Abstract
Activated cannabinoid 1 receptor (CB1R) signaling has been implicated in the development of phenotypes associated with fatty liver, insulin resistance, and impaired suppression of hepatic glucose output. Endoplasmic reticulum stress-associated liver-specific transcription factor CREBH is emerging as a critical player in various hepatic metabolic pathways and regulates hepatic gluconeogenesis in diet-induced obese settings. In this study, we elucidated the critical role of CREBH in mediating CB1R signaling to regulate glucose homeostasis in primary rat and human hepatocytes. mRNA and protein levels and glucose production were analyzed in primary rat and human hepatocytes. ChIP assays were performed together with various transcriptional analyses using standard techniques. CB1R activation by 2-arachidonoylglycerol (2-AG) specifically induced CREBH gene expression via phosphorylation of the JNK signaling pathway and c-Jun binding to the AP-1 binding site in the CREBH gene promoter. 2-AG treatment significantly induced hepatic gluconeogenic gene expression and glucose production in primary hepatocytes, and we demonstrated that the CREBH binding site mutant significantly attenuated 2-AG-mediated activation of the gluconeogenic gene promoter. Endogenous knockdown of CREBH led to ablation of 2-AG-induced gluconeogenic gene expression and glucose production, and the CB1R antagonist AM251 or insulin exhibited repression of CREBH gene induction and subsequently inhibited gluconeogenesis in both rat and human primary hepatocytes. These results demonstrate a novel mechanism of action of activated CB1R signaling to induce hepatic gluconeogenesis via direct activation of CREBH, thereby contributing to a better understanding of the endocannabinoid signaling mechanism involved in regulating the hepatic glucose metabolism.
Keywords: AP-1 Transcription Factor, ER Stress, Gene Regulation, Gluconeogenesis, Hepatocyte, Jun N-terminal Kinase (JNK), Metabolic Regulation, Signal Transduction, CREBH, Cannabinoid Receptor Type 1 (CB1R)
Introduction
Endogenous cannabinoids (endocannabinoids) are lipid mediators that interact with cannabinoid receptors, the two main endocannabinoids being arachidonoyl ethanolamide (AEA,2 anandamide) and 2-arachidonoylglycerol (2-AG). The endocannabinoid system comprises the cannabinoid receptors type 1 (CB1R), which are expressed at high levels in the brain but are also present at much lower concentrations in peripheral tissues, and CB2R, which are expressed predominantly in immune and hematopoietic cells (1). Reports from animal studies and clinical investigations in humans have shown that in the obese state, the endocannabinoid system is hyperactivated because of impaired energy balance (2–4). In obese or hyperglycemic type 2 diabetic patients, circulating levels of AEA and 2-AG are increased, and elevated levels of 2-AG are found in visceral adipose tissue (2, 5, 6). Mice deficient in CB1R are resistant to diet-induced obesity and steatosis, and in wild-type mice, chronic treatment with a CB1R antagonist reversed both of these diet-induced effects. Both genetically and diet-induced obese animal models show elevated levels of endocannabinoids in the hypothalamus and peripheral tissues (7–10). A recent study using a liver-specific CB1R knockout mice model demonstrated that peripheral CB1R could be selectively targeted for the treatment of fatty liver, impaired glucose homeostasis, and dyslipidemia to reduce the neuropsychiatric side effects of a nonselective CB1R signaling blockade in the treatment of obesity-associated conditions (11). Overall, both clinical (12) and animal data regarding the CB1R blockade (8–11) overwhelmingly suggest the beneficial actions of CB1R antagonism on glucose metabolism and insulin sensitivity.
Cyclic AMP response element-binding protein H (CREBH) is an endoplasmic reticulum (ER) stress-associated liver-specific transcription factor that has been reported to be induced by the acute inflammatory response-inducing factor LPS and proinflammatory cytokines IL-6 and TNFα (13). A recent study from our group has demonstrated that CREBH plays an important mediatory role in the hormonal regulation of hepatic gluconeogenesis under fasting or insulin-resistant conditions in the rodent liver (14). Our results demonstrated that CREBH is dramatically induced and activated in diet-induced obese rodent models. Upon induction and activation, CREBH directly regulates the expression of the key hepatic gluconeogenic enzyme genes Pepck (phosphoenolpyruvate carboxykinase) and G6pc (glucose-6-phosphatase catalytic subunit) (14). CREBH has also been demonstrated to mediate ER stress-induced regulation of hepatic iron metabolism (15). Hepatocytes, being professional secretory cells, are likely to be under a subtle but continuous ER stress condition because of the enormous requirement for protein folding in the ER lumen (16). Because of its liver-specific expression and stress-sensory activation, CREBH is emerging to be a key player in regulating various hepatic metabolic pathways.
Although CB1R signaling has been known to affect glucose metabolism, the molecular mechanism behind this phenomenon is still unclear. On the basis of this observation, in this study we investigated whether the CB1R signaling pathway regulates CREBH gene expression and delineated the unprecedented link by which the CB1R signaling pathway initiates an ER stress condition, leading to the activation of CREBH to regulate hepatic gluconeogenesis.
EXPERIMENTAL PROCEDURES
Reagents and Plasmids
2-AG and AM251 were from Tocris Bioscience. Wortmannin and U0126 were from Sigma. Tunicamycin, SB203580, SP600125, and compound C were from Calbiochem. Insulin (Norvolin R) was from Green Cross (Korea). The reporter plasmids Pepck-Luc (-490/+72), G6pc-Luc (-231/+57), Pepck-Luc (CREBH-RE mutant), G6pc-Luc (CREBH-RE mutant), Gal4-Luc (GAL4DBD (UAS)-TK-Luc vector for mammalian one-hybrid assay), and Crebh-Luc were described previously (14, 17). A series of human CREBH promoter luciferase reporter deletion constructs were described previously (14). The AP-1 mutant luciferase reporter construct was subcloned into the pGL3 vector using the XhoI and HindIII restriction sites, and all constructs were confirmed by DNA sequencing. Gal4DBD, Gal4CREBH, dominant negative CREBH (DN-CREBH), and c-Fos plasmids were described previously (14, 17). HA-tagged c-Jun and c-Jun KD (kinase dead mutant) were kind gifts from Dr. Dirk Bohmann (University of Rochester Medical Center).
