Abstract
The enzyme acyl-CoA:lysophosphatidylcholine acyltransferase (Lpcat1) is a critical cytosolic enzyme needed for lung surfactant synthesis that catalyzes an acyltransferase reaction by adding a palmitate to the sn-2 position of lysophospholipids. Here we report that histone H4 protein is subject to palmitoylation catalyzed by Lpcat1 in a calcium-regulated manner. Cytosolic Lpcat1 was observed to shift into the nucleus in lung epithelia in response to exogenous Ca2+. Nuclear Lpcat1 colocalizes with and binds to histone H4, where it catalyzes histone H4 palmitoylation. Mutagenesis studies demonstrated that Ser47 within histone H4 serves as a putative acceptor site, indicative of Lpcat1-mediated O-palmitoylation. Lpcat1 knockdown or expression of a histone H4 Ser47A mutant protein in cells decreased cellular mRNA synthesis. These findings provide the first evidence of a protein substrate for Lpcat1 and reveal that histone lipidation may occur through its O-palmitoylation as a novel post-translational modification. This epigenetic modification regulates global gene transcriptional activity.
Keywords: Calcium, Lung, Nuclear Translocation, Phosphatidylcholine, Phospholipid
Introduction
The enzyme acyl-CoA:lysophosphatidylcholine acyltransferase (Lpcat1) was recently cloned from lung epithelia and is indispensable for generation of pulmonary surfactant. Lpcat1 catalyzes an O-acyltransferase reaction by covalently adding saturated acyl-CoAs (palmitoyl groups, 16:0) to its acceptor lysophospholipids. Specifically, in the lung, it catalyzes an oxyester linkage between palmitate and lysophosphatidylcholine to generate dipalmitoylphosphatidylcholine, the major surface tension-lowering component of pulmonary surfactant in the remodeling pathway (1, 2). However, Lpcat1 appears to be somewhat promiscuous and acts on substrates that include lysophosphatidylcholine, lysoplasmanylcholine, and lysophosphatidylglycercol (1–3). Notably, in addition to lipid substrates, O-acyltransferases can also lipidate some protein substrates (4–7). These enzymes share a highly conserved histidine residue within a catalytic core that is essential for their functionality.
Protein palmitoylation, a prototypical form of lipidation, is an important post-translational modification that occurs ubiquitously in eukaryotes. Protein palmitoylation is generally categorized as S-palmitoylation, N-palmitoylation, and O-palmitoylation based on the chemical linkage between donor and acceptor substrates. Among these, S-palmitoylation is best characterized and involves generation of a reversible thioester bond between a cysteine residue and a palmitate group (8). As many as 250 proteins were found to be modified by S-palmitoylation in mammalian neurons, and these reactions are largely catalyzed by aspartate-histidine-histidine-cysteine (DHHC) palmitoyl acyltransferase family members (8, 9). The biochemistry of N-palmitoylation and O-palmitoylation, however, is less studied. One known substrate for N-palmitoylation is the Sonic Hedgehog protein because its NH2-terminal cysteine is palmitoylated via an amide linkage by Hedgehog acyltransferase (Hhat) (5). O-Palmitoylation is exemplified by Wnt/Wg proteins that harbor an oxyester linkage between monounsaturated palmitate and a serine residue. Another recently described O-palmitoylation target, the peptide hormone preghrelin, is modified by octanoylation at a serine residue. The enzymes that catalyze the oxyester linkages within Wnt/Wg proteins and preghrelin are porcupine (6) and ghrelin O-acyltransferase (7), respectively. The physiological consequences of palmitoylation are diverse; by increasing substrate hydrophobicity, these lipidation reactions appear to modulate interactions of substrates with other biomolecules, often affecting signal transduction, protein stability, intracellular trafficking, and localization (10–12).
Histone family proteins are the protein components of the nucleosome that act as a scaffold for DNA packaging in the nucleus. The protein components in a nucleosome contain two sets of histone H2A, H2B, H3, and H4. Histone proteins are highly vulnerable to post-translational modification at specific amino acids. The covalent post-translational modifications include but are not limited to methylation (13), acetylation (14), phosphorylation (15), ubiquitination (16), sumoylation (17), biotinylation (18), and ADP-ribosylation (19). To date, only one study has recently demonstrated by mass screening histone S-palmitoylation; however, the identity of the acyltransferase that catalyzes this reaction was not investigated (20). We showed previously that intracellular Ca2+ concentrations increase after bacterial infection in lung epithelia (21). We report here that the acyltransferase Lpcat1 shifts into the nucleus in response to pathophysiological levels of Ca2+ observed in lung lining fluid. Once within the nucleus, Lpcat1 interacts with histone H4 protein, thereby catalyzing its O-palmitoylation in a site-specific manner. Lpcat1-mediated histone H4 palmitoylation subsequently regulates global transcriptional activity.
