Abstract
Collagen IX containing the N-terminal noncollagenous domain 4 (NC4) is unique to cartilage and a member of the family of fibril-associated collagens with both collagenous and noncollagenous domains. Collagen IX is located at the surface of fibrils formed by collagen II and a minor proportion of collagen XI, playing roles in tissue stability and integrity. The NC4 domain projects out from the fibril surface and provides sites for interaction with other matrix components such as cartilage oligomeric matrix protein, matrilins, fibromodulin, and osteoadherin. Fragmentation of collagen IX and loss of the NC4 domain are early events in cartilage degradation in joint diseases that precedes major damage of collagen II fibrils. Our results demonstrate that NC4 can function as a novel inhibitor of the complement system able to bind C4, C3, and C9 and to directly inhibit C9 polymerization and assembly of the lytic membrane attack complex. NC4 also binds the complement inhibitors C4b-binding protein and factor H and enhances their cofactor activity in degradation of activated complement components C4b and C3b. NC4 interactions with fibromodulin and osteoadherin inhibited binding to C1q and complement activation by these proteins. Taken together, our results suggest that collagen IX and its interactions with matrix components are important parts of a machinery that protects the cartilage from complement activation and chronic inflammation seen in diseases like rheumatoid arthritis.
Keywords: Collagen, Complement, Extracellular Matrix Proteins, Inflammation, Protein-Protein Interactions, C4b-binding Protein, Factor H, Membrane Attack Complex
Introduction
The complement system represents a central component of the innate immune system and is activated via three different pathways depending on the initiating agent (1). The classical pathway was first found to be triggered by antibodies bound to their targets, whereas the lectin pathway is initiated by specific carbohydrates on bacterial surfaces. In contrast, the alternative pathway is started by autoactivation of the unstable complement component C3 and its subsequent deposition on activating pathogen surfaces. Triggered enzymatic complement cascades result in the assembly of C3- and C5-convertases, which leads to opsonization of activating surfaces with C3b, release of pro-inflammatory anaphylatoxins C3a and C5a, and finally formation of a lytic membrane attack complex (MAC),3 which disrupts membrane integrity (1).
To prevent complement-mediated tissue injury, a fine balance between activation and inhibition of the system is established by several soluble as well as membrane-bound inhibitors (2). C4b-binding protein (C4BP) is the major soluble inhibitor of the classical and lectin pathways, whereas factor H (FH) inhibits the alternative route. Most inhibitors act in two ways as follows: by accelerating the decay of complement convertases or by promoting enzymatic cleavage of the activated complement factors C3b and C4b by a serine proteinase, factor I (FI). Any shift of the balance might provoke development of inflammatory diseases, such as rheumatoid arthritis (RA), glomerulonephritis, multiple sclerosis, Alzheimer disease, and many more (3, 4). There is also an increasing understanding that osteoarthritis (OA) has a significant inflammatory component.
OA and RA involve damage to the articular cartilage in the affected joint and often influence many different joints in the body. Joint replacement in these diseases has been found to attenuate inflammation, which implies that cartilage-derived components might have a role in the inflammation. A large body of evidence shows that complement contributes to the development of RA in man and experimental animals (5). It has been shown that fibromodulin (FM) and osteoadherin (OSAD), constituents of the extracellular matrix, activate complement via the classical pathway and may enhance inflammation in joint disease (6, 7). Fragments of these cartilage components are released by proteases during cartilage destruction to come in contact with complement in synovial fluid. The involvement of these molecules in inflammation may be applied to develop novel approaches for early diagnosis and treatment of arthritic diseases.
Cartilage is composed of a low number of cells surrounded by a dense matrix of the aggrecan proteoglycan bound to hyaluronan and is entangled in a network of collagen fibrils. The collagen fibrils are built by collagens type II and XI with collagen IX attached at their surface. The collagen IX molecule is a heterotrimer (composed of polypeptide chains α1, α2, and α3) and comprises three collagenous domains (COL1–COL3) flanked by four short noncollagenous (NC1–NC4) domains (8). The NC4 domain is larger than the other three NC domains and is formed only by the α1-chain (Fig. 1A). Collagen IX molecules cover the surface of collagen II/XI fibrils in a periodic fashion (9) and act as molecular bridges connecting collagen fibrils with other cartilage matrix components to ensure stiffness and tensile strength of the cartilage (10). The COL3 and NC4 domains project out from the fibril surface, thus providing sites for interactions with matrix proteins like cartilage oligomeric matrix protein (COMP), FM, and others, although matrilin-3 can interact with neighboring collagen domains (11, 12). These interactions are crucial for tissue integrity and stability. The NC4 domain appears to be particularly sensitive to enzymatic attack in cartilage breakdown (13). The process appears to be mediated by matrix metalloproteinase 13 (MMP-13), and one of the fragments released corresponds to almost the entire NC4 domain (13). It has been shown that the loss of the NC4 domain occurs both in cartilage treated with interleukin 1 (IL-1) and in RA4 and precedes major release of collagen II (13, 14). Synovial fluid from patients with arthritic disease contains fragments of collagen IX (15).
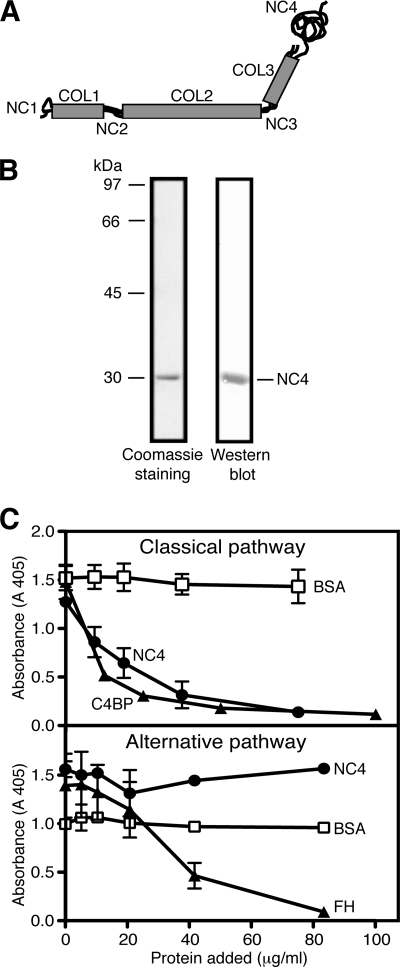
FIGURE 1.
Structure of collagen IX and inhibition of complement-mediated lysis of erythrocytes by NC4. A, schematic illustration of the domain organization of the collagen IX molecule, composed of three polypeptide chains (α1, α2, and α3) and three collagenous (COL1–COL3) domains, separated by four noncollagenous (NC1–NC4) regions. The NC4 domain of collagen IX is larger and consists only of the α1-chain. B, Coomassie staining of SDS-PAGE and Western blot of purified NC4 domain. The NC4 domain migrates at about 30 kDa. C, inhibition of classical and alternative pathway-mediated lysis of erythrocytes by NC4. Sheep or rabbit erythrocytes were used for the classical or alternative pathway-mediated lysis, respectively. C4BP and FH were used as positive controls for the classical and alternative pathways, respectively, and BSA was used as negative control. The graphs represent mean values from three independent experiments performed in duplicate ± S.D., except for the alternative pathway (two experiments in duplicate).
We aimed to investigate if the NC4 domain is able to modulate complement, and we found that it can act as a local inhibitor. NC4 binds both C4BP and FH and enhances their cofactor activities in degradation of C4b and C3b. Furthermore, NC4 attenuates complement directly due to inhibition of C9 polymerization and MAC formation. Moreover, we show that NC4 interactions with other key matrix components have an impact on complement regulation. Taken together, our results suggest that collagen IX might be a component of a mechanism protecting the cartilage from joint-specific autoimmune attack, complement activation, and chronic inflammation seen in diseases such as RA and OA.
EXPERIMENTAL PROCEDURES
Proteins
The NC4 protein was expressed and purified as described previously (16). Briefly, the expression vector pAM 104-2 containing NC4 was transformed into Escherichia coli strain BL21 (Stratagene), and a single colony was grown in LB media containing kanamycin sulfate (50 μg/ml, Sigma). After induction of protein expression with 1 mm isopropyl 1-thio-β-d-galactopyranoside (Saveen Werner), bacterial cells were lysed, and the clarified extract was applied onto a nickel-nitrilotriacetic acid column (GE Healthcare). Protein was eluted using buffer containing 0.5 m imidazole, and NC4-containing fractions were analyzed with SDS-PAGE followed by Western blotting and staining with Coomassie. The fractions containing NC4 were pooled and dialyzed against phosphate-buffered saline (PBS).
