, Arrestin
Introduction
Arrestins were first discovered as negative regulators of G protein-coupled receptor (GPCR)3 signaling (reviewed in Ref. 1). Arrestins specifically bind active phosphorylated forms of their cognate receptors, blocking further G protein activation by steric exclusion (2, 3). The finding that arrestins recruit the complex to the coated pit via interactions with clathrin (4) and adaptor protein AP2 (5) extended their functional role to receptor internalization. Subsequent discoveries that arrestins also initiate the second, G protein-independent round of receptor signaling, recruiting kinases, phosphodiesterases, and ubiquitin ligases to the complex, revealed the role of arrestins as receptor-regulated signaling scaffolds (reviewed in Refs. 1 and 6).
Recent findings suggest that arrestins have multiple functions that do not depend on their association with GPCRs. Free arrestin-24 was shown to regulate histone acetylation and gene transcription (7). Arrestin-1, arrestin-2, and arrestin-3 bind the E3 ubiquitin ligases Mdm2 (murine double minute 2, which functions as negative regulator of the p53 tumor suppressor) (8, 9) and parkin (10), calmodulin (11), and microtubules (12, 13) and recruit Mdm2 and MAPK ERK2 to the cytoskeleton (13). Arrestin-3 binds JNK3 (c-Jun N-terminal kinase 3) (14) and scaffolds the ASK1 (apoptosis signal-regulating kinase 1)-MKK4 (MAPK kinase 4)-JNK3 cascade in a receptor-independent fashion (15, 16), whereas arrestin-2 binds γ-tubulin and regulates centrosome function (17). Thus, arrestins act as versatile organizers of multiprotein signaling complexes, with and without collaboration with GPCRs (reviewed in Refs. 1 and 18).
Although arrestin interactions with numerous partners have been reported, the structural basis of these interactions and the relationships between the binding to signaling proteins and regulation of their functional state remains a largely unexplored area. Here, we took advantage of the ability of arrestin-3 to promote ASK1-dependent JNK3 activation, in contrast to highly homologous arrestin-2, to identify a limited number of arrestin-3-specific residues on the non-receptor-binding surface that are absolutely required for this function. We found that substitutions of one to six residues prevented arrestin-3-dependent JNK3 phosphorylation without an appreciable effect on arrestin-3 binding of JNK3 or upstream kinases. Replacement of homologous elements of arrestin-2 with the corresponding arrestin-3 residues only marginally enhanced its ability to facilitate JNK3 activation. Swapping the β-strand that is part of a contiguous β-sheet in arrestin-2 (19, 20) but is distorted in arrestin-3 (21), significantly reducing arrestin-3 selectivity for active phosphorylated GPCRs (21), did not appreciably change the relative ability of arrestin-2 and arrestin-3 to promote JNK3 activation.
EXPERIMENTAL PROCEDURES
Materials
All restriction enzymes were from New England Biolabs. All other chemicals were from sources described previously (22).
Plasmid Construction
Expression constructs for bovine arrestin-2 and arrestin-3 with a C-terminal FLAG tag in pcDNA3 were described previously (9, 13, 16). Long and short splice variants of arrestin-2 and arrestin-3, respectively, were used because these variants are prevalent in most cells (23–25). Chimeras were generated using restriction sites engineered in homologous positions of arrestin-2 and arrestin-3 coding sequences as described (10, 26). Mutations were introduced by PCR, and all constructs were verified by dideoxy sequencing. Expression constructs for GFP-JNK3, HA-JNK3, and HA-MKK4 were gifts from Drs. Louis Luttrell (Medical University of South Carolina), Robert J. Lefkowitz (Duke University), and Jia Le Dai (The University of Texas MD Anderson Cancer Center), respectively.
Cell Culture and Transient Transfection
COS-7 African green monkey cells were maintained in DMEM supplemented with 10% heat-inactivated FBS (Invitrogen), penicillin, and streptomycin at 37 °C in a humidified incubator with 5% CO2. The cells were plated at 80–90% confluence and transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Before all experiments, cells were serum-starved overnight and used 48 h post-transfection.
Western Blotting
COS-7 cells were incubated with phosphatase inhibitors (50 mm NaF and 10 mm Na3VO4) in serum-free medium for 15 min at 37 °C; washed with cold PBS; and lysed with SDS sample buffer containing 10 mm NaF, 100 μm Na3VO4, 2 mm EDTA, 2 mm EDTA, and 1 mm PMSF. Whole cell lysates were boiled for 5 min and centrifuged at 10,000 × g for 10 min, and the supernatants were used for Western blotting. The proteins were resolved by 10% SDS-PAGE and transferred to PVDF membrane (Millipore, Bedford, MA). Mouse monoclonal antibodies against FLAG (Sigma), HA (Sigma), and phospho-JNK (Cell Signaling) were used at 1:1000 or 1:2000 dilution, followed by HRP-conjugated anti-mouse secondary antibody. Protein bands were detected by enhanced chemiluminescence (West Pico, Pierce), followed by exposure to x-ray film.
Immunoprecipitation
Cells (on 60-mm plates) were lysed in 0.75 ml of lysis buffer (50 mm Tris, 2 mm EDTA, 250 mm NaCl, 10% glycerol, 0.5% Nonidet P-40, 20 mm NaF, 1 mm sodium orthovanadate, 10 mm N-ethylmaleimide, 2 mm benzamidine, and 1 mm PMSF) for 30–60 min at 4 °C. After centrifugation, supernatants were precleared with 20 μl of protein G-agarose. Then, 600 μl of supernatant was incubated with primary antibodies for 2 h, followed by the addition of 12 μl of protein G-agarose beads for 2 h or overnight. The beads were washed three times with 1 ml of lysis buffer, and the proteins were eluted with 50 μl of SDS sample buffer, boiled for 5 min, and analyzed by Western blotting as described above.