EXPERIMENTAL PROCEDURES
Cell Cultures and Reagents
Murine lung epithelial (MLE-12)2 cells were maintained with HITES medium supplemented with 10% FBS in a 37 °C incubator and 5% CO2 as described previously (22). Human embryonic kidney (HEK) 293FT cells were cultured in modified Eagle's medium supplemented with 10% fetal bovine serum, 100 units/ml penicillin/streptomycin, and 2 mm l-glutamine. Anti-histone H4 antibody was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Active RNA polymerase II antibody was from Millipore (Billerica, MA). Histone H4 recombinant protein, palmitic acid, and palmitoyl-CoA were from Sigma. Lpcat1 and serine palmitoyltransferase long chain base subunit 2 (SPTLC2) recombinant proteins and Lpcat1 small hairpin RNA (shRNA) plasmid were from Origene (Rockville, MD). [14C]Palmitoyl-CoA, [3H]palmitic acid, and [3H]oleic acid were from PerkinElmer Life Sciences.
Fluorescent Immunostaining
MLE cells at a concentration of 105 cells/ml were inoculated into glass-bottomed 35-mm plates for 48 h. Cells were cultured with different concentrations of Ca2+ for 2 h, and the treated cells were washed with ice-cold PBS twice and fixed with 4% paraformaldehyde for 1 h. To penetrate the membrane, we incubated the fixed cells with wash buffer (0.5% of Triton X-100 in PBS) for 30 min. The cells were then blocked with 5% BSA for 1 h, followed by rabbit anti-Lpcat1 antibody (1:500) and mouse anti-histone H4 antibody (1:200) double immunostaining for 2 h. Plates were washed three times and incubated with fluorescence-conjugated secondary antibodies for another 1 h. Plates were then washed three times for 10 min. To visualize the nucleus of the cells, we added one drop of DAPI solution (Invitrogen) on plates before microscopy. The images were acquired by a combination laser-scanning microscope system (Nikon A1, Nikon (Mellville, NY)), and the results were analyzed through Nikon NIS-Elements software.
Extraction of Nuclear and Cytosolic Proteins
MLE cells in their exponent growth stage were treated with 2 mm CaCl2 for 2 h. Cells were washed with PBS buffer twice and collected by scraping. Harvested cells were washed with PBS, and the cell pellets were collected by centrifuging at 1500 rpm for 10 min. The pelleted cells were suspended in 1 ml of cytoplasmic membrane lysis buffer (10 mm Hepes, pH 8.0, 1.5 mm MgCl2, 10 mm KCl, 1 mm DTT, 0.1% Igepal® CA-630, and 1:1000 protease inhibitor mixture). The cell suspension was centrifuged at 2000 rpm for 5 min, and the supernatant was collected as the cytosolic fraction and condensed by Amicon Ultra filters. The pellet was lysed with 150 ml of nuclear envelope lysis buffer (20 mm Hepes, pH 8.0, 1.5 mm MgCl2, 420 mm NaCl, 1 mm DTT, 0.2 mm EDTA, 25% (v/v) glycerol, and 1:1000 protease inhibitor mixture) and cleared at 13,000 rpm for 10 min; the supernatant was collected as the nuclear fraction. The fractions were subjected to immunoblot analysis.
Immunoprecipitation, Immunoblotting, and Microsome Isolation
MLE cells during exponential growth were treated with 2 mm Ca2+ for 2 h, and the cells were lysed with lysis buffer (0.3% Triton X-100 (v/v) in PBS and 1:1000 protease inhibitor mixture). Cells were subject to sonication, and samples were spun at 13,000 rpm for 10 min. Cell lysates (containing 1 mg of protein) were incubated and rotated with 2 μg of anti-histone H4 antibody at 4 °C overnight. After incubation with 30 μl of protein A/G-agarose beads for another 3 h, beads were spun down and washed with lysis buffer three times. The washed beads were mixed with SDS-PAGE loading dye prior to SDS-PAGE and immunoblot analysis. Immunoblotting was performed as described previously (22). Microsomes were isolated as described previously (22).
In Vitro Palmitoylation
For the in vitro palmitoylation assay, we used 14C-labeled palmitoyl-CoA to label histone H4 recombinant protein in the presence or absence of Lpcat1. [14C]Palmitoyl-CoA was dried and solubilized in 5× palmitoylation buffer by sonication (final working concentration 65 mm Tris-HCl, pH 7.4, 10 mm MgCl2, 12.5 mm fatty acid-free BSA, and 2 mm EDTA) as we described previously (22). Each reaction contained 1 μg of histone H4 recombinant protein, 2 μCi of [14C]palmitoyl-CoA, and 10 ng of either recombinant Lpcat1, heat-inactivated Lpcat1, or SPTLC2. The reaction was conducted at 37 °C for 30 min in a 50-μl volume. Reactions were terminated by the addition of 1:1 volumes of SDS-PAGE loading dye containing 20 mm DTT. The samples were separated using 10–20% Tris-Tricine gels followed by autoradiography. The corresponding spots in the gels were cut, and the radioactivity was quantitatively measured by scintillation counting. In selected studies, autoradiograms were developed by exposing blots to using high intensity film.