Pooled normal human serum was prepared from the blood of 11 healthy volunteers (17), and aliquots were heat-inactivated by incubation at 56 °C for 30 min. C2- and C6-depleted sera, as well as control human serum, were purchased from Quidel. C4 was purified as described previously (18). C3, C3b, and C4b were purchased from Complement Technology. C4met and C3met, which correspond functionally to C4b and C3b, respectively, were prepared by treatment with methylamine as described previously (19). C4b and C3b were labeled with 125I using the chloramine-T method (20).
C9 component was purified as described previously (21) with some modifications. DEAE-Sephacel was used instead of DEAE-Sephadex, and hydroxyapatite chromatography was followed by binding of proteins to an octyl-Sepharose column and elution with a linear gradient of 1.5–0 m NaCl in 50 mm Tris, pH 8.5. C9-containing fractions were identified by Western blotting and silver staining and stored at −80 °C.
FH was isolated from human plasma (22), and recombinant FI was expressed and purified from eukaryotic cells (23). C4BP-PS complex was purified from plasma (24). To construct the Q109A/D110N/R111Q/G112S and ΔVal-108/Gln-109/Asp-110 mutants, the following primers were used in the site-directed mutagenesis reactions (QuickChange, Stratagene): 5′-ACC ACT AGT CGT TGT GAA AGA GGA GTT GGC TGG AGT-3′and 5′-CGT TGT GAA GTC GCA AAT CAA TCA GTT GGC TGG AGT, respectively. Successful mutagenesis was confirmed by sequencing, and the DNA constructs were transfected into 293 HEK cells using Lipofectamine (Invitrogen). After selection of stable clones with G418, media were collected, and C4BP mutants were purified using affinity chromatography with monoclonal antibody (mAb 104) as described in detail previously (25). Recombinant wild type C4BP, the R39Q/R64Q/R66Q mutant (26), and mutant C4BP proteins lacking single complement control protein (CCP) domains (25), as well as those in which two alanine residues were introduced in the linker regions between CCP domains (25), were expressed and purified in the same way. The C4BP core fragment was prepared as described previously (27) and consists of C-terminal extensions of α-chains together with CCP8 and a small fragment of CCP7. FM (28), OSAD (29), and COMP (30) were prepared as described.
Western Blot Analysis
NC4 or C9 were subjected to 7.5% linear SDS-PAGE and 2.5–10% gradient SDS-PAGE, respectively, and transferred onto a PVDF membrane. The membrane was blocked with 50 mm Tris-HCl, 150 mm NaCl, 0.1% Tween 20, and 30% fish gelatin (Nordic), pH 8.0. NC4 was detected with a rabbit anti-NC4 polyclonal antibody (raised in-house), followed by horseradish peroxidase (HRP)-conjugated swine anti-rabbit secondary antibody (Dako) and visualized with a 3,3′-diaminobenzidine tetrahydrochloride colorimetric substrate system (Sigma). C9 was detected with a goat anti-C9 antibody (Complement Technology) followed by HRP-conjugated mouse anti-goat secondary antibody (Dako) and visualized with the 3,3′-diaminobenzidine tetrahydrochloride system.
Hemolytic Assay
Sheep erythrocytes were used for assays of the classical pathway of complement, although rabbit erythrocytes were used for the alternative pathway.
Sheep erythrocytes (SVA, Uppsala, Sweden) were washed three times with ice-cold DGVB++ buffer (2.5 mm veronal buffer, pH 7.4, 70 mm NaCl, 1 mm MgCl2, 0.15 mm CaCl2, 140 mm glucose, and 0.1% gelatin) and incubated with a complement-fixing antibody (Amboceptor, Behringwerke, Germany) for 20 min at 37 °C. Erythrocytes were then washed twice in ice-cold DGVB++ buffer and incubated in a 96-well plate together with 0.2% serum (diluted in DGVB++ buffer) and increasing concentrations of test protein (in DGVB++ buffer) for 1 h at 37 °C.
Rabbit erythrocytes were washed twice with the Mg2+EGTA buffer (2.5 mm veronal buffer, pH 7.4, 70 mm NaCl, 7 mm MgCl2, 10 mm EGTA, 140 mm glucose, and 0.1% gelatin) and incubated with 2% serum (in Mg2+EGTA) and increasing concentrations of test protein (in Mg2+EGTA) for 1 h at 37 °C.
Plates were centrifuged after the incubation, and the lysis of erythrocytes was measured at 405 nm using a UV-visible spectrophotometer (Cary 50 Bio, Varian).
Deposition of Complement Components from Serum
Incubation steps in the assay were performed in a total volume of 50 μl for 1 h at room temperature unless stated otherwise. Microtiter plates (Maxisorp, Nunc) were coated overnight at 4 °C with NC4 (5 μg/ml), aggregated human IgG (2.5 μg/ml, Immuno), for the classical pathway, mannan (100 μg/ml, Sigma) or acetylated BSA (both for the lectin pathway), zymosan (20 μg/ml, (Sigma), for the alternative pathway), or 1% BSA (as negative control) diluted in phosphate-buffered saline (PBS). The plates were washed with 50 mm Tris-HCl, 150 mm NaCl, 2 mm CaCl2, and 0.1% Tween 20 between each step. Plates were blocked for 2 h with 1% BSA in PBS and incubated with dilutions of human serum in GVB++ buffer (5 mm veronal buffer, pH 7.35, 144 mm NaCl, 1 mm MgCl2, 0.15 mm CaCl2 and 0.1% gelatin). To test if NC4 would inhibit deposition of complement factors initiated by aggregated IgG, NC4 at increasing concentrations was preincubated for 15 min at room temperature with serum diluted in GVB++ (0.2% serum for C4b deposition and 0.3% serum for C3b deposition). Plates were incubated at 37 °C for 20 min (for C4b and C3b detection) or 45 min (for C1q, C9, MBL, and ficolin detection). Complement components were detected using specific polyclonal antibodies (Dako; R&D Systems for MBL detection; anti-ficolin-2 and anti-ficolin-3 kindly provided by P. Garred, University of Copenhagen, Copenhagen, Denmark) followed by incubation with HRP-conjugated antibodies (Dako). The plates were developed with o-phenylenediamine substrate (Dako) and H2O2, and the absorbance at 490 nm was measured.
For FH and C4BP binding from serum, plates were incubated with NC4 (5 μg/ml), C3met (5 μg/ml, positive control for FH binding), C4met (5 μg/ml, positive control for C4BP binding), and 1% BSA (negative control). Plates were incubated as described above, and binding was detected after 1 h of incubation with heat-inactivated serum using anti-C4BP (rabbit anti-C4BP antibody, raised in-house) and anti-FH (sheep anti-FH, Abcam) antibodies, followed by HRP-conjugated secondary antibodies (Dako).
Inhibition of C9 Polymerization
Purified human C9 (0.8 μm, corresponding to 54 μg/ml) was incubated with or without NC4 (at concentrations of 0–2.5 μm, corresponding to 0–75 μg/ml) in 20 mm Tris buffer, pH 7.2, overnight at 37 °C. The samples were mixed with reducing sample buffer, incubated at 95 °C for 5 min, run on 2.5–10% gradient SDS-PAGE, and analyzed by Western blotting as described above.
For inhibition of C9 polymerization on cells, sheep erythrocytes were sensitized with complement-fixing antibody as described in the hemolytic assay. NC4 (200 μg/ml) and α1-antitrypsin (300 μg/ml, negative control) were preincubated with 0.2% human serum for 30 min at 37 °C and incubated with the sensitized erythrocytes for 1 h at 37 °C. The cells were washed twice with TBS buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl) and sedimented for 20 min at 5000 rpm at 4 °C, followed by washing with 0.5 mm phosphate buffer, pH 8.0 (sedimentation for 20 min at 15,000 rpm at 4 °C). Erythrocyte membranes were dissolved in 30 μl of reducing sample buffer, heated for 5 min at 95 °C, and analyzed by 2.5–10% gradient SDS-PAGE followed by Western blotting for C9.