Quantification and Statistical Analysis
The bands on the x-ray film were quantified using Quantity One software (Bio-Rad). StatView software (SAS Institute) was used for statistical analysis of quantitative data; p < 0.05 was considered significant.
RESULTS
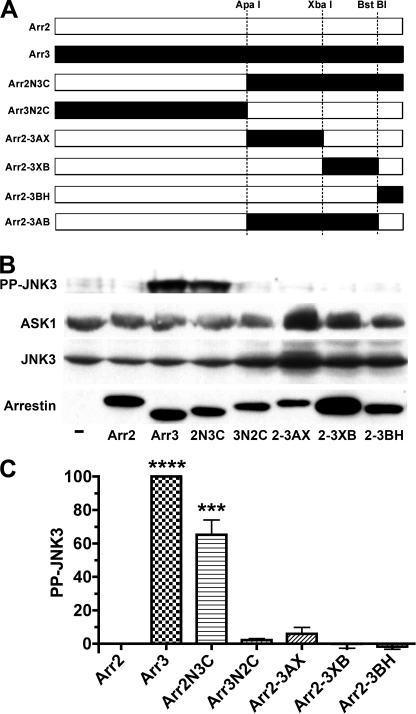
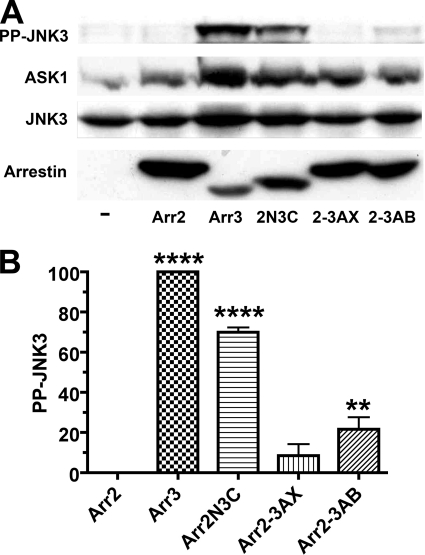
Both non-visual arrestins bind JNK3 (8, 9, 14) and upstream kinases MKK4 and ASK1 (16), yet only arrestin-3 promotes receptor-independent JNK3 phosphorylation (15, 16). A previous study using rat arrestins suggested that the arrestin-3-specific sequence RRS is solely responsible for JNK3 activation (15). However, the RRS motif is unique for rodent arrestin-3 (supplemental Fig. S1) (1, 27). The finding that bovine arrestin-3, which is a natural “mutant” with the deletion of one arginine in this sequence (a feature shared by human, chimpanzee, and zebrafish proteins) (supplemental Fig. S1), effectively activates JNK3 (16) indicated that this issue must be reinvestigated using a more common form of arrestin-3. Because of high structural similarity (19, 21, 28, 29), arrestin chimeras usually fold properly and remain functional (10, 13, 26, 30, 31). Therefore, first we used the same approach as in the previous study (15) and exchanged elements between bovine arrestin-2 and arrestin-3 (Fig. 1). We confirmed the previous finding that a chimera with the arrestin-2 N-terminal domain and the arrestin-3 C-terminal domain (Arr2N3C) activates JNK3, albeit less effectively than arrestin-3, whereas the opposite chimera (Arr3N2C) is completely inactive (Fig. 1, B and C). Interestingly, none of the three smaller C-terminal domain elements of arrestin-3 (Fig. 1A) conferred the ability to activate JNK3 (Fig. 1, B and C). Even the chimera Arr2-3AB, containing 180 residues (85%) of the arrestin-3 C-terminal domain (Fig. 1A), showed much weaker activity than Arr2N3C (Fig. 2, A and B).
FIGURE 1.
Arrestin ability to activate JNK3 is largely determined by the C-terminal domain. A, schematic of the arrestin-2/3 chimeras, with arrestin-2- and arrestin-3-derived elements shown in white and black, respectively. The positions of the restriction sites engineered in homologous places in coding sequences and used to swap elements are indicated. Arr2, arrestin-2; Arr3, arrestin-3. Arr2N3C is a chimera with the N-terminal domain (residues 1–180) of arrestin-2 and the C-terminal domain (residues 182–408) of arrestin-3. Arr3N2C is the reverse chimera with the N- and C-terminal domains of arrestin-3 and arrestin-2, respectively. Arr2-3AX is arrestin-2 with the arrestin-3 element between the ApaI and XbaI sites (residues 182–290). Arr2-3XB is arrestin-2 with the arrestin-3 element between the XbaI and BstBI sites (residues 291–389). Arr2-3BH is arrestin-2 with the arrestin-3 element downstream of the BstBI site (residues 390–408). Arr2-3AB is arrestin-2 with the arrestin-3 element between the ApaI and BstBI sites (residues 182–389). B, COS-7 cells were transfected with HA-ASK1, HA-JNK3, and the indicated arrestins with a C-terminal FLAG tag; harvested 48 h post-transfection; and lysed. The amount of active phosphorylated JNK3 (PP-JNK3; upper blot) and the expression of ASK1, JNK3, and arrestins were determined by Western blotting. C, quantification of the level of JNK3 phosphorylation in cells expressing the indicated arrestins. Phosphorylated JNK3 bands from three to four independent experiments were quantified. The data were analyzed by analysis of variance (ANOVA) with arrestin as a main factor, followed by the Bonferroni-Dunn post-hoc test. The significance of the difference from arrestin-2 is shown. ***, p < 0.001; ****, p < 0.0001. Band intensity is expressed as a percent of the difference between phosphorylated JNK3 in arrestin-3- and arrestin-2-expressing cells.