In Vivo Palmitoylation
MLE cells were pulsed with radiolabeled fatty acids in the presence of 2 mm Ca2+ for 2 h. Prior to cellular labeling, [3H]palmitic acid or [3H]oleic acid was dried under nitrogen gas and solubilized in culture medium by sonication. Cells were incubated in serum-free medium for 1 h with the inclusion of radiolabeled fatty acids at a final concentration of 5 μCi and 2 mm Ca2+. The labeled cells were then lysed, and cell lysates were subjected to anti-histone H4 immunoprecipitation followed by SDS-PAGE, autoradiography, or scintillating counting.
Mutagenesis
Histone H4 cDNA was purchased from Open Biosystems (Huntsville, AL), and the coding region of the gene was cloned into pcDNA 3.1 by using the following primers: forward (5′-atgtcaggacgaggaaaagg-3′) and reverse (5′-ttagccgccgaagccgtacag-3′). NH2-terminal and COOH-terminal truncations of histone H4 were generated by PCR using the following primers: forward (5′-atgatccagggcattaccaag-3′) and reverse (5′-ttagccgccgaagccgtacag-3′) and forward (5′-atgtcaggacgaggaaaagg-3′) and reverse (5′-catagccgtaaccgtcttg-3′), respectively. Histone H4 mutants of T30A, S47A, T71A, and T73A were generated by site-directed mutagenesis (Stratagene, La Jolla, CA) with the following primers: H4 T30/A (forward, 5′-catccagggcattgccaagcccgctatccg-3′; reverse, 5′-cggatagcgggcttggcaatgccctggatg-3′) S47/A (forward, 5′-cgtgaagcgcatcgcgggtctcatctacg-3′; reverse, 5′-cgtagatgagacccgcgatgcgcttcacg-3′), T71/A (forward, 5′-ccgcgacgccgtcgcctacaccgagcacg-3′; reverse, 5′-cgtgctcggtgtaggcgacggcgtcgcgg-3′), and T73/A (forward, 5′-cgccgtcacctacgccgagcacgccaagcg-3′; reverse, 5′-cgcttggcgtgctcggcgtaggtgacggcg-3′), respectively. A Lpcat1 H135A mutant was generated by site-directed mutagenesis with the following primers: forward, 5′-caccttggctccagcttcctcctactttg-3′; reverse, 5′-caaagtaggaggaagctggagccaaggtg-3′.
Pulse-Chase Labeling of RNA and RNA Isolation
MLE cells in exponential growth were pulse-chase-labeled with 1 μCi of [32P]UTP for 2 h in the presence or absence of 2 mm Ca2+. Total RNA was extracted from the cells using TRIzol. The radioactivity of the isolated RNA was measured by scintillation counting.
Statistics
All data were statistically analyzed by Student's t test and presented as means ± S.E.
RESULTS
Lpcat1 Shifts into the Nucleus in the Presence of Ca2+
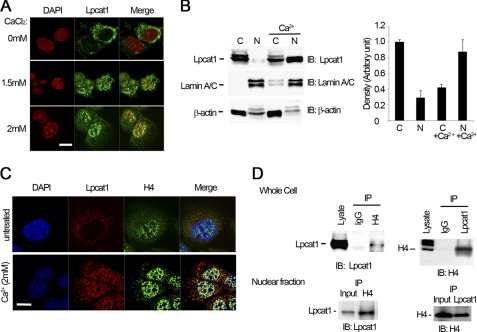
Calcium in lung fluid and within distal lung epithelia can reach high levels, especially during bacterial infection (21, 23). Thus, we treated cells with differing concentrations of Ca2+, and the cells were subjected to anti-Lpcat1 fluorescent immunostaining (Fig. 1A). At 1.5 mm, exogenous Ca2+ triggered a shift of Lpcat1 from the cytosol into the nucleus. A substantial portion of cytosolic Lpcat1 moved into the nucleus when Ca2+ concentrations reached 2 mm, which approximates the levels in normal epithelial lining fluid (24). To confirm these observations, we fractionated the cytosolic and the nuclear portions of cells and analyzed the fractions by immunoblotting. We observed at least two prominent bands that reacted with Lpcat1 antibody in cytosolic fractions suggestive of a post-translationally modified enzyme. However, only one band was detected in the nuclear portion by immunoblotting, suggesting that Lpcat1 exists perhaps as a reversibly modified protein or has isoforms with selectivity with regard to nuclear entry. Further, as expected under native cultured conditions with medium containing 1 mm Ca2+, only small amounts of Lpcat1 were detected in the nuclear fraction (<10%). After treatment with 2 mm Ca2+ for 1 h, the bulk (∼60%) of Lpcat1 was observed in the nucleus (Fig. 1B). Thus, a physiologic inflammatory stimulus is sufficient to relocate a large amount of a soluble lipogenic enzyme into the nuclear compartment.