Cell Culture and Transient Transfection Assay
AML12 and H4IIE cells were obtained from the American Type Culture Collection. Maintenance of cell lines and transient transfections were performed as described previously (17). Total cDNA used for each transfection was adjusted to 1 μg/well by adding the appropriate amount of empty vector and CMV-β-galactosidase plasmids as an internal control. Cells were harvested 40–48 h post-transfection for the luciferase and β-galactosidase assays. The luciferase activity was normalized to β-galactosidase activity and expressed as relative luciferase units.
Preparation of Recombinant Adenovirus
For endogenous knockdown of Crebh gene expression, an adenoviral delivery system was used. Adenoviruses for unspecific (USi) control RNAi and Crebh RNAi were described previously (14).
Isolation and Culture of Primary Rat Hepatocytes
Primary hepatocytes were prepared from 200- to 300-g Sprague-Dawley rats by the collagenase perfusion method, as described previously (14, 17). The viability of cells was analyzed using trypan blue staining. Cells were maintained in (Medium 199, IX with Earle's salts and l-glutamine) M199 medium (Mediatech) overnight for attachment, and experiments were performed as indicated.
Primary Human Hepatocyte Culture
PHHs were obtained from the Liver Tissue and Cell Distribution System of the National Institutes of Health (S. Strom, University of Pittsburgh, PA). Hepatocytes were cultured as described previously (17).
RNA Isolation and Analysis
Total RNA was isolated for northern hybridization using probes for Pepck, G6pc, Crebh, and GAPDH, as described previously (14, 17). Semiquantitative and qPCR analysis in primary rat hepatocytes and primary human hepatocytes were performed using primers for Pepck, G6pc, Crebh, Atf6, Srebf-1, Ppargc1α, Cb1r, and β-actin (primer sequences are available on request), as described previously (14, 17).
Western Blot Analysis
Cell lysate preparation and Western blot analysis in primary rat hepatocytes using rabbit polyclonal JNK (t-JNK), rabbit polyclonal phospho-JNK (P-JNK), rabbit polyclonal c-Jun, rabbit polyclonal phospho-c-Jun (Cell Signaling Technology, Inc.), β-tubulin (Santa Cruz Biotechnology, Inc.), and mouse polyclonal CREBH antibodies were described previously (14, 17). To confirm the CB1R protein level, a ProteoExtract subcellular proteome extraction kit (Calbiochem) and CB1R antibody (Sigma, C1233) were purchased.
ChIP assay
The ChIP assay was performed according to the manufacturer's protocol (Upstate). Briefly, AML12 cells were transfected with reporter plasmids, and treatments were performed as indicated. Cells were then fixed with 1% formaldehyde and harvested. Soluble chromatin was immunoprecipitated with rabbit polyclonal c-Jun, rabbit polyclonal phospho-c-Jun, mouse polyclonal CREBH antibody, and mouse monoclonal HNF-4α antibody (Santa Cruz Biotechnology, Inc.). After recovering DNA, qPCR was performed using primers encompassing human CREBH promoter (-900/-750) forward, 5′-GCGAGCAGGTGAGGTAGG; and reverse, 5′-AAACCAGCTTGGACTCCG; or rat Pepck promoter (-500/-270) and human G6pc promoter (-300/+57), as described previously (14). Endogenous ChIP assays were performed using primers encompassing mouse Crebh promoter (-900/-750) forward, 5′-CCTCAATGAGCAAGTATCAATCGA; and reverse, 5′-GCTTCCTGCTTTTCTTTCATTTGC; or mouse Pepck promoter (-500/-270) forward, 5′-GATGGCCAGAGAATCCACCACACA; and reverse, 5′-TAGCCGAGACGCCTCTTGGACTT; or mouse G6pc promoter (-300/+57) forward, 5′-TAATTGGCTCTGCCAATGGCGATC; and reverse, 5′-ATCAGTCTGTGCCTTGCCCCTGT.
Glucose Production Assay
Glucose production from primary rat hepatocytes was measured according to the manufacturer's protocol using a colorimetric glucose oxidase assay (Sigma). Briefly, after the experimental time period as indicated, the cells were washed three times with PBS, and cells were incubated for 3 h at 37 °C, 5% CO2 in glucose production buffer (glucose-free DMEM (pH 7.4) containing 20 mmol/liter sodium lactate, 1 mmol/liter sodium pyruvate, and 15 mmol/liter HEPES, without phenol red). The glucose assays were performed in triplicate, and the intra-assay coefficient of variation was <5%.
Statistical Analyses
Data are expressed as mean ± S.D. Statistical analysis was performed using Student's t test or analysis of variance followed by Duncan's multiple comparison tests. All experiments were performed at least three times. Differences were considered significant at p < 0.05.
RESULTS
CB1R Signaling Induces and Activates CREBH Gene Expression
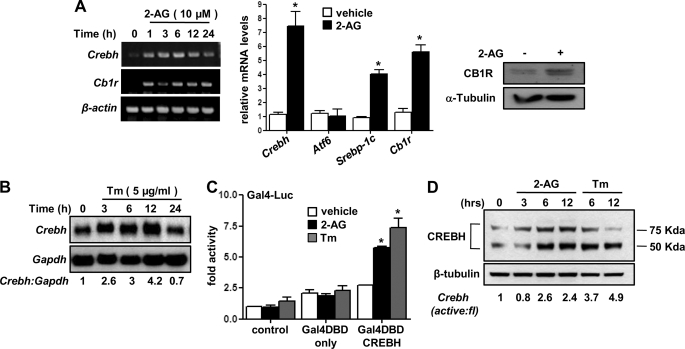
Previous studies have demonstrated that CB1R signaling regulates glucose metabolism (7, 11). In addition, we recently demonstrated that CREBH plays an important role in regulating key hepatic gluconeogenic enzyme gene expression (14). On the basis of those observations, we first examined the role of CB1R in Crebh gene expression (Fig. 1). Treatment of rat primary hepatocytes with the CB1R agonist 2-AG demonstrated a significant increase of CREBH gene expression in a time-dependent manner, along with the CB1R mRNA level (Fig. 1A, left panel). Real-time PCR (qPCR) analysis of gene expression showed that 2-AG treatment specifically induces CREBH but has no significant effect on activating transcription factor 6 (Atf6) gene expression (Fig. 1A, center panel), an ER stress-activated transcription factor belonging to the CREB/ATF family (13, 18). Induction of the sterol response element binding protein (Srebp)-1c mRNA level (Fig. 1A, right panel) and CB1R mRNA and protein levels (Fig. 1A, right panel) served as positive controls to ensure proper working of the experimental conditions. ER stress inducer Tunicamycin (Tm) showed a similar induction pattern of the CREBH mRNA level (Fig. 1B) in a time-dependent manner. Next, we examined the effect of 2-AG and Tm on activation of CREBH. Both 2-AG and Tm significantly activated Gal4DBD-fused CREBH (Fig. 1C), suggesting that both CB1R signaling and ER stress activates CREBH. ER-bound CREBH is cleaved upon activation to produce a 75-kDa (full-length) and 50-kDa (N-terminal active form) protein. Therefore, we examined whether 2-AG induces cleavage of CREBH (Fig. 1D). 2-AG- and, as expected, Tm-treated primary hepatocytes demonstrated cleavage of endogenous CREBH protein to generate its active form in a time-dependent manner. Overall, these initial observations suggest that CB1R signaling induces and activates CREBH gene expression.