FIGURE 2.
Parts of the arrestin-3 C-terminal domain confer lower activity than the complete C-terminal domain. A, COS-7 cells were transfected with HA-ASK1, HA-JNK3, and the indicated arrestins with a C-terminal FLAG tag. The amount of active phosphorylated JNK3 (PP-JNK3) 48 h post-transfection and the expression of ASK1, JNK3, and arrestins were determined by Western blotting. B, quantification of the level of JNK3 phosphorylation in cells expressing the indicated arrestins. Phosphorylated JNK3 bands from three independent experiments were quantified. The data were analyzed by ANOVA with arrestin as a main factor, followed by the Bonferroni-Dunn post-hoc test. The significance of the difference from arrestin-2 is shown. **, p < 0.01; ****, p < 0.0001. Band intensity is expressed as a percent of the difference between phosphorylated JNK3 in arrestin-3- and arrestin-2-expressing cells. Arr2, arrestin-2; Arr3, arrestin-3; 2N3C, Arr2N3C; 2-3AX, Arr2-3AX; 2-3AB, Arr2-3AB (detailed chimera descriptions are in the legend to Fig. 1).
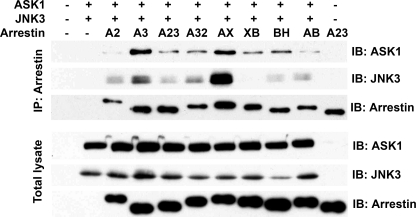
To investigate whether differential binding of JNK3 and/or ASK1 underlies the activity, we coexpressed FLAG-tagged WT and chimeric arrestins with HA-tagged ASK1 and JNK3 (which are very different sizes and do not interfere with each other's detection on the same blot); immunoprecipitated arrestins using anti-FLAG antibody; and detected immunoprecipitated arrestin, JNK3, and ASK1 by Western blotting (Fig. 3). Surprisingly, we found no correlation between the affinity of these interactions and JNK3 activation. It is important to keep in mind that low-affinity complexes dissociate in the process, so that only relatively high-affinity partners are retained. This makes co-immunoprecipitation a one-way experiment: a positive outcome is meaningful, whereas a negative outcome suggests that the proteins either do not interact or associate with low affinity. As far as binding detectable by immunoprecipitation is concerned, completely inactive proteins range from very poor interactors bringing down only small amounts of ASK1 and JNK3 (e.g. arrestin-2) to those that co-immunoprecipitate with even more ASK1 and JNK3 that arrestin-3 (e.g. Arr2-3AX chimera), with everything in between (Fig. 3). Importantly, the same is true for active constructs: arrestin-3, Arr2N3C, and Arr2-3AB chimeras co-immunoprecipitated with similar amount of JNK3 (Fig. 3), yet their activities differed dramatically (Fig. 2). Thus, there does not appear to be a meaningful correlation between binding of these kinases by different arrestins and their ability to activate JNK3. This is hardly surprising: effective scaffolds, just like effective enzymes, must have rapid turnover, which cannot be achieved with high-affinity interactions.
FIGURE 3.
Strength of binding of arrestin to ASK1 and JNK3 does not predict its ability to promote JNK3 phosphorylation. COS-7 cells were transfected with HA-ASK1, HA-JNK3, and the indicated arrestins with a C-terminal FLAG tag or empty vector; harvested 48 h post-transfection; and lysed. Arrestins were immunoprecipitated (IP) with anti-FLAG antibody, and the amount of arrestins and co-immunoprecipitated HA-ASK1 and HA-JNK3 was determined by Western blotting (upper three blots). The expression of these proteins was determined in cell lysates (lower three blots). Cells transfected with empty vector only (first lane), HA-ASK1 + HA-JNK3 only (second lane), or arrestin only (last lane) served as controls. The results of a representative experiment of three performed are shown. Note that the level of JNK3 or ASK1 co-immunoprecipitated with arrestins does not correlate with their ability to promote JNK3 phosphorylation (Figs. 1 and 2): the virtually inactive Arr2-3AX (AX) chimera brought down more ASK1 and JNK3 than fully active arrestin-3, whereas the Arr2N3C (A23) and Arr2-3BH (BH) chimeras, which showed ∼70% and none of arrestin-3 (A3) activity, respectively, co-immunoprecipitated virtually the same amounts of both kinases. A2, arrestin-2; A32, Arr3N2C; XB, Arr2-3XB; AB, Arr2-3AB (detailed chimera descriptions are in the legend to Fig. 1). IB, immunoblot.