Direct Binding Assays
For C4, C4b, C3, and C3b binding, microtiter plates were coated with NC4 (5 μg/ml), FH (5 μg/ml, positive control for C3b binding), C4BP (5 μg/ml, positive control for C4b binding), and 1% BSA (as negative control) diluted in PBS. Plates were blocked as described above, and increasing concentrations of C4, C4met, C3, and C3met in binding buffer (50 mm Hepes, 100 mm NaCl, 2 mm CaCl2, 50 μg/ml BSA, pH 7.4) were added to the wells and incubated for 2 h at room temperature. The plates were then incubated with polyclonal antibodies against C4c and C3d (Dako) followed by incubation with HRP-conjugated secondary antibodies and developed as described for the deposition assay.
For direct binding of purified FH and C4BP, plates were coated with NC4 (5 μg/ml), C3met (5 μg/ml), C4met (5 μg/ml), and 1% BSA and incubated for 2 h at 37 °C. Increasing concentrations of purified FH and C4BP in binding buffer were added to the wells and allowed to bind for 2 h at room temperature. Detection of binding was performed using anti-FH and anti-C4BP antibodies followed by secondary HRP-conjugated antibodies. To assess the effect of Ca2+ and other divalent ions on the interaction between NC4 and complement components, the binding buffer was supplemented with 5 mm EDTA. For investigations of the effect of ionic strength on NC4-C4BP and NC4-FH interactions, the binding buffer was supplemented with NaCl to concentrations ranging from 150 to 500 mm.
To assess the binding of NC4 to C4BP-PS, recC4BP, its recombinant mutants, and C4BP core fragment, 40 nm of each C4BP variant were incubated with NC4 (5 μg/ml) immobilized on a plate. Bound C4BP was detected with an anti-C4BP antibody, followed by a goat anti-rabbit HRP conjugate (Dako).
For binding of NC4 to COMP, FM, and OSAD, microtiter plates were coated with COMP (5 μg/ml), FM (10 μg/ml), OSAD (5 μg/ml), and BSA (1%) overnight at 4 °C. Increasing concentrations of NC4 in binding buffer were added. After incubation for 1 h at room temperature, NC4 binding was detected using a rabbit polyclonal antibody against NC4, followed by HRP-conjugated secondary antibody.
Binding of C1q and C4b from Serum to COMP, FM, and OSAD in the Presence of NC4
Microtiter plates were coated with COMP (5 μg/ml), OSAD (5 μg/ml), FM (10 μg/ml), and aggregated IgG or 1% BSA overnight at 4 °C. After blocking, plates were incubated for 1 h at room temperature with 1% serum in GVB++ (for C1q and C4b binding to FM and OSAD) and 10% heat-inactivated serum in GVB++ (for C1q binding to COMP) and increasing NC4 concentrations. C1q and C4b binding was detected using polyclonal antibodies to the respective protein (Dako), followed by incubation with HRP-conjugated secondary antibodies (Dako).
C4b and C3b Degradation Assay
For measuring C4b degradation, FI (20 μg/ml) was mixed with C4met (50 μg/ml) and trace amounts of 125I-labeled C4b and NC4 at increasing concentrations. C4BP (100 μg/ml) was used as cofactor in the positive control, and FI was omitted in the negative control. The C3b degradation assay was performed in a similar way except that FI was mixed with 150 μg/ml C3met, trace amounts of 125I-labeled C3b, and increasing amounts of NC4. As a positive control for C3b degradation, FH (20 μg/ml) was used as cofactor, and FI was omitted in the negative control. In the experiment testing whether NC4 inhibited cofactor activity of C4BP and FH, these two proteins were used in the same concentrations as in the positive control samples. To test the ability of NC4 to enhance C4BP and FH cofactor activities, these were used at 15 and 1.25 μg/ml, respectively. Plasma purified C4BP as well as WT recC4BP and the ΔVal-108/Gln-109/Asp-110 mutant were used, both at 5 μg/ml. The samples were incubated at 37 °C, and the reactions were stopped by adding reducing SDS-PAGE sample buffer, followed by incubation at 95 °C for 5 min. The proteins were separated by 10–15% gradient SDS-PAGE, and radiolabel was visualized using Fluorescent Image Analyzer (FLA-300, Fujifilm, Tokyo, Japan). Intensities of protein bands were analyzed using ImageGauge version 4.1 (Fuji Photo Film).
RESULTS
NC4 Inhibits the Complement System
The NC4 domain of collagen IX containing the His tag was purified from bacterial cells using a nickel-nitrilotriacetic acid column, dialyzed against PBS, and analyzed by Coomassie staining and Western blotting (Fig. 1B).
To investigate the ability of the NC4 domain to inhibit complement, we used hemolytic assays for the classical and alternative pathways. NC4 inhibited only the classical pathway (Fig. 1C). When compared with the major inhibitor of the classical pathway C4BP, NC4 was less potent. At about 0.8 μm, NC4 inhibited the classical pathway by 50%, although 0.03 μm C4BP was required to obtain the same effect. The concentration of NC4 in the sera of OA and RA patients is around 4 μg/ml with ∼1 μg/ml in healthy controls, although NC4 levels can reach up to 250 μg/ml in the synovial fluid of patients.4 The finding that NC4 inhibits the classical pathway by 50% at ∼24 μg/ml is in accordance with the physiological concentrations of NC4 that can be found in synovial fluid. The lack of NC4 activity in the alternative pathway assay may be due to the much higher serum concentration required than in the classical pathway.
NC4 Binds C4, C3, and C9
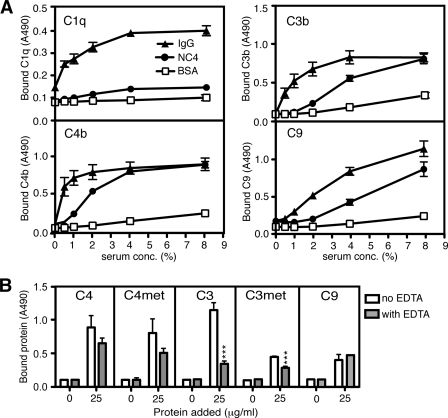
Several cartilage proteins have been found to interact with C1q and to activate (6, 7) or inhibit complement (31–33). To investigate the ability of NC4 to activate complement, we used immobilized NC4 on the surface of a microtiter plate. As positive controls, we immobilized aggregated human IgG (classical pathway), mannan, or acetylated BSA (lectin pathway) and zymosan (alternative pathway). Deposition of complement components from pooled human serum was detected with specific antibodies. We did not detect deposition of C3b in the conditions allowing only alternative pathway activation (data not shown), but there was NC4-dependent deposition of C3b/C3, C4b/C4, and C9 under conditions allowing classical and lectin pathway activation (Fig. 2A). However, we did not detect C1q (Fig. 2A), MBL, ficolin-2, or ficolin-3 deposition on NC4 (data not shown), indicating that NC4 does not interact with any known activators of the classical and lectin pathways. Furthermore, NC4 bound purified C4, C3, and C9 (Fig. 2B) displaying lower binding to C3met and C9 than to C3, C4, and C4met (Fig. 2B). In contrast to the interaction with C4, C4met, and C9, NC4 binding to C3 and C3met was dependent on calcium and other divalent ions as it decreased significantly in the presence of EDTA (Fig. 2B).
FIGURE 2.
NC4 binds C4/C4b, C3/C3b, and C9. A, NC4 binds C4b, C3b, and C9 from serum. NC4 and controls (IgG and BSA as positive and negative controls, respectively) were coated onto microtiter plates and incubated with increasing amounts of human serum. C1q, C4b, C3b, and C9 deposition was detected using specific antibodies. Data were collected from three independent duplicate experiments. B, NC4 binds to purified C4/C4b, C3/C3b, and C9. Microtiter plates were coated with NC4. Purified C4, C4met (corresponds functionally to C4b), C3, C3met (corresponds functionally to C3b), and C9 at increasing concentrations were allowed to bind for 1 h at room temperature. Binding of complement components was detected with specific antibodies. For assessing dependence of the interaction on calcium and other divalent ions, 5 mm EDTA was added to the reaction buffer. Data presented are from three independent duplicate experiments ± S.D. One-way ANOVA test was used to calculate statistical significance between the binding in the presence and absence of EDTA. ***, p < 0.001.