FIGURE 1.
Induction and activation of Crebh by cannabinoid receptor 1 signaling. A and B, primary rat hepatocytes were treated with cannabinoid receptor 1 (CB1R) agonist 2-AG (A) or ER stress inducer Tunicamycin (Tm) (B) for the indicated time period, and total RNA was isolated for semiquantitative RT-PCR (A, left panel) or qPCR analysis for the 12-h time point samples (A, center panel) and northern hybridization B, H4IIE cells were treated with 2-AG for 12 h, and cell lysates were prepared for Western blot analysis using the indicated antibodies (A, right panel). Data represent mean ± S.D. of three individual experiments. *, p < 0.05 versus vehicle-treated cells. C, AML12 cells were cotransfected with the Gal4-Luc (50 ng) and Gal4DBD (200 ng) or Gal4DBD-CREBH (200 ng) expression vectors for 24 h followed by treatment with 2-AG and Tm for a further 12 h in serum-free media. Data are expressed in relative luciferase units and as the fold activation relative to the control. Data represent mean ± S.D. of three individual experiments.*, p < 0.05 versus control. D, primary rat hepatocytes were treated with 2-AG or Tm for the indicated time period, and cell lysates were prepared for Western blot analysis using the indicated antibodies. Data are representative of two individual experiments.
CB1R Signaling Induces CREBH Gene Expression via the JNK and AP-1 Pathways
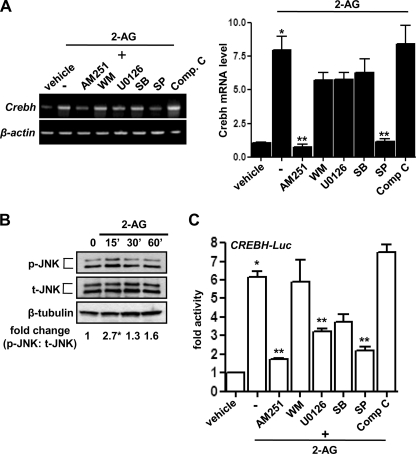
To evaluate the potential signaling pathways involved in CB1R signaling-mediated induction of CREBH gene expression, primary hepatocytes were pretreated with several specific inhibitors of cell signaling pathways before 2-AG treatment. Semiquantitative PCR analysis indicated that pretreatment of AM251 (a CB1R antagonist), U0126 (an inhibitor of ERK), and SP600125 (SP, an inhibitor of JNK) prevented CB1R-mediated induction of CREBH gene expression (Fig. 2A). However, there was no significant effect of Wortmannin (WM, an inhibitor of PI3 kinase), SB-203580 (SB, an inhibitor of p38MAPK), and compound C (Comp. C, an inhibitor of AMPK) on CREBH gene expression. 2-AG treatment also demonstrated an acute but transient phosphorylation of JNK (P-JNK) (Fig. 2B). Similarly, treatment of cell signaling inhibitors on CREBH gene promoter-preceding 2-AG treatment (Fig. 2C) demonstrated a similar inhibition pattern of the promoter activity, comparable with the CREBH mRNA level. These results suggest that CB1R signaling induces CREBH gene expression via the ERK1/2 and JNK pathways.
FIGURE 2.
The JNK signaling pathway mediates induction of Crebh by 2-AG. A, primary rat hepatocytes were pretreated with the Cb1r-specific antagonist AM251 (10 μm) or protein kinase inhibitors Wortmannin (WM, 0.1 μm), U0 (U0126, 10 μm), SB203580 (SB, 25 μm), SP600125 (SP, 25 μm), and compound C (Comp C, 10 μm) for 1 h followed by treatment with 2-AG for 12 h. Total RNA was isolated for semiquantitative RT-PCR analysis (left panel) and qPCR analysis (right panel) of Crebh mRNA expression and was normalized to β-actin expression. Data are representative of three individual experiments. B, primary rat hepatocytes were treated with 2-AG for the indicated time period, and cell lysates were prepared for Western blot analysis using the indicated antibodies. Data are representative of three individual experiments. C, AML12 cells were transfected with Crebh-Luc (100 ng). 24 h after transfection, cells were pretreated with the indicated inhibitors for 1 h preceding 2-AG treatment for a further 12 h in serum-free media. Data are expressed as the fold activation relative to the control, representing mean ± S.D. of three individual experiments.*, p < 0.05; **, p < 0.05 versus vehicle and 2-AG treated cells, respectively.