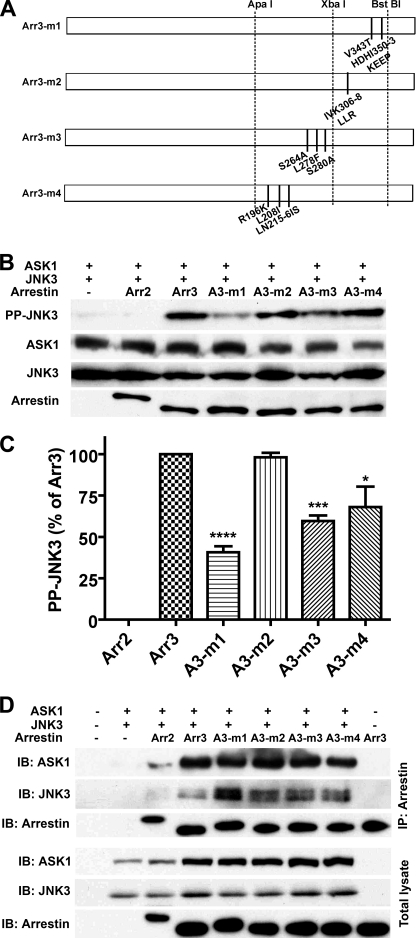
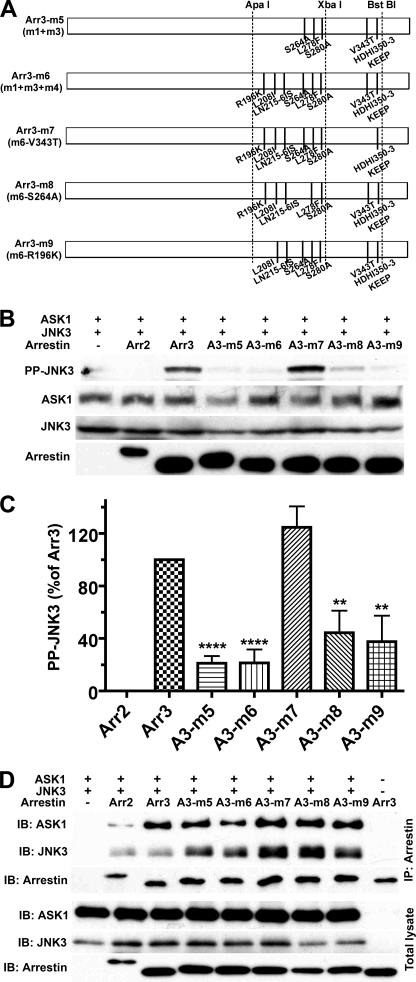
These data suggest that multiple elements of arrestin-3 are required for productive scaffolding of the ASK1-MKK4-JNK3 cascade and that the strength of the binding of individual kinases does not predict arrestin activity in this regard. Therefore, we changed strategy and replaced groups of arrestin-3 residues with their arrestin-2 homologs (Fig. 4A), focusing on the C-terminal domain identified as a key determinant of this function (Fig. 1). Of four mutations tested, I306L/V307L/K308R had no effect, whereas V343T + H350K/D351E/H352E/I353P (m1), S264A + L278F + S280A (m3), and R196K + L208I + L215I/N216S (m4) reduced the ability of arrestin-3 to promote JNK3 activation to varying degrees (Fig. 4, B and C). In this series, the strength of interaction also did not correlate with activity, e.g. the m1 mutant bound ASK1 just like arrestin-3 and bound JNK3 noticeably better (Fig. 4D) but showed only half of the ability to promote JNK3 phosphorylation (Fig. 4, B and C). Next, we combined effective substitutions, creating m5 (m1 + m3) and m6 (m1 + m3 + m4) mutants, and then eliminated individual substitutions V343T, S264A, and R196K from m6 (Fig. 5A). Both m5 and m6 mutations were significantly less effective than parental m1, m3, and m4 mutations (Fig. 5, B and C). Interestingly, the addition of m4 to the m1 + m3 combination (compare m5 and m6) had no further deleterious effect, and the restoration of an arginine in the RS motif homologous to RRS in rat arrestin (supplemental Fig. S1) by eliminating the R196K substitution (m9) did not improve the performance of the m6 mutant (Fig. 5). Elimination of the S264A substitution (m8) also had no significant effect on the m6 combination mutant, whereas elimination of the V343T substitution (m7) greatly improved its activity (Fig. 5, B and C). Similar to the other series (Figs. 3 and 4), the strength of the interactions with JNK3 and ASK1 did not correlate with activity. For example, both m7 and m8 mutants appeared to bind ASK1 similar to WT arrestin-3 and bound JNK3 even tighter (Fig. 5D), but their abilities to activate JNK3 were very different: m7 was indistinguishable from WT arrestin-3, whereas m8 was significantly impaired (Fig. 5).
FIGURE 4.
Replacement of selected residues of arrestin-3 with arrestin-2 homologs reduces its ability to promote JNK3 phosphorylation. A, schematic of the arrestin-3 (Arr3) mutants. The positions of the same restriction sites as in Fig. 1 are indicated. m1 is arrestin-3 with V343T + H350K/D351E/H352E/I353P (HDHI350–3KEEP). m2 is arrestin-3 with I306L/V307L/K308R (IVK306–8LLR). m3 is arrestin-3 with S264A + L278F + S280A. m4 is arrestin-3 with R196K + L208I + L215I/N216S (LN215–6IS). B, COS-7 cells expressing HA-ASK1, HA-JNK3, and the indicated arrestins with a C-terminal FLAG tag were lysed, and the amount of active phosphorylated JNK3 (PP-JNK3; upper blot) and the expression of ASK1, JNK3, and arrestins were determined by Western blotting. C, quantification of the level of JNK3 phosphorylation in cells expressing the indicated arrestins. Phosphorylated JNK3 bands from three to four independent experiments were quantified. The data were analyzed by ANOVA with arrestin as a main factor, followed by the Bonferroni-Dunn post-hoc test. The significance of the difference from fully active arrestin-3 is shown. *, p < 0.05; ***, p < 0.001; ****, p < 0.0001. Intensity is expressed as a percent of the difference between phosphorylated JNK3 in arrestin-3- and arrestin-2-expressing cells. D, COS-7 cells were transfected with the indicated proteins and lysed 48 h post-transfection. Arrestins were immunoprecipitated (IP) with anti-FLAG antibody, and the amount of arrestins and co-immunoprecipitated HA-ASK1 and HA-JNK3 was determined by Western blotting (upper three blots). The expression of these proteins was determined in cell lysates (lower three blots). Cells transfected with empty vector only (first lane), HA-ASK1 + HA-JNK3 only (second lane), or arrestin only (last lane) served as controls. The results of a representative experiment of three performed are shown. IB, immunoblot.