FIGURE 1.
Lpcat1 association with histone H4 in the nucleus is Ca2+-regulated. A, subcellular localization of Lpcat1 in response to exogenous Ca2+. MLE cells were treated with different concentrations of Ca2+ for 1 h, and cells were processed for anti-Lpcat1 fluorescent immunostaining. Cells were also stained with DAPI to visualize the nucleus. B, cells were treated with 2 mm Ca2+ for 1 h, and nuclear and cytosolic fractions were isolated and used for immunoblotting (IB) as indicated. The right panel displays densitometric results of immunoblots using cytosolic and nuclear fractions before and after Ca2+ treatment. C, colocalization of nuclear Lpcat1 and histone H4 protein. Cells were incubated and treated with 2 mm Ca2+. Anti-Lpcat1 and histone H4 protein fluorescent immunostaining was conducted, and the nucleus was visualized by DAPI. The merged color of colocalized nuclear Lpcat1 and histone H4 is yellow in the nucleus. D, coimmunoprecipitation. Ca2+-treated cells were harvested, and cell lysates or nuclear fractions were processed for immunoprecipitation (IP) with histone H4 protein antibodies followed by Lpcat1 immunoblotting (left). In the right panel, the antibodies were reversed using anti-Lpcat1 immunoprecipitates that were analyzed by histone H4 immunoblotting. The above representative results are from three independent experiments. Scale bar (A and C), 10 μm. Error bars, S.E.
Lpcat1 Colocalizes and Interacts with Histone H4 Protein
Because histones are the most abundant nuclear proteins in eukaryotic cells, we hypothesized that Lpcat1 might interact with this family of proteins. Histone family proteins are decorated with DNA, where they assemble as basic structural units as nucleosomes to maintain nuclear architecture. Notably, Lpcat1 (Fig. 1A) scattered in the nucleus, where it neither associated with the nuclear membrane nor resided in the nucleolus; this raised the possibility that the enzyme might colocalize with the nucleosome. To test this hypothesis, we fluorescently immunostained cells with Lpcat1 and an integral component, histone protein H4. As shown in Fig. 1C (top panel), double immunostaining showed that these proteins are highly compartmentalized because Lpcat1 resides in the cytosol and histone H4 localizes in the nucleus. However, Lpcat1 and histone H4 are highly colocalized in Ca2+-treated cells (Fig. 1C, lower panels). To demonstrate if these proteins interact, we performed immunoprecipitation with Lpcat1 antibody and analyzed the precipitates with histone H4 immunoblotting. In related studies, the antibodies were reversed. The results demonstrate for the first time that Lpcat1 is associated with histone H4 both in total cell lysates and from nuclear preparations after a Ca2+ stimulus (Fig. 1D).
Lpcat1 Catalyzes Histone H4 Protein Palmitoylation in Vitro
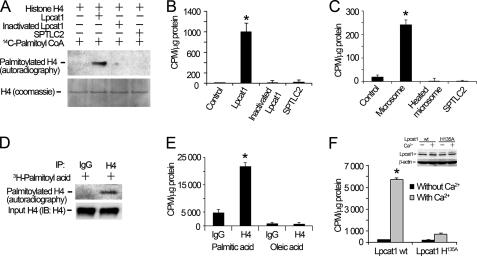
To assess the biochemical significance of Lpcat1 interaction with histone H4, we tested if the histone protein was a new acyltransferase substrate. To evaluate this possibility, we performed in vitro palmitoylation assays using Lpcat1 as the catalyzing enzyme and using [14C]palmitoyl-CoA and histone H4 recombinant protein as the donor and acceptor substrate, respectively. As showed in autoradiography experiments in Fig. 2A, histone H4 was acylated in the presence of recombinant Lpcat1. Further, histone H4 palmitoylation is specific and dependent on Lpcat1 activity because heat-inactivated Lpcat1 and a related palmitoyltransferase, SPTLC2, did not execute this reaction. Hence, an intact catalytic core of Lpcat1 is essential and sufficient for histone H4 protein palmitoylation. In addition, the resolution of histone H4 acylation products under SDS-PAGE used DTT, yet the linkage between histone H4 and the palmitate group was preserved under these reducing conditions; thus, the data suggest a reaction other than S-palmitoylation. Quantitative analysis of radioactive bands corresponding to palmitoylated histone H4 were consistent with the results from autoradiography (Fig. 2B). Because Lpcat1 is ER-bound in microsomes, we also used this fraction as an enzyme source and observed that it was sufficient to catalyze palmitoylation of histone H4 protein (Fig. 2C), albeit activities were lower than that from purified recombinant Lpcat1 protein. Hence, these results demonstrate that Lpcat1 is required and sufficient to catalyze histone H4 protein palmitoylation in vitro.