NC4 Inhibits the Classical Pathway at the C9 Level
To study at which step in the classical pathway the NC4 inhibits the complement, we first tested the inhibition of C4b and C3b deposition on plates coated with aggregated IgG in the presence of NC4 added to the serum. We did not observe a statistically significant decrease in the deposition of C4b and C3b (Fig. 3A). These findings indicate that NC4 might inhibit the complement activation at the MAC level. To organize MAC, the C5b fragment forms a stable dimer with C6, which upon C7 binding forms a C5b-7 complex with increased binding to the cell surface. One molecule of C8 then binds to form a C5b-8 complex followed by binding and circular polymerization of up to 16 molecules of C9 and the formation of MAC. C9 polymerization also occurs in the absence of C5b-8 upon prolonged incubation with formation of SDS-resistant tubular complexes (34).
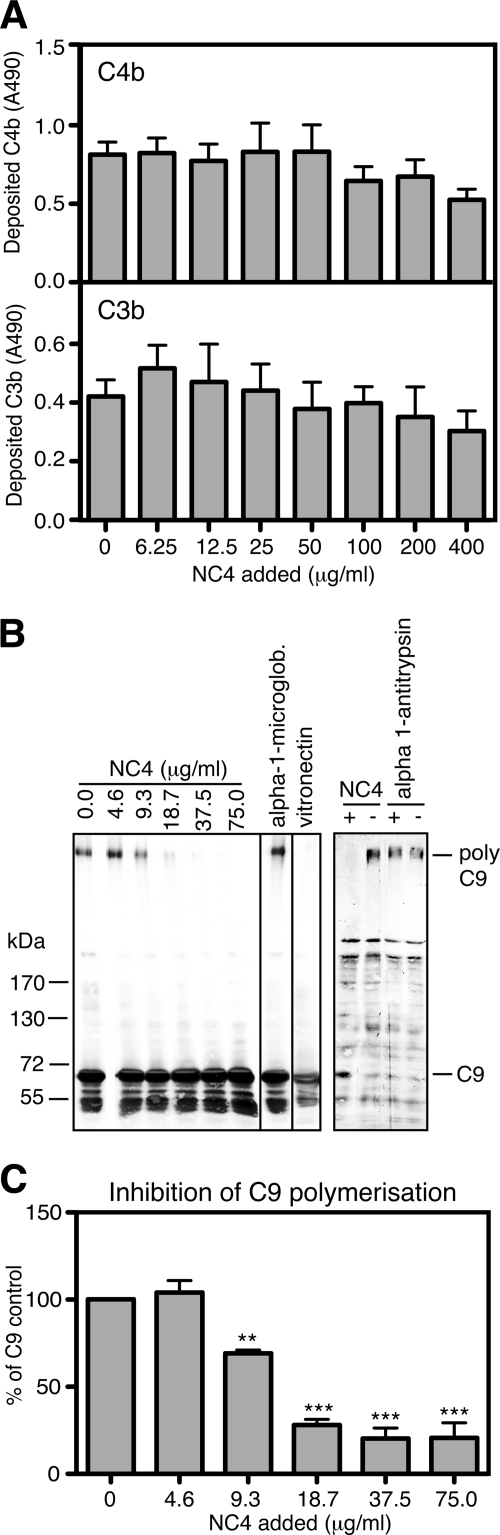
FIGURE 3.
Inhibition of the complement system by NC4. A, NC4 does not inhibit the complement system at the C4b and C3b levels. To study the ability of NC4 to inhibit C4b and C3b deposition, aggregated IgG and BSA as positive and negative control, respectively, were coated onto microtiter plates. Increasing amounts of NC4 were preincubated with human serum (0.2% serum for C4b and 0.3% for C3b deposition) and added to the plate. Deposition of C4b and C3b was detected with specific antibodies. B, NC4 inhibits C9 polymerization. C9 was mixed with different amounts of NC4 (0–75 μg/ml, corresponding to 0–2.5 μm), α1-microglobulin (130 μg/ml, corresponding to 5 μm) as negative control, and vitronectin (380 μg/ml, corresponding to 5 μm) as positive control (left panel). The samples were separated on 2.5–10% gradient SDS-PAGE and transferred onto PVDF membrane, and C9 was detected using specific antibodies. To assess the inhibition of C9 polymerization by NC4 on the cell surface, human serum was preincubated with or without NC4 (200 μg/ml, corresponding to 6 μm) and α1-antitrypsin (300 μg/ml, corresponding to 6 μm) as negative control. Sensitized sheep erythrocytes were then incubated with the serum samples at 37 °C for 1 h. Erythrocyte membranes were sedimented, and C9 was detected by Western blot analysis (right panel). C, intensities of the bands for polymerized C9 (from left panel) were determined by densitometry and presented as mean value from three independent experiments ± S.D. One-way ANOVA test was performed to test significant differences between C9 polymerization in the absence and in the presence of increasing NC4 concentrations. **, p < 0.01; ***, p < 0.001.
To examine whether NC4 can inhibit C9 polymerization, we used in addition to the method requiring MAC assembly on the plastic surface methods which allowed analysis of C9 polymerization in solution and on the cell membranes. C9 (53 μg/ml, corresponding to 0.8 μm) was incubated with increasing amounts of NC4 (0–75 μg/ml, corresponding to 0–2.5 μm) overnight at 37 °C, and the samples were analyzed by SDS-PAGE under reducing conditions followed by Western blotting with anti-C9 antibodies. As a positive control, we used vitronectin (380 μg/ml corresponding to 5 μm), and α1-microglobulin (130 μg/ml, corresponding to 5 μm) was used as the negative control. NC4 inhibited C9 polymerization in a concentration-dependent manner (Fig. 3B, left panel). The intensities of the bands for polymerized C9 were determined by densitometry (Fig. 3C).
To further evaluate whether NC4 can inhibit C9 polymerization on cells, we used antibody-sensitized sheep erythrocytes. NC4 was preincubated for 30 min at 37 °C with 0.2% human serum and then incubated with erythrocytes for 60 min at 37 °C. Erythrocyte membranes were washed, dissolved in sample buffer, analyzed by 2.5–10% gradient SDS-PAGE, followed by Western blotting for polymerized C9. NC4 inhibited deposition of polymeric C9 on erythrocyte membranes (Fig. 3B, right panel). α1-Antitrypsin was used as negative control.
NC4 Binds C4BP and FH
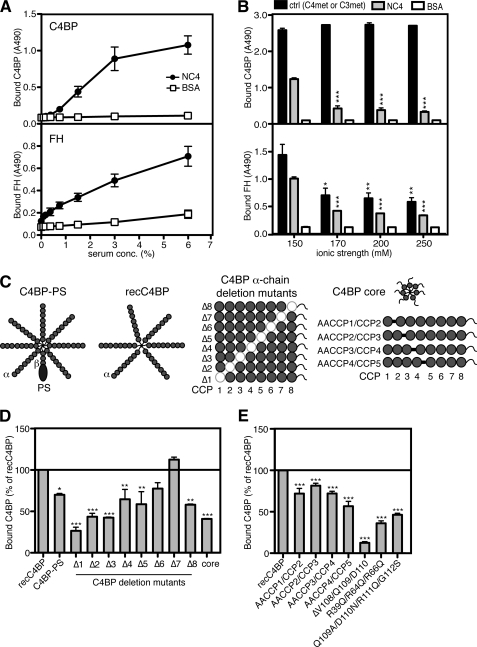
Because matrix proteins such as FM, OSAD, and chondroadherin bind complement inhibitors (7, 17), we investigated if the NC4 domain binds C4BP and/or FH. Indeed, by assessing the binding of C4BP and FH derived from heat-inactivated human serum to immobilized NC4, we observed concentration-dependent binding of C4BP and FH (Fig. 4A). NC4 interacted also with purified C4BP and FH proteins in a concentration-dependent manner (data not shown). The binding between NC4 and either C4BP or FH is based on ionic interactions because the presence of increasing NaCl concentrations in the reaction inhibited the NC4-C4BP and NC4-FH interactions (Fig. 4B). As a positive control for C4BP binding, we used C4met, while C3met was used as a control for FH binding. The interactions were not dependent on calcium or other divalent ions, as the results were the same in the presence of EDTA (data not shown).
FIGURE 4.