FIGURE 5.
Combinations of arrestin-3/arrestin-2 substitutions identify Val-343 as the key residue required for JNK3 phosphorylation. A, schematic of the arrestin-3 (Arr3) mutants. The positions of the same restriction sites as in Fig. 1 are indicated. m5 is m1 + m3 (arrestin-3 with V343T + H350K/D351E/H352E/I353P (HDHI350–3KEEP) + S264A + L278F + S280A). m6 is m1 + m3 + m4 (arrestin-3 with V343T + H350K/D351E/H352E/I353P + S264A + L278F + S280A + R196K + L208I + L215I/N216S (LN215–6IS)). m7 is m6 without the V343T mutation. m8 is m6 without the S264A mutation. m9 is m6 without the R196K mutation. B, COS-7 cells expressing HA-ASK1, HA-JNK3, and the indicated arrestins with a C-terminal FLAG tag were lysed, and the amount of active phosphorylated JNK3 (PP-JNK3; upper blot) and the expression of ASK1, JNK3, and arrestins were determined by Western blotting. C, quantification of the level of JNK3 phosphorylation in cells expressing the indicated arrestins. Phosphorylated JNK3 bands from three independent experiments were quantified. The data were analyzed by ANOVA with arrestin as a main factor, followed by the Bonferroni-Dunn post-hoc test. The significance of the difference from fully active arrestin-3 is shown. **, p < 0.01; ****, p < 0.0001. Band intensity is expressed as a percent of the difference between phosphorylated JNK3 in arrestin-3- and arrestin-2-expressing cells. D, COS-7 cells were transfected with the indicated proteins and lysed 48 h post-transfection. Arrestins were immunoprecipitated (IP) with anti-FLAG antibody, and the amount of arrestins and co-immunoprecipitated HA-ASK1 and HA-JNK3 was determined by Western blotting (upper three blots). The expression of these proteins was determined in cell lysates (lower three blots). Cells transfected with HA-ASK1 + HA-JNK3 only (first lane) or arrestin only (last lane) served as controls. The results of a representative experiment of three performed are shown. IB, immunoblot.
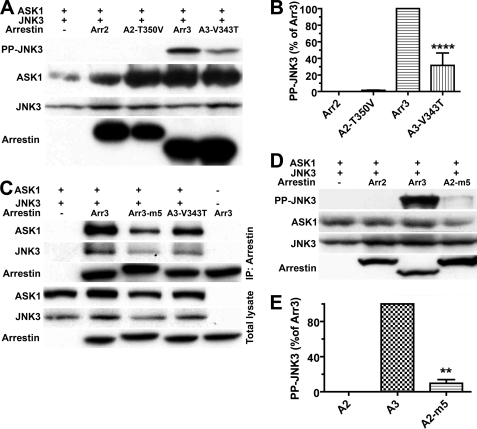
These data identify Val-343 as one of the major players in JNK3 activation, as reversal of the V343T mutation essentially restored the ability of the m6 multiple mutant to activate JNK3 (compare m6 and m7 in Fig. 5). Therefore, we tested the effect of the single V343T mutation and found that it reduced arrestin-3 ability to facilitate JNK3 activation by ∼70% (Fig. 6, A and B). Interestingly, the reverse mutation T350V in arrestin-2 did not appreciably increase its activity (Fig. 6, A and B). This is consistent with the idea that multiple arrestin elements are necessary for productive scaffolding of the ASK1-MKK4-JNK3 cascade, so the elimination of a single one can destroy the activity of arrestin-3, whereas the introduction of one cannot create arrestin-2 with the ability to activate JNK3. To further explore this, we constructed arrestin-2 with multiple arrestin-3-like residues: A263S + F277L + A279S + T350V + K357H/E358D/E359H/P360I (A2-m5 in Fig. 6). Although the activity of this mutant increased relative to WT arrestin-2, it still reached only ∼10% of that of arrestin-3 (Fig. 6, D and E). This is hardly surprising, considering that the Arr2-3AB chimera, containing 180 arrestin-3 residues (including all introduced in A2-m5), showed only ∼20% of arrestin-3 activity, and even the Arr2N3C chimera, with the whole arrestin-3 C-terminal domain, was ∼30% less active than WT arrestin-3 (Figs. 1 and 2).
FIGURE 6.
A single substitution significantly reduces the activity of arrestin-3, but even multiple substitutions do not bring the activity of arrestin-2 to the level of arrestin-3. A and D, COS-7 cells expressing HA-ASK1, HA-JNK3, and the indicated arrestins with a C-terminal FLAG tag were lysed, and the amount of active phosphorylated JNK3 (PP-JNK3; upper blot) and the expression of ASK1, JNK3, and arrestins were determined by Western blotting. B and E, quantification of the level of JNK3 phosphorylation in cells expressing the indicated arrestins. Phosphorylated JNK3 bands from three independent experiments were quantified. The data were analyzed by ANOVA with arrestin as a main factor, followed by the Bonferroni-Dunn post-hoc test. The significance of the difference from fully active arrestin-3 (Arr3, A3; B) or from inactive arrestin-2 (A2; E) is shown; **, p < 0.01; ****, p < 0.0001. Band intensity is expressed as a percent of the difference between phosphorylated JNK3 in arrestin-3- and arrestin-2-expressing cells. C, COS-7 cells were transfected with the indicated proteins and lysed 48 h post-transfection. Arrestins were immunoprecipitated (IP) with anti-FLAG antibody, and the amount of arrestins and co-immunoprecipitated HA-ASK1 and HA-JNK3 was determined by Western blotting (upper three blots). The expression of these proteins was determined in cell lysates (lower three blots). Cells transfected with HA-ASK1 + HA-JNK3 only (first lane) or arrestin only (last lane) served as controls. The results of a representative experiment of three performed are shown.