FIGURE 2.
Lpcat1 catalyzes histone H4 protein palmitoylation in vitro and in vivo. A, in vitro palmitoylation. Histone H4 protein palmitoylation reactions were conducted in the presence of Lpcat1, heat-inactivated (denatured) Lpcat1, and a related palmitoyltransferase, SPTLC2, using [14C]palmitoyl-CoA as a donor and recombinant histone H4 substrate. The lower panel shows histone H4 protein input controls. B, the relevant bands on nitrocellulose membranes were cut, and the radioactivity was counted using a scintillation counter. *, p = 0.0017, radioactivity of Lpcat1 versus dpm of heat-inactivated Lpcat1. C, histone H4 protein palmitoylation reactions were conducted in the presence of lung microsomes, heat-inactivated Lpcat1, and SPTLC2. *, p = 0.0002, radioactivity of microsome versus dpm of heat-inactivated microsome. D, in vivo palmitoylation. MLE cells were pulse-labeled with [3H]palmitoyl acid in the presence of 2 mm Ca2+ for 2 h. Cell lysates were immunoprecipitated (IP) with anti-H4 antibody or IgG to detect palmitoylated H4 followed by autoradiography. Cell lysates were analyzed by V5 immunoblotting as an input control in the lower panel. E, MLE cells were pulse-labeled with [3H]palmitoyl acid or [3H]oleic acid in the presence or absence of 2 mm Ca2+ for 2 h. Cell lysates were immunoprecipitated with H4 antibody, and the radioactivity of the precipitates was measured by scintillation counting. *, p = 0.017, radioactivity of H4 in palmitic acid group versus oleic acid group. F, HEK 293 cells were transfected with pcDNA3.1/Lpcat1 or a Lpcat1 catalytically inactive mutant (Lpcat1 H135A) for 24 h. Cells were pulse-labeled with [3H]palmitoyl acid with or without Ca2+ as above. Histone radioactivity was determined as described in E. The inset shows the protein expression levels of Lpcat1 wt and Lpcat1 H135A mutant and β-actin. *, p = 0.0005, radioactivity of wild type Lpcat1 versus dpm of H135A Lpcat1. The data represent three independent experiments. Error bars, S.E.
Lpcat1 Catalyzes Histone H4 Protein Palmitoylation in Vivo
To assess histone acylation by Lpcat1 physiologically, cells were pulse-chase-labeled with [3H]palmitic acid in the presence of Ca2+, the cell lysates were immunoprecipitated with histone H4 antibody, and the precipitates were separated by SDS-PAGE and then analyzed by autoradiography. Autoradiography results showed that histone H4 was labeled with [3H]palmitate by palmitoylation (Fig. 2D). To evaluate lipidation donor specificity, we pulse-labeled cells with [3H]oleic acid but did not detect histone H4 acylation activity under similar conditions (Fig. 2E). It is widely believed that the Lpcat1 His135 residue is crucial for Lpcat1 catalytic activity for lysophosphatidylcholine acylation, and Lpcat1 protein levels are almost undetectable in HEK 293 cells compared with MLE cells (data not shown). Thus, we replaced Lpcat1 His135 with Ala and overexpressed this mutant in HEK 293 cells. Both wild type and H135A mutant Lpcat1 constructs expressed at comparable levels (Fig. 2F, inset). Further, the ability of the Lpcat1 H135A mutant to palmitoylate histone H4 was markedly reduced compared with wild type Lpcat1 (Fig. 2F). As a whole, these data suggest that Lpcat1 catalytic activity for histone H4 is relatively selective and donor-dependent and requires an intact catalytic structural component essential for histone H4 palmitoylation. These results demonstrate that histone H4 is modified by palmitoylation in vitro and in vivo and suggest that this reaction is nuclear Lpcat1-dependent.