Interaction of NC4 with C4BP and FH. A, NC4 binds C4BP and FH from serum. NC4 was coated on microtiter plates, and increasing amounts of heat-inactivated serum were added. Bound C4BP and FH were detected with specific antibodies. BSA was used as a negative control. B, NC4 binds purified C4BP and FH, and these interactions were ionic in character. C4BP and FH were allowed to bind to immobilized NC4 in binding buffer supplemented with increasing concentrations of NaCl (150–250 mm). C4met and C3met were used as positive controls for C4BP and FH binding, respectively, and BSA was used as negative control. Bound C4BP and FH were detected with polyclonal antibodies. The graph represents data from two independent experiments done in duplicate. One-way ANOVA test was performed to calculate statistical difference of the groups, compared with the binding at 150 mm NaCl. C, C4BP variants and fragments used to determine binding sites for NC4 are as follows: C4BP-PS; recC4BP; C4BP α-chain deletion mutants lacking single CCP domains; C4BP α-chain mutants in which two alanine residues were introduced in linker regions between CCP domains; and C4BP core. The main isoform of C4BP in the blood circulation contains seven identical α-chains and one β-chain. The β-chain containing C4BP in circulation was bound to anticoagulant PS, forming a C4BP-PS complex. The recC4BP lacks the β-chain and the associated PS and was used as control. C4BP α-chain deletion mutants lack single CCP domains (represented by white circles in each α-chain), and C4BP core fragment contains the C-terminal extensions of the α-chains together with CCP8 and a small fragment of CCP7. D and E, NC4 has several binding sites on C4BP α-chain. The C4BP variants were allowed to bind to NC4 immobilized on a plate. Bound C4BP was detected with polyclonal antibody. The graph represents data from three independent experiments done in duplicate ± S.D. Statistical significance was calculated using one-way ANOVA test. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
The main isoform of C4BP protein contains seven identical α-chains and one β-chain. Each α-chain is composed of eight CCP domains, and the β-chain contains three CCP domains. Additionally, both chains have 60 amino acids at their C terminus that are involved in polymerization (35). The β-chain-containing C4BP in the circulation is bound to vitamin K-dependent anticoagulant protein S, forming a C4BP-PS complex (Fig. 4C). To identify the domains of C4BP that bind the NC4 domain, we assessed binding of C4BP-PS, recombinant C4BP (recC4BP), C4BP deletion mutants lacking single CCP domains from the α-chains, and C4BP core fragment (Fig. 4D) to immobilized NC4. The C4BP-PS did not bind to NC4 as well as the recC4BP (Fig. 4D), which could be explained by a certain degree of sterical hindrance at the core where the β-chain and PS are bound. Additionally, this finding demonstrates that the C4BP β-chain is not necessary for the interaction with NC4. The polymeric nature of C4BP is important for the interaction with NC4 apparently due to avidity effects. This was demonstrated when we investigated polymeric wild type recC4BP composed of six α-chains compared with single α-chain protein that was produced after introduction of a STOP codon after CCP8 (25). Monomeric α-chain composed of CCP1–8 did not bind NC4 in comparison with polymeric recC4BP, which is the form found in vivo, showing that several α-chains can be engaged simultaneously in the interaction with NC4 (data not shown). Deletion of CCP1–5 and -8 resulted in a significantly decreased binding of C4BP to NC4, indicating several possible binding sites along the α-chain (Fig. 4D). The effect was most pronounced when CCP1–3 were removed. Deletion of CCP6 and CCP7 did not affect binding to NC4, although binding of the C4BP core fragment containing the 60-amino acid C-terminal residues of the α-chains together with CCP8 and a small fragment of CCP7 was lower than to the whole C4BP. The binding site was further localized to a sequence of amino acids forming a loop in CCP2 as the mutants Q109A/D110N/R111Q/G112S and ΔVal-108/Gln-109/Asp-110 showed impaired binding to NC4 (Fig. 4E). Furthermore, positively charged amino acids located at the interface between CCP1 and CCP2 were also involved in binding as shown by impaired interaction between the R39Q/R64Q/R66Q mutant and NC4 (Fig. 4E). Mutants in which two alanine residues were introduced in the linker regions between CCP domains showed statistically significant impaired binding to NC4, indicating that binding sites for NC4 stretch over more than one CCP domain (Fig. 4E). Taken together, the NC4 domain appears to bind to several sites along the α-chain, with the main binding region at the periphery of the C4BP protein (CCP1–3).
NC4 Is Not a Cofactor for C4b and C3b Degradation but Enhances the Cofactor Activities of C4BP and FH
Using degradation assays, we studied if NC4 may act as a cofactor for FI-mediated degradation of C4b and C3b, as observed for several proteins able to interact with C4/C4b and C3/C3b. This method detects the presence or absence of specific degradation products (such as C4d, iC3b, and C3d) by using 125I-labeled C4b or C3b proteins.
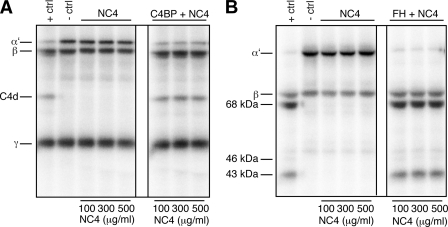
Increasing amounts of NC4 were mixed with FI, C4b/C3b, and 125I-labeled C4b/C3b, and the samples were incubated at 37 °C for 5 h. As a positive control for C4b degradation, C4BP (100 μg/ml) was used as cofactor. In the presence of this cofactor, FI degrades the C4b α′-chain to C4c and C4d products. The C4b β-chain and γ-chain remain unchanged (Fig. 5A, +ctrl). As a negative control, the same reaction was performed in the absence of FI.
FIGURE 5.
NC4 is not a cofactor in degradation of C4b and C3b. A, NC4 does not support FI-mediated C4b degradation and does not inhibit C4BP cofactor activity. For C4b degradation, different amounts of NC4 were incubated with FI, C4met, trace amounts of 125I-labeled C4b (left panel), and with 100 μg/ml C4BP (right panel) for 5 h at 37 °C and then separated on a 10–15% gradient SDS-PAGE. In the positive control (+ctrl), C4BP was mixed with FI, C4met, and trace amounts of 125I-labeled C4b, and in the negative control (−ctrl), FI was omitted. B, NC4 is not a cofactor for FI-mediated C3b degradation and does not inhibit FH cofactor activity. For C3b degradation, different amounts of NC4 were incubated with FI and C3met, trace amounts of 125I-labeled C3b (left panel), and with 20 μg/ml FH (right panel) for 5 h at 37 °C and then separated on a 10–15% gradient SDS-PAGE. In the positive control, FH was mixed with FI and C3met, and trace amounts of 125I-labeled C3b, and in the negative control, FI was omitted.
For the C3b degradation, FH (20 μg/ml) was used as cofactor in the positive control. In the presence of FH, the C3b α′-chain is cleaved by FI to 68- and 43-kDa products, although the C3b β-chain is not degraded (Fig. 5B, +ctrl). The absence of any of these fragments in the presence of NC4 shows that NC4 is not able to act as a cofactor for FI-mediated C4b and C3b degradation (Fig. 5, A and B, left panels).
To assess if NC4-C4BP and NC4-FH interactions have functional consequences, we used two strategies. In the first strategy, we tested if NC4 might inhibit C4BP and FH cofactor activities, and in the second strategy, we tested if NC4 might accelerate C4BP and FH cofactor activities, leading to more efficient C4b and C3b cleavage by FI. In the first strategy, C4BP (100 μg/ml) and 20 μg/ml FH were incubated with increasing NC4 concentrations in the presence of FI, C4b/C3b, and 125I-labeled C4b/C3b. NC4 interactions with C4BP and FH did not inhibit FI-mediated C4b and C3b cleavage under these conditions (Fig. 5, A and B, right panels, marked with C4BP + NC4 and FH + NC4, respectively).