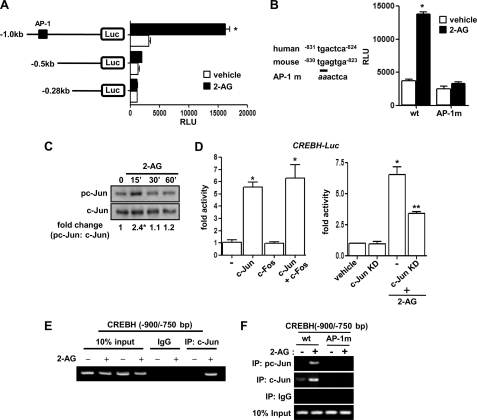
Next, we tried to delineate the molecular mechanism for transcriptional activation of CREBH by CB1R signaling. Treatment of 2-AG to several serial deletion constructs of the CREBH promoter revealed a putative AP-1 binding site that renders 2-AG responsiveness to the CREBH promoter (Fig. 3A). A point mutation of the AP-1 site (AP-1 m) dramatically abolished 2-AG-mediated activation of the CREBH gene promoter (Fig. 3B). Moreover, Western blot analysis revealed that 2-AG treatment significantly increased c-Jun phosphorylation (Fig. 3C). c-Jun homodimerization significantly activated the CREBH gene promoter, whereas c-Jun/c-fos heterodimerization showed no significant activation of the CREBH promoter compared with c-Jun alone (Fig. 3D, left panel). Furthermore, a kinase-dead mutant of c-Jun (c-Jun KD, S63/73A) cotransfection significantly inhibited 2-AG-mediated activation of the CREBH gene promoter (Fig. 3D, right panel). Finally, an endogenous ChIP assay in AML12 cells demonstrated c-Jun occupancy in the CREBH promoter region upon 2-AG treatment (Fig. 3E). This result was further confirmed by using wild-type and AP-1 mutant CREBH promoter constructs. 2-AG treatment significantly enhanced c-Jun phosphorylation and binding to the AP-1 site in the wild-type CREBH promoter but not in the AP-1 mutant promoter (Fig. 3F), thereby suggesting that the AP-1 site confers 2-AG responsiveness in the CREBH promoter. Overall, these data suggest that the JNK signal transduction pathway is a critical component for CB1R-mediated induction of CREBH.
FIGURE 3.
c-Jun mediated activation of the Crebh gene promoter by Cb1r signaling. A and B, AML12 cells were transfected with several deletion constructs (A) or the AP-1 mutant (AP-1 m) construct of Crebh-Luc (100 ng) (B) and treated with 2-AG as indicated. Data are expressed as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.001 versus control. C, primary rat hepatocytes were treated with 2-AG for the indicated time period, and cell lysates were prepared for Western blot analysis using the indicated antibodies. Data are representative of three individual experiments. D, AML12 cells were cotransfected with Crebh-Luc (100 ng) and c-Jun and c-Fos expression vectors (400 ng each) (left panel) or treated with 2-AG in the absence or presence of cotransfected c-Jun KD (400 ng each) (right panel). Data are expressed as the fold activation relative to the control, representing mean ± S.D. of three individual experiments.*, p < 0.05; **, p < 0.05 versus control and 2-AG treated cells, respectively. E, endogenous ChIP assay. AML12 cells were treated with vehicle or 2-AG for 12 h. Soluble chromatin was prepared and immunoprecipitated with monoclonal antibody against c-Jun or IgG only as indicated. 10% of the soluble chromatin was used as input. A semiquantitative RT-PCR analysis was performed to determine the binding of c-Jun to the endogenous Crebh gene promoter. F, ChIP assay. AML12 cells were transfected with Crebh-Luc (-1000/+1) (200 ng) wild-type (wt), or AP-1 mutant, and following transfection and serum starvation, cells were treated with vehicle or 2-AG for 12 h. Soluble chromatin was prepared and immunoprecipitated with monoclonal antibody against c-Jun, phospho-c-Jun, or IgG only as indicated. 10% of the soluble chromatin was used as input. A semiquantitative RT-PCR analysis was performed to determine the binding of c-Jun and phospho-c-Jun to the endogenous or to the transfected Crebh promoter constructs. Data are representative of three independent experiments.
CB1R Signaling Induces Hepatic Gluconeogenesis in Primary Hepatocytes
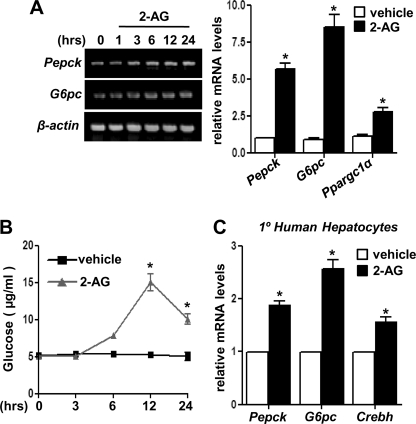
Several studies have demonstrated a correlation between CB1R signaling and glucose metabolism (7, 11). However, to date, no clear evidence has been presented to elucidate the role of CB1R signaling on hepatic gluconeogenesis. Therefore, we addressed the effect of CB1R signaling on hepatic gluconeogenesis. 2-AG (Fig. 4A) treatment in primary rat hepatocytes significantly induced mRNA levels of the key gluconeogenic enzyme genes Pepck and G6pc as well as peroxisome proliferator-activated receptor-γ coactivator 1α (Ppargc1α) in a time-dependent manner. 2-AG treatment also demonstrated a significant increase in glucose production in hepatocyte culture media (Fig. 4B). Furthermore, 2-AG treatment in primary human hepatocytes showed a significant induction of CREBH along with Pepck and G6pc mRNA levels (Fig. 4C), thereby suggesting that CB1R signaling induces hepatic gluconeogenesis in primary hepatocytes.
FIGURE 4.
Induction of hepatic gluconeogenesis by Cb1r signaling. A–C, primary rat hepatocytes (A and B) or primary human hepatocytes (C) were treated with 2-AG, and total RNA was isolated (A and C) for semiquantitative RT-PCR (A, left panel) or qPCR analysis after 12 h treatment (A, right panel, and C). Measurement of glucose production (B) was performed using glucose-free media supplemented with gluconeogenic substrate sodium lactate (20 mm) and sodium pyruvate (1 mm) after 12-h 2-AG treatment. Data represent mean ± S.D. of three individual experiments. *, p < 0.05 versus vehicle treated cells.