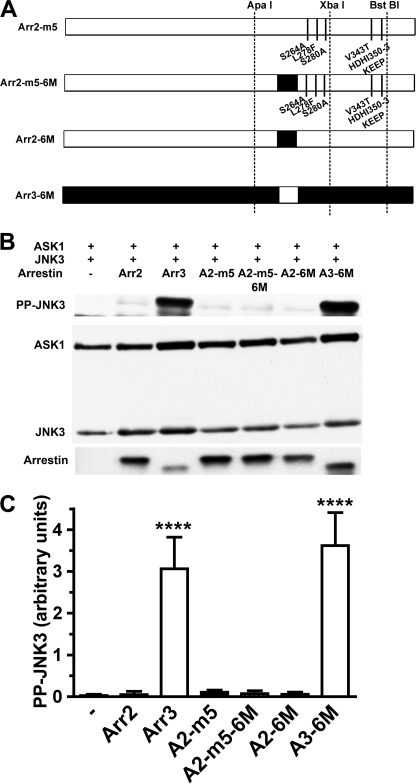
Another known peculiarity of arrestin-3 is significantly lower selectivity for active phosphorylated receptors than other subtypes (30). The recently solved crystal structure of arrestin-3 revealed that, in contrast to arrestin-1 (28), arrestin-2 (19, 20), and arrestin-4 (29), part of β-strand XVI in the C-terminal domain is distorted and does not form a contiguous β-sheet with neighboring β-strands (21). Follow-up mutagenesis showed that swapping this element between arrestin-3 and arrestin-2 partially reverses their receptor-binding characteristics (21). Because key determinants of the arrestin-3 ability to promote JNK3 activation are localized in the C-terminal domain (Fig. 1), we introduced six mutations into β-strand XVI in arrestin-2 and arrestin-3 (I233V + N245S + M255Q + E256V + A258Q + T261Q and V234I + S246N + Q256M + V257E + Q259A + Q262T, respectively). These sets of mutations converted β-strand XVI from arrestin-2-like into arrestin-3-like and vice versa. They were shown previously to change the receptor-binding profile of both non-visual arrestins (21). In arrestin-2, this set of mutations was also introduced on the background of the A2-m5 mutant (Fig. 7). We found that the exchange of this element did not affect the ability of either arrestin to facilitate JNK3 activation: arrestin-2-based A2-6M and A2-m5-6M both remained ineffective, whereas arrestin-3-based A3-6M promoted JNK3 phosphorylation to the same extent as WT arrestin-3 (Fig. 7). Thus, receptor binding and JNK3 activation are independent functions of arrestin proteins that can be individually manipulated by appropriate mutations.
FIGURE 7.
JNK3 activation is not affected by mutations that change receptor-binding characteristics of arrestins. A, schematic of the arrestin-2 (Arr2) and arrestin-3 (Arr3) mutants. The positions of the same restriction sites as in Fig. 1 are indicated. m5 is arrestin-2 with A263S + F277L + A279S + T350V + K357H/E358D/E359H/P360I (KEEP357–360HDHI). m5-6M is arrestin-2 m5 with additional mutations I233V, N245S, M255Q, E256V, A258Q, and T261Q, converting β-strand XVI into arrestin-3-like (shown as a black rectangle). A2-6M is arrestin-2 with I233V + N245S + M255Q + E256V + A258Q + T261Q. A3-6M is arrestin-3 with V234I + S246N + Q256M + V257E + Q259A + Q262T with arrestin-2-like β-strand XVI (shown as a white rectangle). B, COS-7 cells expressing HA-ASK1, HA-JNK3, and the indicated arrestins with a C-terminal FLAG tag were lysed, and the amount of active phosphorylated JNK3 (PP-JNK3; upper blot) and the expression of ASK1, JNK3, and arrestins were determined by Western blotting. C, quantification of the level of JNK3 phosphorylation in cells expressing the indicated arrestins. Phosphorylated JNK3 bands from four independent experiments were quantified. The data were analyzed by ANOVA with arrestin as a main factor, followed by Dunnett's post-hoc test. The significance of the difference from inactive WT arrestin-2 is shown. ****, p < 0.0001. Note that none of the arrestin-2-based proteins was different from WT arrestin-2.
DISCUSSION
At the protein level, bovine arrestin-2 and arrestin-3 share 78% identity and 88% similarity (23, 32). Rat proteins show virtually the same levels of homology (33), yet in both species, arrestin-3 promotes JNK3 activation robustly, whereas arrestin-2 does not (15, 16). The first reports suggested that arrestin-2 does not bind JNK3 (34) because it does not have a MAPK-docking motif (15), which would have provided a simple explanation for this functional difference. However, subsequent studies using different methods detected comparable interaction of the two non-visual arrestins with JNK3 (8, 9, 14) and both upstream kinases MKK4 and ASK1 (16). These data suggested that the difference is more subtle and led to the idea that the two arrestins hold the kinases in a distinct relative orientation, which yields a productive complex in the case of arrestin-3 and a silent one in the case of arrestin-2 (16). This model does not exclude the possibility that a few arrestin-3 residues are largely responsible for the unique ability of arrestin-3 to promote JNK3 activation.