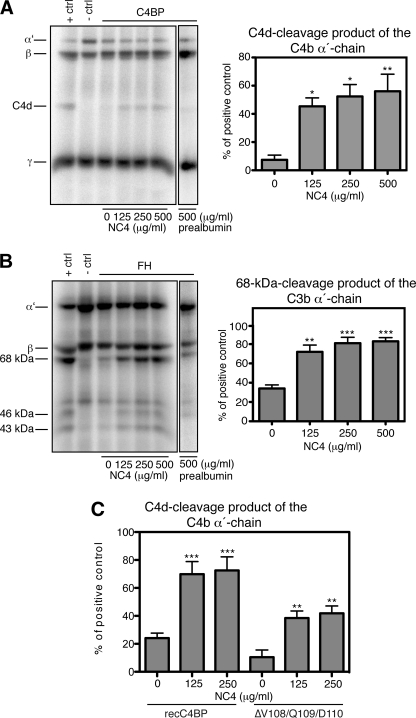
For a second strategy, we first titrated C4BP and FH to find the concentration of the cofactors at which the C4b and C3b α-chains are only partially degraded. This would facilitate the detection of any enhancement of C4b and C3b degradation in the presence of NC4. C4BP at 15 and 1.25 μg/ml FH were incubated with or without increasing amounts of NC4 for 30 min at 37 °C. FI, C4b/C3b, and 125I-labeled C4b/C3b were then added to the reactions, and finally proteins were analyzed by SDS-PAGE. Under these conditions the addition of NC4 resulted in more efficient degradation of C4b and C3b (Fig. 6, A and B). This effect was specific to NC4, and we could not observe it when prealbumin was used in the study as control (Fig. 6, A and B). The intensities of the C4d cleavage product and of the 68-kDa cleavage product of C3b α-chain were quantified by densitometry (Fig. 6, A and B, bar diagrams).
FIGURE 6.
NC4 enhances degradation of C4b and C3b in the presence of FI and cofactors. A, NC4 enhances C4BP cofactor activity. Different amounts of NC4 were mixed with 15 μg/ml C4BP, FI, C4met, and trace amounts of 125I-labeled C4b for 20 h at 37 °C. In the positive control, 100 μg/ml C4BP was mixed with FI, C4met, and trace amounts of 125I-labeled C4b, and in the negative control FI was omitted. Prealbumin (500 μg/ml) was used as negative control. The intensities of the C4d-cleavage products were calculated from three independent experiments and shown as mean values ± S.D. (bar diagram). B, NC4 enhances FH cofactor activity. Different amounts of NC4 were mixed with 1.25 μg/ml FH, FI, C3met, and trace amounts of 125I-labeled C3b for 5 h at 37 °C. In the positive control, 20 μg/ml FH was mixed with FI, C3met, and trace amounts of 125I-labeled C3b, and in the negative control, FI was omitted. The intensities of the 68-kDa cleavage products of the C3b α-chains were calculated from three independent experiments and shown as mean values ± S.D. C, enhancing by NC4 depends on its ability to bind C4b. The degradation assay was performed as in A but with 5 μg/ml of two forms of recC4BP: wild type that interacts well with NC4 and the ΔVal-108/Gln-109/Asp-110 mutant, which binds NC4 poorly. Both C4BP variants bind C4b. NC4-enhanced degradation of C4b in the presence of either form of C4BP implied that the enhancing effect depends on NC4-C4b interaction. One-way ANOVA test was used to calculate statistical significance in A–C. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
To address whether the enhancing effect of NC4 was due to the ability of NC4 to bind C4b or C4BP, we studied degradation using the ΔVal-108/Gln-109/Asp-110 C4BP mutant, which binds poorly to NC4 but retains its cofactor activity. The mutant contributed to enhanced degradation of C4b in a manner similar to WT recC4BP implying that the NC4-C4b interaction is responsible for the enhancing effect (Fig. 6C). Furthermore, we found no interaction between NC4 and FI, neither in purified form nor from serum (data not shown).
NC4 Interacts with Other Matrix Proteins
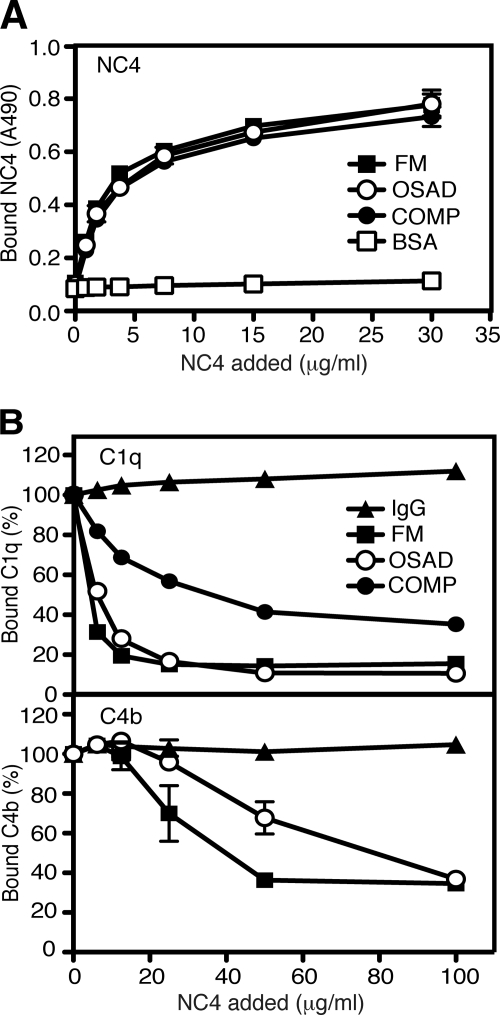
The NC4 domain by projecting out from the collagen fibril surface provides sites for interaction with other matrix components. We aimed to investigate if NC4 interacts with matrix proteins already known to modulate complement, such as FM, OSAD, and COMP, and if these interactions have an impact on the complement modulation. COMP, FM, and OSAD were coated on a microtiter plate, and NC4 was allowed to bind at increasing concentrations. NC4 bound strongly to COMP, FM, and OSAD (Fig. 7A), corroborating published data (16, 36).
FIGURE 7.
Interaction of NC4 with FM, OSAD, and COMP. A, NC4 binds to FM, OSAD, and COMP. NC4 at increasing concentrations was allowed to bind to immobilized FM, OSAD, and COMP in binding buffer. BSA was used as negative control. Bound NC4 was detected with polyclonal antibody. B, NC4 inhibits binding of serum-derived C1q to FM, OSAD, and COMP (upper panel) and serum-derived C4b to FM and OSAD (bottom panel). FM, OSAD, and COMP were coated on a microtiter plate. Aggregated IgG was used as positive control. For C1q and C4b binding to FM and OSAD, increasing amounts of NC4 were mixed with 1% serum and added to the plate, and 10% heat-inactivated serum was used to mix with increasing amounts of NC4 for C1q binding to COMP. Bound C1q and C4b were detected with specific antibodies. The data represent the percent of bound C1q and C4b from three independent duplicate experiments ± S.D.
The classical pathway of complement is triggered by the multimolecular C1 complex formed by C1q and two copies of the serine proteases C1r and C1s (37). C1q is the recognition subunit of the complex and is composed of collagen-like regions (stalks) and C-terminal globular regions, involved in binding to various targets. C1s and C1r bind to the stalks of C1q (38) and undergo conformational changes upon target binding. Both FM and OSAD activate the classical pathway by binding to the globular heads of C1q (6, 7), and COMP inhibits the classical pathway via binding to C1q stalk region (33), thereby preventing activation of the C1r and C1s proteases.
We assessed C1q binding from serum to FM, OSAD, and COMP in the presence of NC4. Because COMP interacts directly with the C1q stalk regions, where C1s and C1r are bound, we used heat-inactivated serum to assess binding of free C1q to COMP. Increasing the concentration of NC4 to 100 μg/ml in the reaction inhibited the C1q binding to FM and OSAD by 85 and 90%, respectively. This may depend on competition by the known interactions of the N-terminal tyrosine sulfate domain of these proteins with NC4 (16). C1q binding to COMP was inhibited by 65% under the same conditions (Fig. 7B). COMP is a homopentameric molecule composed of five identical subunits (39), each able to bind NC4, which could explain the lower inhibition level of C1q binding to coated COMP than to FM and OSAD. Importantly, NC4 did not inhibit binding of C1q to IgG confirming that its activity in the assay is related to binding to cartilage molecules and not C1q. Consequently, C4b deposition on FM and OSAD was decreased by NC4 (Fig. 7B), indicating that NC4 can decrease or abolish activation of the complement system by these proteins.