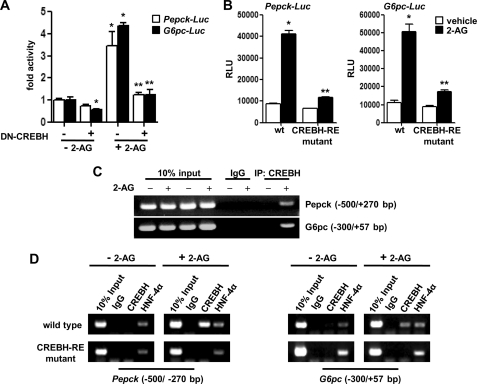
CREBH Is Required for CB1R Signaling-mediated Activation of the Pepck and G6pc Gene Promoters
Next, we tried to elucidate the molecular mechanism of 2-AG-mediated induction of hepatic gluconeogenic enzyme genes and the role of CREBH in mediating this effect. 2-AG treatment demonstrated a significant induction of the Pepck and G6pc gene promoters. Interestingly, cotransfection with the dominant negative mutant of CREBH (DN-CREBH) impaired the 2-AG-mediated transcriptional activation of the Pepck and G6pc promoters by ∼ 85% (Fig. 5A). In our previous study, we characterized the binding site of CREBH in both Pepck and G6pc gene promoters (14). Accordingly, wild-type promoters of Pepck and G6pc were significantly activated upon 2-AG treatment, whereas 2-AG responsiveness in the CREBH-binding site mutant (CREBH-RE mutant) promoters were blunted significantly (∼80% decrease compared with the wild-type constructs) (Fig. 5B). Next, we assessed the binding of CREBH to the endogenous Pepck and G6pc gene promoters upon 2-AG treatment (Fig. 5C), and finally, endogenous CREBH binding to the CREBH-response elements in the Pepck and G6pc gene promoters upon activation of CB1R signaling was confirmed by ChIP assay with CREBH-specific antibody. Hepatocyte Nuclear Factor (HNF)-4α binding was demonstrated as a positive control (Fig. 5D).
FIGURE 5.
Crebh is required for Cb1r signaling-mediated activation of Pck1 and G6pc gene promoters. A and B, AML12 cells were cotransfected with Pepck-luc or G6pc-Luc (100 ng each) in the absence or presence of DN-CREBH expression vector (A) or transfected with wild-type and CREBH-RE mutant promoter constructs for Pck1 or G6pc (B), as indicated. 24 h post-transfection, cells were exposed to 2-AG treatment for a further 12 h in serum-free media. Data are expressed as the fold activation relative to the control, representing mean ± S.D. of three individual experiments.*, p < 0.05; **, p < 0.05 versus control and 2-AG treated cells in the absence of cotransfected DN-CREBH (A) or CREBH-RE mutant promoters (B), respectively. C, endogenous ChIP assay. AML12 cells were treated with vehicle or 2-AG for 12 h. Soluble chromatin was prepared and immunoprecipitated with monoclonal antibody against CREBH or IgG only as indicated. 10% of the soluble chromatin was used as input. A semiquantitative RT-PCR analysis was performed to determine the binding of c-Jun to the endogenous Pepck and G6pc gene promoters. D, ChIP assay. AML12 cells were transfected with Pepck-Luc or G6pc-Luc (200 ng) wild-type or CREBH-RE mutant. Following transfection and serum starvation, cells were treated with vehicle or 2-AG for 12 h. Soluble chromatin was prepared and immunoprecipitated with monoclonal antibody against CREBH, HNF-4α, or IgG only as indicated. 10% of the soluble chromatin was used as input. A semiquantitative RT-PCR analysis was performed using primers amplifying the indicated regions of the Pepck and G6pc promoter to determine the binding of CREBH and HNF-4α after 2-AG treatment. Data are representative of three independent experiments.
As expected, 2-AG treatment significant enhanced CREBH binding to the endogenous (Fig. 5C) as well as to the wild-type promoters but no binding was observed in the mutant promoter constructs (Fig. 5D). Endogenous HNF-4α binding to the Pepck and G6pc promoters (wild-type and CREBH-RE mutant) remained unaffected in the absence or presence of 2-AG treatment. These data indicate that CB1R signaling-mediated CREBH occupancy plays a key role in the activation of gluconeogenic gene promoters.
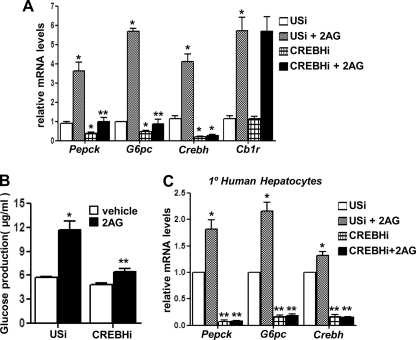
Knockdown of CREBH Attenuates CB1R Signaling-mediated Up-regulation of Hepatic Gluconeogenesis
To clarify the role of CREBH in gluconeogenic gene expression during CB1R signaling activation in primary rat hepatocytes, we used adenoviral-mediated knockdown of CREBH expression (CREBH shRNA, CREBHi). 2-AG treatment led to a marked induction of gluconeogenic enzyme genes Pepck and G6pc as well as CREBH and CB1R mRNA levels in USi-infected hepatocytes but failed to significantly induce Pepck or G6pc expression in CREBH shRNA (CREBHi)-infected hepatocytes (Fig. 6A) or any significant increase of glucose production in hepatocytes (B). Furthermore, 2-AG treatment demonstrated a similar significant induction pattern of Pepck, G6pc, and CREBH expression in primary human hepatocytes, and knockdown of endogenous CREBH expression dramatically abolished the 2-AG-mediated induction of Pepck and G6pc (Fig. 6C). Interestingly, knockdown of CREBH alone led to significant reduction of basal Pepck and G6pc mRNA levels in primary human hepatocytes. Overall, these results suggest a key role of CREBH in mediating CB1R signaling and ER stress-induced hepatic gluconeogenesis.
FIGURE 6.
Knockdown of Crebh expression attenuates Cb1r signaling-mediated up-regulation of hepatic gluconeogenesis. A and B, primary rat hepatocytes were infected with adenovirus USi (50 multiplicity of infection) or CREBH shRNA (CREBHi, 50 multiplicity of infection) for 48 h followed by treatment with 2-AG for 12 h, and total RNA was isolated for qPCR analysis (A). Measurement of glucose production (B) was performed using glucose-free media supplemented with gluconeogenic substrate sodium lactate (20 mm) and sodium pyruvate (1 mm). Data represent mean ± S.D. of three individual experiments. *, p < 0.05; **, p < 0.001 versus vehicle and 2-AG treated cells in the absence of CREBHi overexpression, respectively. C, primary human hepatocytes were infected with adenovirus USi (50 multiplicity of infection) or CREBH shRNA (CREBHi, 50 multiplicity of infection) for 48 h followed by treatment with 2-AG for 12 h, and total RNA was isolated for qPCR analysis. Data represent mean ± S.D. of three individual experiments. *, p < 0.05; **, p < 0.001 versus vehicle and 2-AG treated cells in the absence of CREBHi overexpression, respectively.