Characterizing Lpcat1 Enzymatic Activity against Histone H4
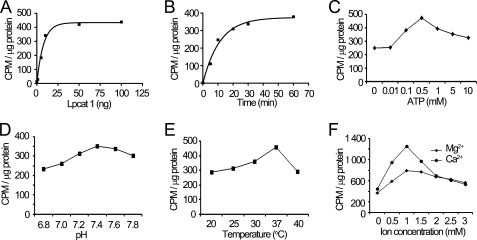
We next optimized Lpcat1 enzymatic activity against histone H4. The data show that palmitoylation of histone H4 is an energy-dependent reaction that occurs using low concentrations of recombinant Lpcat1 and is nearly maximal by 30 min; reactions occur at a pH optima of 7.4 at 37 °C and are sensitive to the presence of divalent cations (Fig. 3).
FIGURE 3.
Characterization of Lpcat1 enzymatic activity for histone H4 protein palmitoylation. In vitro palmitoylation experiments were carried out in the presence of different amounts of Lpcat1 recombinant protein (A), optimized for reaction time (B), ATP concentration (C), pH (D), temperature (E), and cation dependence (Ca2+ and Mg2+) (F). The data represent three independent experiments.
Ser47 within Histone H4 Protein Is the Putative O-Palmitoylation Site
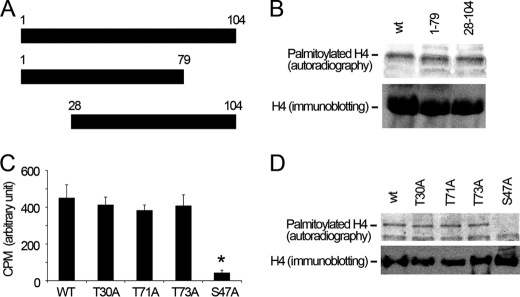
We next investigated the palmitoylation site in histone H4 protein using deletional and point mutagenesis. We truncated histone H4 protein (Fig. 4A). We expressed these truncations using an in vitro translation system and performed [14C]palmitoyl-CoA palmitoylation assays in the presence of recombinant Lpcat1. All translated proteins were expressed at similar levels as demonstrated by anti-V5 immunoblotting (Fig. 4B). Results showed that the full-length, NH2-terminal truncated, and COOH-terminal truncated fragments are effectively palmitoylated, indicating that the palmitoylation site is in the middle, hinge region of the primary histone H4 sequence. Sequence analysis reveals that histone H4 is devoid of cysteine residues, suggesting that Ser or Thr residues are potential targets for O-palmitoylation. Thus, we substituted these candidate residues with Ala in the mid-region of histone H4. The wild type and mutated Lpcat1 proteins were expressed comparably in vitro as above. Of the point mutants tested, only the S47A construct was observed to be resistant to in vitro palmitoylation (Fig. 4, C and D). These results suggest that the putative histone H4 O-palmitoylation site is Ser47.
FIGURE 4.
Ser47 is the putative O-palmitoylation site for Lpcat1 within histone H4. A, schematic presentation of truncated mutants of histone H4 protein. B, wild type and truncated histone H4 mutants were in vitro translated and palmitoylated with [14C]palmitoyl-CoA. The lower panel shows the expression levels of the wild type and mutant constructs. C, histone H4 protein Ser and Thr mutants were in vitro translated and palmitoylated with [14C]palmitoyl-CoA. The reactions were incubated with 1 μg of H4 antibody and 30 μl of protein A/G-agarose beads overnight, and the radioactivity of the washed beads was determined by scintillation counting. p = 0.0019, radioactivity in S47A-H4 versus T30A H4. D, the washed beads in C were separated by SDS-PAGE and analyzed by autoradiography and H4 immunoblotting. The data represent three independent experiments. Error bars, S.E.