DISCUSSION
Rheumatic diseases such as RA and OA are chronic diseases causing damage to the cartilage, synovium, and bone in the joints. These diseases affect a significant proportion of the population. Currently, the diagnosis and monitoring of RA and particularly OA are based on methods limited in that they detect clinical symptoms at advanced stages of the diseases. Thus, it is important to identify novel targets for early diagnosis and prediction of disease evolution. Inflammation has a role in disease development, and identification of novel regulatory interactions between cartilage components and the immune system may lead to development of new treatments.
Collagen type IX is bound at the surface of the collagen II/XI fibrils and with its COL3 and NC4 domains exposed provides sites for interaction with other cartilage proteins, ensuring the formation of macromolecular networks important for tissue functions. Upon cartilage breakdown, proteins normally retained in the cartilage are released into the synovial fluid and serum and have been shown to have the potential to be used as markers for diagnosis and prognosis of joint diseases (40). Some of these proteins have been found to interact with components of complement. FM, OSAD, chondroadherin (6, 7), and COMP (33) are able to either activate or inhibit complement. All complement components can be found in the synovial fluid, and it has been well recognized that complement activation is an important initiator as well as sustainer of inflammation in arthritic diseases (5).
Fragmentation of collagen IX and loss of its NC4 domain from cartilage can be a result of cytokine-induced protease activation and appears to be an event in cartilage breakdown (13). Collagen IX fragments have been found in synovial fluids from patients with arthritic diseases (41). Additionally, mutant mice lacking collagen IX show matrix destabilization and are more susceptible to inflammatory arthritis (42). This could be explained by an enhanced accessibility of collagen-specific antibodies and complement components to cartilage and raises the possibility that the NC4 domain of collagen IX and its interactions with other matrix components might have a role in the modulation of complement, thereby protecting cartilage from damage. In this study, we investigated this in detail and found that the NC4 domain of collagen IX is a local inhibitor of complement. It inhibits complement by preventing C9 polymerization and attenuation of MAC formation.
Proteins able to interact with C4/C4b and C3/C3b sometimes play a role as cofactors for FI-mediated C4b and C3b degradation, but we now show that this is not the case for NC4. Upon cartilage breakdown, several cartilage proteins with complement-activating potential are released into synovial fluid. In this situation, NC4 by binding C4/C4b and C3/C3b might prevent progress to the next step of the complement cascade initiated by other cartilage proteins and thus slowing down the process of complement activation.
We found that the NC4 domain interacts with the complement inhibitors C4BP and FH, which suggests a physiological role in the local regulation of complement. These interactions are ionic in character, and NC4 has several bindings sites along the C4BP α-chain, albeit possibly with different affinities. In healthy cartilage, NC4 is an integral part of the CIX molecule. Unfortunately, the full-length heterotrimer has not been expressed recombinantly. This represents a major endeavor, in view of the extensive post-translational modifications such as formation of hydroxyproline and lysine as well glycosylations. Purification of CIX from cartilage is not a viable alternative except from lathyritic animals because the collagen largely occurs covalently cross-linked to the collagen II fibers. Therefore, we can only speculate what would be the role of the intact CIX. However, two scenarios are possible; first, in the healthy cartilage NC4 might localize the inhibitors to the collagen fibrils to protect them from complement attack. As cartilage has a dense matrix poorly permeable to most serum proteins, NC4 may play a role in controlling complement mainly at the surface of the tissue. Second, upon collagen IX fragmentation and release of the NC4 domain into synovial fluid of patients with active disease, this fragment may locally inhibit complement by binding C4BP and FH. Moreover, C4BP and FH bound to NC4 not only maintain their cofactor activity, but also promote the enzymatic cleavage of C3b and C4b by FI more effectively than unbound. This may be related to the fact that NC4 interacts with C3b and C4b and may either affect their conformation making them more susceptible for cleavage by FI or acts as cross-link between C4BP/FH and C4b/C3b increasing the affinity of their interactions. By accelerating cofactor activities of C4BP and FH, NC4 might serve to maintain the complement activation at a low level even when initiated by other cartilage proteins or protein fragments, preventing the release of C5a and MAC formation. This could be an important mechanism protecting intact cartilage from complement being activated. It appears that these mechanisms are more relevant for disease progression/chronicity than to disease initiation. Because RA has a much more prominent inflammatory component than OA, these mechanisms of regulation of the pro-inflammatory complement cascade may be more relevant in RA than OA.
We aimed to investigate if the observed C4, C3, and C9 deposition from serum on NC4 results from a direct interaction, rather than from complement activation. For this purpose, we used heat-inactivated and C2- and C6-depleted sera. Surprisingly, strong binding of C4, C3, and C9 to NC4 could not be detected from C2- and C6-depleted sera nor control normal human serum from the same vendor, all prepared from single donors (data not shown). Binding observed with our pooled serum that was heat-inactivated was also weak indicating a certain level of complement activation. Furthermore, we studied deposition of C4, C3, and C9 on NC4 from individual sera of 11 healthy donors and found that 30% of the sera did not support heat-sensitive deposition of C4, C3, and C9 on NC4 (data not shown). In the remaining sera, binding of C4, C3, and C9 was significant but varied in intensity. These pooled sera did not show any significant binding of MBL, C1q, or ficolin-2/3 to immobilized NC4, although these proteins bound well from serum to positive controls, IgG, mannan, and acetylated BSA (data not shown). There is a concern that immobilization of NC4 masks binding epitope for C1 or MBL, but such immobilized NC4 triggers classical pathway activation in serum. Therefore, it appears that the epitope binding complement initiator is exposed under these conditions. Additionally, in 30% of our tested sera, we could not detect MBL deposition on mannan implying MBL deficiency. This was not correlated with the ability to deposit complement on NC4. These findings indicate that a yet unknown molecule able to induce the classical pathway might be involved in interaction with NC4 and that not all of the healthy donor sera might contain this protein. The NC4 domain is a positively charged molecule, capable of binding heparin (43). One may speculate that it binds acidic molecules and glycosaminoglycans present in serum. Thus, NC4 might cause some activation of complement by binding a yet unknown complement initiator, which warrants further investigation.
The cartilage proteins FM, OSAD, and COMP interact in complex ways with complement. FM and OSAD bind to C1q globular heads and activate complement. COMP binds C1q stalks and inhibits the classical pathway, although it simultaneously binds properdin and activates the alternative pathway. Interestingly, complexes between COMP and C3b resulting from COMP-mediated complement activation in vivo were found in sera from RA patients (33). Because the NC4 domain interacts with FM, OSAD, and COMP, a question arises how the interactions between the different cartilage proteins effect complement regulation. In an attempt to shed some light on this, we studied the ability of FM, OSAD, and COMP to bind C1q in the presence of NC4. The interactions with NC4 led to inhibition of C1q binding to FM, OSAD, and to lesser extent COMP, suggesting that when released into the synovial fluid NC4 might inhibit the complement activating potential of FM and OSAD, and the inhibitory potential of COMP. The outcome of this regulatory process would depend on the levels of cartilage proteins released into the synovial fluid upon cartilage damage and might be an important factor for monitoring and predicting disease development. FM and OSAD both contain tyrosine sulfate-rich domains that mimic heparin and can interact with a variety of heparin-binding proteins (16). The inhibition of C1q binding to these proteins by NC4 seems to depend on the competition for the same binding site between C1q and NC4. Taken together, the results from this study show that the NC4 domain of collagen IX inhibits complement by preventing C9 polymerization and enhancing cofactor activities of the major soluble complement inhibitors C4BP and FH.
This work was supported, in whole or in part, by a National Institutes of Health grant from NIAMS. This work was also supported by Swedish Research Council Grants K2009-68X-14928-06-3 and K2011-52X-05668-32-3, Swedish Foundation for Strategic Research, Foundations of Österlund, Greta and Johan Kock, Inga-Britt, and Arne Lundberg, King Gustav Vth's 80th Anniversary, Knut and Alice Wallenberg Foundation, and grants from the University Hospital in Malmö/Lund. Conflict of interest: Dr. Heinegård owns stock in AnaMar Medical and has filed a patent application concerning type IX collagen as marker of joint destruction.
M. Danfelter, P. Önnerfjord, and D. Heinegård, unpublished observations.
- MAC
- membrane attack complex
- NC4
- noncollagenous domain 4
- COMP
- cartilage oligomeric matrix protein
- FM
- fibromodulin
- OSAD
- osteoadherin
- FH
- factor H
- FI
- factor I
- RA
- rheumatoid arthritis
- OA
- osteoarthritis
- CCP domain
- complement control protein domain
- ANOVA
- analysis of variance
- PS
- protein S
- recC4BP
- recombinant C4BP
- MBL
- mannan-binding lectin.