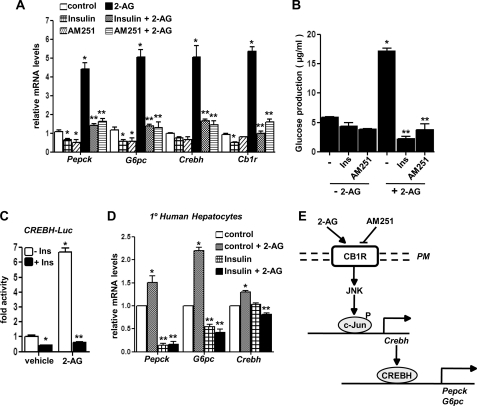
AM251, a CB1R-specific Antagonist, and Insulin Reverse CB1R Signaling-mediated Induction of Hepatic Gluconeogenesis
Finally, we tried to investigate the effect of CB1R antagonist AM251 and insulin on CB1R signaling-mediated activation of CREBH and the subsequent increase in hepatic gluconeogenesis. Interestingly, treatment of either AM251 or insulin significantly down-regulated 2-AG-mediated induction of Pepck and G6pc gene expression along with the CREBH mRNA level in primary rat hepatocytes (Fig. 7A), and as a consequence of this decrease in gene expression, we observed a dramatic decrease in glucose production in primary hepatocytes upon AM251 and insulin treatment (Fig. 7B). Interestingly, insulin showed significant inhibition of the 2-AG-activated CREBH gene promoter (Fig. 7C). Consistent with these data, insulin significantly decreased mRNA levels of the 2-AG-induced Pepck, G6pc, and CREBH genes in primary human hepatocytes (Fig. 7D). Overall, these findings, in both rodent and human primary hepatocytes, strongly indicate CREBH to play a critical role in mediating CB1R signaling and ER stress-induced hepatic gluconeogenesis.
FIGURE 7.
Cb1r-specific antagonist AM251 and insulin reverses 2-AG mediated induction of hepatic gluconeogenesis. A and B, primary rat hepatocytes were pretreated with insulin (Ins, 10 nm) or AM251 for 1 h followed by treatment with 2-AG in the continued presence or absence of insulin or AM251 for 12 h, and total RNA was isolated for qPCR analysis. Measurement of glucose production (B) was performed using glucose-free media supplemented with gluconeogenic substrate sodium lactate (20 mm) and sodium pyruvate (1 mm). Data represent mean ± S.D. of three individual experiments. *, p < 0.05; **, p < 0.001 versus control and 2-AG, respectively. C, AML12 cells were transfected with Crebh-Luc (100 ng) for 24 h. Cells were then pretreated with insulin for 1 h followed by treatments with 2-AG for 12 h in serum-free media as indicated. Data are expressed as the fold activation relative to the control, representing mean ± S.D. of three individual experiments. *, p < 0.05; **, p < 0.001 versus vehicle and 2-AG treated cells, respectively. D, primary human hepatocytes were pretreated with insulin (Ins, 10 nm) for 1 h followed by treatment with 2-AG in the continued presence or absence of insulin for 12 h, and total RNA was isolated for qPCR analysis. Data represent mean ± S.D. of three individual experiments. *, p < 0.05; **, p < 0.05 versus control and 2-AG, respectively. E, schematic depiction of CB1R-mediated induction of hepatic gluconeogenesis. CB1R signaling phosphorylates JNK, leading to phosphorylation and activation of c-Jun. Phosphorylated c-Jun activates and induces Crebh gene expression, which in turn induces hepatic gluconeogenesis via induction of Pepck and G6pc gene expression and increases hepatic glucose production.
DISCUSSION
The current findings delineate a novel mechanism by which the hepatic endocannabinoid-CB1R signaling system induces and activates ER-bound stress-sensory transcription factor CREBH via the JNK signaling pathway. Activation of CREBH leads to the induction of the key hepatic gluconeogenic enzyme genes Pepck and G6pc and increases glucose production in primary hepatocyte culture (Fig. 7E). Several findings have demonstrated the role of CB1R signaling in diet-induced obesity and insulin resistance conditions, suggesting a critical role of cannabinoid signaling in regulating glucose metabolism (2–7). However, to date, there is no evidence of involvement of CB1R signaling in initiating a stress condition and directly affecting hepatic gluconeogenic enzyme gene expression. Considering the importance of the endocannabinoid system and stress signaling in modulating various metabolic pathways (7, 19, 20), our present findings shed light into the detailed molecular mechanism of action of the hepatic CB1R signaling pathway, which may provide important clues in controlling glucose homeostasis.
Previous studies have demonstrated the endocannabinoid system to be one of the most relevant regulators of energy balance. Blockade of CB1 receptors causes a reduction in food intake and a sustained weight loss and enhances insulin sensitivity in both humans and rodents (7). It is well established that the endocannabinoid system contributes to the control of lipid and glucose metabolism. However, not much information is available regarding the downstream signaling pathway involved in mediating the effects of endocannabinoid and activated CB1R in liver. Our present findings identify the downstream kinase (JNK) and a novel target of CB1R (CREBH) that connects the endocannabinoid system to the dysregulation of hepatic glucose homeostasis. CB1R has been reported previously to activate the ERK, p38MAPK, PI3K, and JNK signaling pathways (21). A recent report demonstrated that CB1R agonist, AEA, treatment in skeletal muscle cells activated ERK and p38MAPK signaling and impaired insulin-stimulated Akt phosphorylation, leading to insulin-resistant conditions in skeletal muscles (22). Our results indicate that 2-AG-mediated induction and activation of CREBH occurs via JNK activation and c-Jun phosphorylation, indicating a broad spectrum of the mode of action of CB1R that may be tissue-specific in nature. However, our results using various signaling pathway inhibitors to determine the specific protein kinase involved in 2-AG-mediated activation of the CREBH promoter (Fig. 2C) suggest the possibility of cross-talk between more than one signaling pathway, although JNK seemed to be the predominant kinase involved in this phenomenon. Moreover, it has also been reported that the biosynthesis and degradation of AEA and 2-AG occur through distinct pathways and likely result in a distinct mode of action of these two agonists (1). Another previous report demonstrated ER stress-mediated induction of hepatic gluconeogenesis and hepatic glucose production (19). However, it was unclear regarding the mechanism by which ER stress induced gluconeogenesis. Our findings indicate CREBH to be the missing link that connects ER stress to hepatic gluconeogenesis.