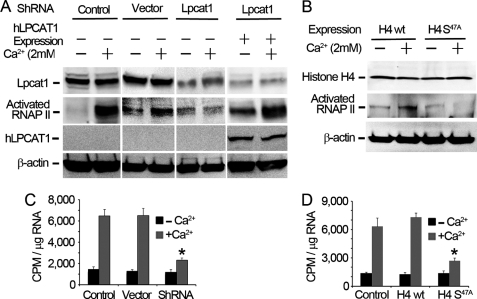
Histone H4 Palmitoylation Regulates Global RNA Synthesis
To assess the physiological consequences of histone lipidation, we evaluated RNA synthesis in cells. Histone post-translational modification regulates RNA polymerase II activity; thus, we measured RNA polymerase II activation in Ca2+-treated cells. Calcium treatment of cells increased levels of the activated form of RNA polymerase II, an effect that is Lpcat1-dependent (Fig. 5A). This latter observation was first demonstrated by depleting endogenous Lpcat1 using shRNAi (Fig. 5A, lanes 5 and 6). Next we knocked down endogenous murine Lpcat1 and performed rescue studies by overexpressing human Lpcat1 (Fig. 5A, lanes 7 and 8). The data show that activation of RNA polymerase II is reversibly regulated by these maneuvers where we modulated Lpcat1 expression. We further observed that expression of a histone H4 S47A mutant construct versus wild type histone H4 in cells substantially reduced RNA polymerase II activation in Ca2+-treated cells (Fig. 5B). Because Ser47 is the palmitoylation site within histone H4, these results suggest that activation of RNA polymerase II is linked to site-specific modification of histone H4. Activation of RNA polymerase II causes global increases in gene transcriptional activity; thus, we investigated if histone H4 protein palmitoylation up-regulates genome-wide transcription. We executed pulse-chase studies by labeling cells with [32P]UTP in the presence or absence of Ca2+ treatment and then harvested newly synthesized RNA. The total RNA radioactivity in Ca2+-treated cells was significantly higher than that of untreated cells, indicative of gene transcriptional activation (Fig. 5C). To determine if up-regulation of gene transcriptional activity is Lpcat1-dependent, we knocked down Lpcat1 by shRNA in cells. Lpcat1 shRNA knockdown significantly reduced total cellular RNA synthesis to approximate levels of untreated (−Ca2+) cells. We next determined if gene transcriptional activity depends on histone H4 palmitoylation. We overexpressed wild type histone H4 and the H4 S47A mutant in cells and observed that RNA synthesis in cells ectopically expressing the S47A mutant was significantly lower than that of cells transfected with wild type histone H4 (Fig. 5D). Collectively, these studies strongly suggest that Lpcat1 may be required for RNA synthesis in lung epithelia via histone palmitoylation.
FIGURE 5.
Histone H4 palmitoylation regulates global RNA synthesis. A, MLE cells were transfected with empty vector or Lpcat1 shRNA plasmids for 48 h, followed by exposure of cells to the presence or absence of 2 mm Ca2+. For rescue, one group of cells was first transfected with Lpcat1 shRNA for 48 h prior to overexpression with human Lpcat1 (hLpcat1) for another 24 h. Cell lysates were subject to Lpcat1, RNA polymerase II, and β-actin immunoblot analysis. B, wild type histone H4 and histone H4 S47A mutant plasmids expressing a COOH-terminal V5 tag were ectopically expressed in MLE cells, and after 24 h, cells were treated with or without 2 mm Ca2+. Cell lysates were then processed for V5, RNA polymerase II, and β-actin immunoblot analysis. C and D, MLE cells in A or B were pulse-chase-labeled with [32P]UTP for 2 h in the presence or absence of 2 mm Ca2+, total RNA was extracted using the TRIzol method, and the radioactivity of the isolated RNA was measured by scintillation counting. The data represent three independent experiments. C, p = 0.0015, radioactivity in Ca2+-treated shRNA versus Ca2+-treated wild type; D, p = 0.0004, radioactivity in Ca2+-treated H4 S47A mutation versus Ca2+-treated wild type H4. Error bars, S.E.
DISCUSSION
The findings of this study are the first demonstration of a protein substrate for Lpcat1 and the identification and characterization of histone O-palmitoylation. We further demonstrate that Lpcat1-induced histone lipidation is linked to cellular control of gene transcriptional activity. An unexpected finding here is that a soluble, ER-bound lipogenic enzyme indispensable for lung surfactant generation relocates to the nuclear compartment in response to calcium signals to execute its binding and covalent lipidation of a histone protein. It is possible that Lpcat1-mediated O-palmitoylation of histone H4 either modulates its interaction with other scaffolding molecules to alter nuclear signaling events or is involved in augmentation of epigenetic events that act in concert with other histone modifications. The biological consequences of O-palmitolyation may be complex but may affect fundamental nuclear processes involving gene expression, cell cycle progression, or mitosis. We observed that Ca2+ availability appears highly relevant to drive nuclear Lpcat1 import and histone H4 O-palmitoylation. The observations that during the prophase of mitosis, the nuclear envelope is disrupted, chromosomal condensation occurs, and nuclear contents are transiently exposed to high calcium concentrations (25, 26) suggest that Lpcat1-mediated histone O-palmitoylation might be highly regulated during cell division. However, these possibilities are highly speculative and await further study.
The ability of Lpcat1 to translocate to the nucleus resembles the enzymatic behavior of other lipogenic enzymes. Several studies indicate that a related enzyme, cholinephosphate cytidyltransferase, also shifts into the nucleus in response to bacterial infection, high concentrations of Ca2+, or extracellular hypertonicity (23, 27, 28). Similar subcellular migratory behavior was demonstrated with carnitine palmitoyltransferase I (29), an enzyme highly expressed in cancer cell lines. Coincidentally, Lpcat1 is ubiquitously expressed in a variety of tissues and cell lines, and its expression is particularly high in colon cancer cell lines (30). These observations suggest that nuclear import of these lipogenic enzymes might be a mechanism to initiate molecular signals that confers a growth advantage for neoplastic cells. The molecular apparatus that aids in Lpcat1 nuclear entry is currently under investigation. The presence of a canonical nuclear localization signal within its primary sequence suggests that the assembly of importin-like components may also play a role in concert with calcium signals for nuclear entry.