Crystal structures of the two proteins also reveal high similarity in the core arrestin fold (19–21), which ensures the viability and functionality of chimeras (10, 13, 26, 30, 31). Therefore, to address this issue, we performed extensive swapping of different elements (Figs. 1–3) and found that the most important determinants of arrestin-3 ability to activate JNK3 are localized in the C-terminal domain. However, even Arr2N3C is less effective in this regard than WT arrestin-3 (Fig. 1), demonstrating that some N-terminal domain residues lost in this chimera are also important for JNK3 activation. These data are consistent with the earlier finding that each of the kinases in the ASK1-MKK4-JNK3 cascade interacts with both arrestin domains (16, 35). Next, to identify key C-terminal domain residues responsible for the unique ability of arrestin-3 to promote JNK3 activation, we replaced arrestin-3-specific residues with the corresponding arrestin-2 homologs (Figs. 4 and 5). Analysis of mutants' ability to activate JNK3 identified Val-343 as the key player, with Leu-278, Ser-280, His-350, Asp-351, His-352, and Ile-353 playing supporting roles (Fig. 5). However, even the introduction of eight key arrestin-3 residues into arrestin-2 (A2-m5 in Fig. 6) does not confer an ability to activate JNK3 comparable with WT arrestin-3 (Fig. 6). Moreover, the addition of the “loose” arrestin-3-derived β-strand, alone (A2-6M) or in the context of A2-m5 (A2-m5-6M), does not create the ability to promote JNK3 phosphorylation in arrestin-2 (Fig. 7). Similarly, the introduction of the homologous element of arrestin-2 into arrestin-3 (A3-6M) does not change its efficiency in JNK3 activation (Fig. 7). These data clearly show that multiple arrestin-3 elements acting in concert are responsible for JNK3 activation, so this function can be easily weakened or destroyed by mutations (Figs. 4–6) but cannot be recreated in arrestin-2 by replacing just a few residues (Figs. 6 and 7).
Comparison of the interactions of individual kinases in the ASK1-MKK4-JNK3 signaling module with arrestin-2 and arrestin-3 suggested that the binding per se does not predict arrestin effectiveness as a JNK3 activator (9, 16, 35). Comparison of the binding of ASK1 and JNK3 to multiple chimeras and mutants with their ability to promote JNK3 activation (Figs. 1–6) strongly supports this conclusion. Several constructed arrestins show remarkable dissociation between these functions. For example, the Arr2-3AX chimera binds ASK1 and JNK3 much better than WT arrestin-3 but does not facilitate JNK3 phosphorylation at all (Fig. 1). This combination of characteristics makes it a promising candidate for dominant-negative action, which is likely to suppress pro-apoptotic signaling and enhance cell survival. This idea needs to be tested experimentally.
The localization of GPCR- and microtubule-binding sites on the concave sides of the two arrestin domains (22, 26, 36–38) and the sites for the majority of non-receptor partners on the opposite convex surface suggested that independent manipulation of different arrestin functions by targeted mutations can be exploited to construct “designer” arrestins that channel the signaling from specific receptors to the pathways of interest for therapeutic purposes (18). Our data provide further support for this idea, demonstrating that GPCR binding, binding of ASK1 and JNK3, and scaffolding of the signaling module that activates JNK3 in the cell can be separated by introducing appropriate substitutions. Several of the mutants and chimeras characterized here are in fact signaling-biased arrestins. These results pave the way to experimental testing of their ability to promote or suppress cell survival.
Supplementary Material
Acknowledgments
We are grateful to Drs. Louis Luttrell, Robert J. Lefkowitz, Jia Le Dai, and Haian Fu (Emory University) for the GFP-JNK3, HA-JNK3, HA-MKK4, and HA-ASK1 expression constructs, respectively.
This work was supported in part by National Institutes of Health Grants GM077561, GM081756, and EY011500 (to V. V. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
We use systematic names of arrestin proteins: arrestin-1 (historic names S-antigen, 48-kDa protein, visual or rod arrestin), arrestin-2 (β-arrestin or β-arrestin-1), arrestin-3 (β-arrestin-2 or hTHY-ARRX), and arrestin-4 (cone or X-arrestin; for unclear reasons, its gene is called “arrestin-3” in the HUGO Database).
- GPCR
- G protein-coupled receptor
- ANOVA
- analysis of variance.