In addition to glucose metabolism, endocannabinoid-activated CB1R signaling has been implicated in dysregulation of the lipid metabolism in the liver (7, 11, 12). Recent studies using global and liver-specific knockdown of CB1R in mice has demonstrated resistance to high-fat diet induced obesity (11) and chronic alcohol-induced hepatic steatosis (23). CB1R activation has been shown to directly stimulate hepatic gene expression of the lipogenic transcription factor Srebp-1c and its downstream targets acetyl-CoA carboxylase-1 (Acc1) and fatty acid synthase (Fas), leading to increased lipogenesis. Interestingly, both Srebp-1c and CREBH are ER-resident transcription factors and are regulated by regulated intramembrane proteolysis in response to ER stress (13) that leads to activation of their downstream targets and metabolic pathways, lipogenesis and gluconeogenesis, respectively. However, our results suggest that CB1R signaling has no observable effect on another ER-bound transcription factor, Atf6. A recent report (24) indicated a negative regulatory role of Atf6 in hepatic gluconeogenesis by interacting with and inhibiting CREB-regulated transcription coactivator 2 (CRTC2) function, whereas our previous study demonstrated that CRTC2 potentiates the CREBH-mediated induction of Pepck and G6pc gene expression. These interesting but opposite observations indicate the possibility of an early response and delayed-response phase under various ER stress condition.
Clinical data has demonstrated that serum levels of both 2-AG and AEA were increased in type II diabetic patients and the circulating 2-AG level was significantly correlated with body fat, visceral fat mass, fasting plasma insulin concentrations and C-reactive protein (CRP) level (7). In this regard, a previous study has reported that CREBH induces CRP gene expression and CREBH knock-out mice demonstrated a significant reduction in CRP mRNA level (13). Our results indicate that CREBH knockdown in primary rat and human hepatocytes led to significant attenuation of CB1R signaling and ER stress-induced gluconeogenic gene expression along with reduction in glucose production. Future studies with liver-specific ablation of CREBH in genetically engineered rodent models exposed to ER stress or diet-induced obese conditions will provide a clear picture regarding the role of CREBH in mediating these effects. The pharmacological blockade of CB1R by Rimonabant (RIO, Rimonabant in Obesity) as well as a Rimonabant diabetes trial demonstrated the efficacy of CB1 receptor antagonism in treating insulin resistance conditions as well as the improvement of glucose homeostasis in humans (25, 26). Similar results have been obtained using AM251, which was also able to decrease circulating plasma glucose and enhance glucose tolerance following glucose load in chronically treated ob/ob mice (9). In accordance with these observations, our results in human and rat primary hepatocytes indicate that blockade of CB1R signaling by insulin or AM251 resulted in a dramatic decrease in CREBH gene expression and subsequently led to decrease in Pepck and G6pc gene expression and glucose production. However, possible neuropsychiatric side effects because of non-selective pharmacological inhibition of CB1R have led to withdrawal of Rimonabant from clinical trials. Therefore, detailed elucidation of the molecular mechanism involved in CB1R-mediated up-regulation of hepatic gluconeogenesis may provide alternative options for potential therapeutic purposes in diet-induced obese conditions. In this regard, in vitro studies have reported that treatment with the CB1 antagonist AM251 increased mRNA levels of the α 1 isoform of AMP-activated protein kinase (AMPKα1) in myotubes from lean and obese patients, an effect that might lead to fatty acid oxidation (27). Recent studies from our group have also demonstrated the beneficial effects of pharmacological activation of AMPKα1 in rodent models by the anti-diabetic drug metformin (28) and the anti-lipidemic drug fenofibrate (29) in the diabetic and non-alcoholic steatohepatitis settings, respectively. Furthermore, our recent study indicates that induction of CREBH gene expression can be attenuated by metformin-induced induction of orphan nuclear receptor small heterodimer partner (SHP, NR0B2) gene expression (14). With detailed elucidation of the molecular mechanism of CB1R signaling action, our current findings might provide interesting clues to further therapeutic options in the treatment of the hyperactivated endocannabinoid system state.
In summary, our study demonstrates the molecular mechanism of endocannabinoid system-mediated regulation of hepatic gluconeogenesis involving the activation of the JNK signaling pathway and the ER-bound transcription factor CREBH and unravels an unprecedented link between hepatic CB1R and stress signaling to initiate a perturbation in glucose homeostasis in hepatocytes.
Acknowledgment
We would like to thank Dr. Seok-Yong Choi for critical reading of the manuscript and helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants DK44442 and DK58379 (to J. Y. L. C.). This work was also supported by Future-based Technology Development Program (BIO Fields) through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology Grant 2010-0019512 (to H.-S. C.); by Korea Healthcare Technology R&D Project, Ministry for Health, Welfare, and Family Affairs, Republic of Korea Grant A100588 (to H.-S. C.); and by National Creative Research Initiatives Grant 20110018305 funded by the Korean Ministry of Education, Science and Technology (to H.-S. C.).
- AEA
- arachidonoyl ethanolamide
- 2-AG
- 2-arachidonoyl glycerol
- CB1R
- cannabinoid receptor type 1
- CREBH
- cAMP-responsive element-binding protein H
- ER
- endoplasmic reticulum
- AP-1
- activator protein 1
- Pepck
- phosphoenolpyruvate carboxykinase
- G6pc
- glucose-6-phosphatase catalytic subunit
- USi
- unspecific RNAi
- qPCR
- quantitative PCR
- Tm
- Tunicamycin
- HNF
- hepatocyte nuclear factor
- RE
- response element
- DBD
- DNA-binding domain
- UAS
- upstream activation sequence
- TK
- thymidine kinase
- ATF
- activating transcription factor.