REFERENCES
- 1. Ricklin D., Hajishengallis G., Yang K., Lambris J. D. (2010) Nat. Immunol. 11, 785–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zipfel P. F., Skerka C. (2009) Nat. Rev. Immunol. 9, 729–740 [DOI] [PubMed] [Google Scholar]
- 3. Morgan B. P., Walport M. J. (1991) Immunol. Today 12, 301–306 [DOI] [PubMed] [Google Scholar]
- 4. Sjöberg A. P., Trouw L. A., Blom A. M. (2009) Trends Immunol. 30, 83–90 [DOI] [PubMed] [Google Scholar]
- 5. Okroj M., Heinegård D., Holmdahl R., Blom A. M. (2007) Ann. Med. 39, 517–530 [DOI] [PubMed] [Google Scholar]
- 6. Sjöberg A., Onnerfjord P., Mörgelin M., Heinegård D., Blom A. M. (2005) J. Biol. Chem. 280, 32301–32308 [DOI] [PubMed] [Google Scholar]
- 7. Sjöberg A. P., Manderson G. A., Mörgelin M., Day A. J., Heinegård D., Blom A. M. (2009) Mol. Immunol. 46, 830–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. van der Rest M., Mayne R., Ninomiya Y., Seidah N. G., Chretien M., Olsen B. R. (1985) J. Biol. Chem. 260, 220–225 [PubMed] [Google Scholar]
- 9. Eyre D. R., Pietka T., Weis M. A., Wu J. J. (2004) J. Biol. Chem. 279, 2568–2574 [DOI] [PubMed] [Google Scholar]
- 10. Shaw L. M., Olsen B. R. (1991) Trends Biochem. Sci. 16, 191–194 [DOI] [PubMed] [Google Scholar]
- 11. Budde B., Blumbach K., Ylöstalo J., Zaucke F., Ehlen H. W., Wagener R., Ala-Kokko L., Paulsson M., Bruckner P., Grässel S. (2005) Mol. Cell. Biol. 25, 10465–10478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mann H. H., Ozbek S., Engel J., Paulsson M., Wagener R. (2004) J. Biol. Chem. 279, 25294–25298 [DOI] [PubMed] [Google Scholar]
- 13. Danfelter M., Onnerfjord P., Heinegård D. (2007) J. Biol. Chem. 282, 36933–36941 [DOI] [PubMed] [Google Scholar]
- 14. Kojima T., Mwale F., Yasuda T., Girard C., Poole A. R., Laverty S. (2001) Arthritis Rheum. 44, 120–127 [DOI] [PubMed] [Google Scholar]
- 15. Wotton S. F., Dieppe P. A., Duance V. C. (1999) Rheumatology 38, 338–345 [DOI] [PubMed] [Google Scholar]
- 16. Tillgren V., Onnerfjord P., Haglund L., Heinegård D. (2009) J. Biol. Chem. 284, 28543–28553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Happonen K. E., Sjöberg A. P., Mörgelin M., Heinegård D., Blom A. M. (2009) J. Immunol. 182, 1518–1525 [DOI] [PubMed] [Google Scholar]
- 18. Andersson M., Hanson A., Englund G., Dahlbäck B. (1991) Eur. J. Clin. Pharmacol. 40, 261–265 [DOI] [PubMed] [Google Scholar]
- 19. Blom A. M., Villoutreix B. O., Dahlbäck B. (2003) J. Biol. Chem. 278, 43437–43442 [DOI] [PubMed] [Google Scholar]
- 20. Greenwood F. C., Hunter W. M., Glover J. S. (1963) Biochem. J. 89, 114–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Biesecker G., Müller-Eberhard H. J. (1980) J. Immunol. 124, 1291–1296 [PubMed] [Google Scholar]
- 22. Blom A. M., Kask L., Dahlbäck B. (2003) Mol. Immunol. 39, 547–556 [DOI] [PubMed] [Google Scholar]
- 23. Nilsson S. C., Trouw L. A., Renault N., Miteva M. A., Genel F., Zelazko M., Marquart H., Muller K., Sjöholm A. G., Truedsson L., Villoutreix B. O., Blom A. M. (2009) Eur. J. Immunol. 39, 310–323 [DOI] [PubMed] [Google Scholar]
- 24. Dahlbäck B. (1983) Biochem. J. 209, 837–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Blom A. M., Kask L., Dahlbäck B. (2001) J. Biol. Chem. 276, 27136–27144 [DOI] [PubMed] [Google Scholar]
- 26. Blom A. M., Webb J., Villoutreix B. O., Dahlbäck B. (1999) J. Biol. Chem. 274, 19237–19245 [DOI] [PubMed] [Google Scholar]
- 27. García de Frutos P., Härdig Y., Dahlbäck B. (1995) J. Biol. Chem. 270, 26950–26955 [DOI] [PubMed] [Google Scholar]
- 28. Heinegård D., Larsson T., Sommarin Y., Franzén A., Paulsson M., Hedbom E. (1986) J. Biol. Chem. 261, 13866–13872 [PubMed] [Google Scholar]
- 29. Onnerfjord P., Heathfield T. F., Heinegård D. (2004) J. Biol. Chem. 279, 26–33 [DOI] [PubMed] [Google Scholar]
- 30. Rosenberg K., Olsson H., Mörgelin M., Heinegård D. (1998) J. Biol. Chem. 273, 20397–20403 [DOI] [PubMed] [Google Scholar]
- 31. Groeneveld T. W., Oroszlán M., Owens R. T., Faber-Krol M. C., Bakker A. C., Arlaud G. J., McQuillan D. J., Kishore U., Daha M. R., Roos A. (2005) J. Immunol. 175, 4715–4723 [DOI] [PubMed] [Google Scholar]
- 32. Krumdieck R., Höök M., Rosenberg L. C., Volanakis J. E. (1992) J. Immunol. 149, 3695–3701 [PubMed] [Google Scholar]
- 33. Happonen K. E., Saxne T., Aspberg A., Mörgelin M., Heinegård D., Blom A. M. (2010) Arthritis Rheum. 62, 3574–3583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Podack E. R., Tschopp J. (1982) J. Biol. Chem. 257, 15204–15212 [PubMed] [Google Scholar]
- 35. Kask L., Hillarp A., Ramesh B., Dahlbäck B., Blom A. M. (2002) Biochemistry 41, 9349–9357 [DOI] [PubMed] [Google Scholar]
- 36. Thur J., Rosenberg K., Nitsche D. P., Pihlajamaa T., Ala-Kokko L., Heinegård D., Paulsson M., Maurer P. (2001) J. Biol. Chem. 276, 6083–6092 [DOI] [PubMed] [Google Scholar]
- 37. Wallis R., Mitchell D. A., Schmid R., Schwaeble W. J., Keeble A. H. (2010) Immunobiology 215, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pflieger D., Przybylski C., Gonnet F., Le Caer J. P., Lunardi T., Arlaud G. J., Daniel R. (2010) Mol. Cell. Proteomics 9, 593–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hedbom E., Antonsson P., Hjerpe A., Aeschlimann D., Paulsson M., Rosa-Pimentel E., Sommarin Y., Wendel M., Oldberg A., Heinegård D. (1992) J. Biol. Chem. 267, 6132–6136 [PubMed] [Google Scholar]
- 40. Crnkic M., Månsson B., Larsson L., Geborek P., Heinegård D., Saxne T. (2003) Arthritis Res. Ther. 5, R181–R185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wotton S. F., Duance V. C. (1991) Biochem. Soc. Trans. 19, 375S. [DOI] [PubMed] [Google Scholar]
- 42. Carlsen S., Nandakumar K. S., Holmdahl R. (2006) Arthritis Res. Ther. 8, R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pihlajamaa T., Lankinen H., Ylöstalo J., Valmu L., Jäälinoja J., Zaucke F., Spitznagel L., Gösling S., Puustinen A., Mörgelin M., Peränen J., Maurer P., Ala-Kokko L., Kilpelaïnen I. (2004) J. Biol. Chem. 279, 24265–24273 [DOI] [PubMed] [Google Scholar]