REFERENCES
- 1. Gurevich V. V., Gurevich E. V. (2006) Pharmacol. Ther. 110, 465–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wilden U., Hall S. W., Kühn H. (1986) Proc. Natl. Acad. Sci. U.S.A. 83, 1174–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Krupnick J. G., Gurevich V. V., Benovic J. L. (1997) J. Biol. Chem. 272, 18125–18131 [DOI] [PubMed] [Google Scholar]
- 4. Goodman O. B., Jr., Krupnick J. G., Santini F., Gurevich V. V., Penn R. B., Gagnon A. W., Keen J. H., Benovic J. L. (1996) Nature 383, 447–450 [DOI] [PubMed] [Google Scholar]
- 5. Laporte S. A., Oakley R. H., Zhang J., Holt J. A., Ferguson S. S., Caron M. G., Barak L. S. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3712–3717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. DeWire S. M., Ahn S., Lefkowitz R. J., Shenoy S. K. (2007) Annu. Rev. Physiol. 69, 483–510 [DOI] [PubMed] [Google Scholar]
- 7. Kang J., Shi Y., Xiang B., Qu B., Su W., Zhu M., Zhang M., Bao G., Wang F., Zhang X., Yang R., Fan F., Chen X., Pei G., Ma L. (2005) Cell 123, 833–847 [DOI] [PubMed] [Google Scholar]
- 8. Wang P., Wu Y., Ge X., Ma L., Pei G. (2003) J. Biol. Chem. 278, 11648–11653 [DOI] [PubMed] [Google Scholar]
- 9. Song X., Raman D., Gurevich E. V., Vishnivetskiy S. A., Gurevich V. V. (2006) J. Biol. Chem. 281, 21491–21499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ahmed M. R., Zhan X., Song X., Kook S., Gurevich V. V., Gurevich E. V. (2011) Biochemistry 50, 3749–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wu N., Hanson S. M., Francis D. J., Vishnivetskiy S. A., Thibonnier M., Klug C. S., Shoham M., Gurevich V. V. (2006) J. Mol. Biol. 364, 955–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nair K. S., Hanson S. M., Mendez A., Gurevich E. V., Kennedy M. J., Shestopalov V. I., Vishnivetskiy S. A., Chen J., Hurley J. B., Gurevich V. V., Slepak V. Z. (2005) Neuron 46, 555–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hanson S. M., Cleghorn W. M., Francis D. J., Vishnivetskiy S. A., Raman D., Song X., Nair K. S., Slepak V. Z., Klug C. S., Gurevich V. V. (2007) J. Mol. Biol. 368, 375–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scott M. G., Le Rouzic E., Périanin A., Pierotti V., Enslen H., Benichou S., Marullo S., Benmerah A. (2002) J. Biol. Chem. 277, 37693–37701 [DOI] [PubMed] [Google Scholar]
- 15. Miller W. E., McDonald P. H., Cai S. F., Field M. E., Davis R. J., Lefkowitz R. J. (2001) J. Biol. Chem. 276, 27770–27777 [DOI] [PubMed] [Google Scholar]
- 16. Song X., Coffa S., Fu H., Gurevich V. V. (2009) J. Biol. Chem. 284, 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shankar H., Michal A., Kern R. C., Kang D. S., Gurevich V. V., Benovic J. L. (2010) J. Biol. Chem. 285, 8316–8329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gurevich V. V., Gurevich E. V. (2010) Expert Rev. Mol. Med. 12, e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Han M., Gurevich V. V., Vishnivetskiy S. A., Sigler P. B., Schubert C. (2001) Structure 9, 869–880 [DOI] [PubMed] [Google Scholar]
- 20. Milano S. K., Pace H. C., Kim Y. M., Brenner C., Benovic J. L. (2002) Biochemistry 41, 3321–3328 [DOI] [PubMed] [Google Scholar]
- 21. Zhan X., Gimenez L. E., Gurevich V. V., Spiller B. W. (2011) J. Mol. Biol. 406, 467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vishnivetskiy S. A., Gimenez L. E., Francis D. J., Hanson S. M., Hubbell W. L., Klug C. S., Gurevich V. V. (2011) J. Biol. Chem. 286, 24288–24299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sterne-Marr R., Gurevich V. V., Goldsmith P., Bodine R. C., Sanders C., Donoso L. A., Benovic J. L. (1993) J. Biol. Chem. 268, 15640–15648 [PubMed] [Google Scholar]
- 24. Gurevich E. V., Benovic J. L., Gurevich V. V. (2002) Neuroscience 109, 421–436 [DOI] [PubMed] [Google Scholar]
- 25. Gurevich E. V., Benovic J. L., Gurevich V. V. (2004) J. Neurochem. 91, 1404–1416 [DOI] [PubMed] [Google Scholar]
- 26. Vishnivetskiy S. A., Hosey M. M., Benovic J. L., Gurevich V. V. (2004) J. Biol. Chem. 279, 1262–1268 [DOI] [PubMed] [Google Scholar]
- 27. Gurevich E. V., Gurevich V. V. (2006) Genome Biol. 7, 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hirsch J. A., Schubert C., Gurevich V. V., Sigler P. B. (1999) Cell 97, 257–269 [DOI] [PubMed] [Google Scholar]
- 29. Sutton R. B., Vishnivetskiy S. A., Robert J., Hanson S. M., Raman D., Knox B. E., Kono M., Navarro J., Gurevich V. V. (2005) J. Mol. Biol. 354, 1069–1080 [DOI] [PubMed] [Google Scholar]
- 30. Gurevich V. V., Dion S. B., Onorato J. J., Ptasienski J., Kim C. M., Sterne-Marr R., Hosey M. M., Benovic J. L. (1995) J. Biol. Chem. 270, 720–731 [DOI] [PubMed] [Google Scholar]
- 31. Gurevich V. V., Richardson R. M., Kim C. M., Hosey M. M., Benovic J. L. (1993) J. Biol. Chem. 268, 16879–16882 [PubMed] [Google Scholar]
- 32. Lohse M. J., Benovic J. L., Codina J., Caron M. G., Lefkowitz R. J. (1990) Science 248, 1547–1550 [DOI] [PubMed] [Google Scholar]
- 33. Attramadal H., Arriza J. L., Aoki C., Dawson T. M., Codina J., Kwatra M. M., Snyder S. H., Caron M. G., Lefkowitz R. J. (1992) J. Biol. Chem. 267, 17882–17890 [PubMed] [Google Scholar]
- 34. McDonald P. H., Chow C. W., Miller W. E., Laporte S. A., Field M. E., Lin F. T., Davis R. J., Lefkowitz R. J. (2000) Science 290, 1574–1577 [DOI] [PubMed] [Google Scholar]
- 35. Song X., Gurevich E. V., Gurevich V. V. (2007) J. Neurochem. 103, 1053–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hanson S. M., Francis D. J., Vishnivetskiy S. A., Klug C. S., Gurevich V. V. (2006) J. Biol. Chem. 281, 9765–9772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hanson S. M., Gurevich V. V. (2006) J. Biol. Chem. 281, 3458–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hanson S. M., Francis D. J., Vishnivetskiy S. A., Kolobova E. A., Hubbell W. L., Klug C. S., Gurevich V. V. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 4900–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.