REFERENCES
- 1. Pacher P., Bátkai S., Kunos G. (2006) Pharmacol. Rev. 58, 389–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matias I., Gonthier M. P., Orlando P., Martiadis V., De Petrocellis L., Cervino C., Petrosino S., Hoareau L., Festy F., Pasquali R., Roche R., Maj M., Pagotto U., Monteleone P., Di Marzo V. (2006) J. Clin. Endocrinol. Metab. 91, 3171–3180 [DOI] [PubMed] [Google Scholar]
- 3. Osei-Hyiaman D., DePetrillo M., Pacher P., Liu J., Radaeva S., Bátkai S., Harvey-White J., Mackie K., Offertáler L., Wang L., Kunos G. (2005) J. Clin. Invest. 115, 1298–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Di Marzo V., Goparaju S. K., Wang L., Liu J., Bátkai S., Járai Z., Fezza F., Miura G. I., Palmiter R. D., Sugiura T., Kunos G. (2001) Nature 410, 822–825 [DOI] [PubMed] [Google Scholar]
- 5. Engeli S., Böhnke J., Feldpausch M., Gorzelniak K., Janke J., Bátkai S., Pacher P., Harvey-White J., Luft F. C., Sharma A. M., Jordan J. (2005) Diabetes 54, 2838–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blüher M., Engeli S., Klöting N., Berndt J., Fasshauer M., Bátkai S., Pacher P., Schön M. R., Jordan J., Stumvoll M. (2006) Diabetes 55, 3053–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nogueiras R., Diaz-Arteaga A., Lockie S. H., Velásquez D. A., Tschop J., López M., Cadwell C. C., Diéguez C., Tschöp M. H. (2009) Pharmacol. Res. 60, 93–98 [DOI] [PubMed] [Google Scholar]
- 8. Ravinet Trillou C., Arnone M., Delgorge C., Gonalons N., Keane P., Maffrand J. P., Soubrie P. (2003) Am. J. Physiol. Regul. Integr. Comp. Physiol. 284, R345–353 [DOI] [PubMed] [Google Scholar]
- 9. Irwin N., Hunter K., Frizzell N., Flatt P. R. (2008) Eur. J. Pharmacol. 581, 226–233 [DOI] [PubMed] [Google Scholar]
- 10. Nogueiras R., Veyrat-Durebex C., Suchanek P. M., Klein M., Tschöp J., Caldwell C., Woods S. C., Wittmann G., Watanabe M., Liposits Z., Fekete C., Reizes O., Rohner-Jeanrenaud F., Tschöp M. H. (2008) Diabetes 57, 2977–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Osei-Hyiaman D., Liu J., Zhou L., Godlewski G., Harvey-White J., Jeong W. I., Bátkai S., Marsicano G., Lutz B., Buettner C., Kunos G. (2008) J. Clin. Invest. 118, 3160–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Després J. P., Golay A., Sjöström L. (2005) N. Engl. J. Med. 353, 2121–2134 [DOI] [PubMed] [Google Scholar]
- 13. Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D. T., Back S. H., Kaufman R. J. (2006) Cell 124, 587–599 [DOI] [PubMed] [Google Scholar]
- 14. Lee M. W., Chanda D., Yang J., Oh H., Kim S. S., Yoon Y. S., Hong S., Park K. G., Lee I. K., Choi C. S., Hanson R. W., Choi H. S., Koo S. H. (2010) Cell Metab. 11, 331–339 [DOI] [PubMed] [Google Scholar]
- 15. Vecchi C., Montosi G., Zhang K., Lamberti I., Duncan S. A., Kaufman R. J., Pietrangelo A. (2009) Science 325, 877–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu J., Kaufman R. J. (2006) Cell Death Differ. 13, 374–384 [DOI] [PubMed] [Google Scholar]
- 17. Chanda D., Li T., Song K. H., Kim Y. H., Sim J., Lee C. H., Chiang J. Y., Choi H. S. (2009) J. Biol. Chem. 284, 28510–28521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haze K., Yoshida H., Yanagi H., Yura T., Mori K. (1999) Mol. Biol. Cell 10, 3787–3799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang D., Wei Y., Schmoll D., Maclean K. N., Pagliassotti M. J. (2006) Endocrinology 147, 350–358 [DOI] [PubMed] [Google Scholar]
- 20. Scheuner D., Kaufman R. J. (2008) Endocr. Rev. 29, 317–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guzmán M. (2003) Nat. Rev. Cancer 3, 745–755 [DOI] [PubMed] [Google Scholar]
- 22. Eckardt K., Sell H., Taube A., Koenen M., Platzbecker B., Cramer A., Horrighs A., Lehtonen M., Tennagels N., Eckel J. (2009) Diabetologia 52, 664–674 [DOI] [PubMed] [Google Scholar]
- 23. Jeong W. I., Osei-Hyiaman D., Park O., Liu J., Bátkai S., Mukhopadhyay P., Horiguchi N., Harvey-White J., Marsicano G., Lutz B., Gao B., Kunos G. (2008) Cell Metab. 7, 227–235 [DOI] [PubMed] [Google Scholar]
- 24. Wang Y., Vera L., Fischer W. H., Montminy M. (2009) Nature 460, 534–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pi-Sunyer F. X., Aronne L. J., Heshmati H. M., Devin J., Rosenstock J. (2006) JAMA 295, 761–775 [DOI] [PubMed] [Google Scholar]
- 26. Van Gaal L. F., Rissanen A. M., Scheen A. J., Ziegler O., Rössner S. (2005) Lancet 365, 1389–1397 [DOI] [PubMed] [Google Scholar]
- 27. Cavuoto P., McAinch A. J., Hatzinikolas G., Cameron-Smith D., Wittert G. A. (2007) Mol. Cell Endocrinol. 267, 63–69 [DOI] [PubMed] [Google Scholar]
- 28. Kim Y. D., Park K. G., Lee Y. S., Park Y. Y., Kim D. K., Nedumaran B., Jang W. G., Cho W. J., Ha J., Lee I. K., Lee C. H., Choi H. S. (2008) Diabetes 57, 306–314 [DOI] [PubMed] [Google Scholar]
- 29. Chanda D., Lee C. H., Kim Y. H., Noh J. R., Kim D. K., Park J. H., Hwang J. H., Lee M. R., Jeong K. H., Lee I. K., Kweon G. R., Shong M., Oh G. T., Chiang J. Y., Choi H. S. (2009) Hepatology 50, 880–892 [DOI] [PMC free article] [PubMed] [Google Scholar]