Of the three types of covalent linkages between the donor (palmitate) and the acceptor substrates, S-palmitoylation and N-palmitoylation use the cysteine -SH side group or amide group, respectively, to form a thioester linkage or amide linkage. In contrast, O-palmitoylation involves a serine -OH group to form an oxyester linkage. Database analysis of the histone H4 primary sequence reveals that it lacks cysteine residues, and thus both S-palmitoylation and N-palmitoylation could be excluded as modifications. Further, the observation that high concentrations of reducing agents included during processing of our reaction products did not eliminate or reduce the levels of palmitoylated histone H4 strongly argues that this covalent linkage is not a thioester bond but a modification that is stable. There are only two Ser residues within the histone H4 sequence, Ser1 and Ser47. Deletion of the NH2 terminus did not alter palmitoylation levels in vitro (Fig. 4B), suggesting that Ser1 within histone H4 is not the acceptor site for palmitoylation. Replacement of Ser47 with Ala greatly reduced histone H4 palmitoylation, indicating that Lpcat1 catalyzes O-palmitoylation. These data are also consistent with the ability of Lpcat1 to catalyze O-palmitoylation of its lipid substrates. Because Ser residues within histone H4 are also phosphorylation acceptor sites, it will be of interest to elucidate the molecular interplay between these post-translation modifications on histone architecture and related functional consequences in vivo.
It is well documented that histone proteins are vulnerable to numerous covalent modifications that in turn dictate transcriptional regulatory activity by controlling chromatin condensation and DNA accessibility. Hence, histone modifications, in part, constitute an epigenetic histone code that is generated by histone-modifying enzymes. This epigenetic code containing these specific covalent marks is sensed by proteins to regulate chromatin behavior. Our studies suggest that perhaps Lpcat1 might partake in epigenetic modification by initiating a new molecular signature on histone proteins. Our biochemical observations may be important because the lipidation of histone H4 might increase its hydrophobicity, altering histone and DNA association to modify chromatin and nucleosomal architecture to regulate genome-wide transcriptional activity. Although the current work shows that histone lipidation alters cellular RNA synthesis, additional work is needed to assess the conformational signatures relating to euchromatin and links with specific genes that may be up-regulated in response to calcium signals that drive Lpcat1-histone H4 lipidation. These are ongoing studies that will increase our understanding of the mechanisms of histone lipidation and its speculative role as a possible epigenetic signal of fundamental physiological significance.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL096376, HL097376, and HL098174 (to R. K. M.). This material is based upon work supported, in part, by the United States Department of Veterans Affairs, Veterans Health Administration, Office of Research and Development, Biomedical Laboratory Research and Development. This work was also supported by a Merit Review Award from the United States Department of Veterans Affairs.
- MLE
- murine lung epithelial
- HEK
- human embryonic kidney
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Nakanishi H., Shindou H., Hishikawa D., Harayama T., Ogasawara R., Suwabe A., Taguchi R., Shimizu T. (2006) J. Biol. Chem. 281, 20140–20147 [DOI] [PubMed] [Google Scholar]
- 2. Soupene E., Fyrst H., Kuypers F. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 88–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheng L., Han X., Shi Y. (2009) Am. J. Physiol. Endocrinol. Metab. 297, E1276–E1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Matsuda S., Inoue T., Lee H. C., Kono N., Tanaka F., Gengyo-Ando K., Mitani S., Arai H. (2008) Genes Cells 13, 879–888 [DOI] [PubMed] [Google Scholar]
- 5. Chamoun Z., Mann R. K., Nellen D., von Kessler D. P., Bellotto M., Beachy P. A., Basler K. (2001) Science 293, 2080–2084 [DOI] [PubMed] [Google Scholar]
- 6. Takada R., Satomi Y., Kurata T., Ueno N., Norioka S., Kondoh H., Takao T., Takada S. (2006) Dev. Cell 11, 791–801 [DOI] [PubMed] [Google Scholar]
- 7. Yang J., Brown M. S., Liang G., Grishin N. V., Goldstein J. L. (2008) Cell 132, 387–396 [DOI] [PubMed] [Google Scholar]
- 8. Greaves J., Chamberlain L. H. (2011) Trends Biochem. Sci. 36, 245–253 [DOI] [PubMed] [Google Scholar]
- 9. Kang R., Wan J., Arstikaitis P., Takahashi H., Huang K., Bailey A. O., Thompson J. X., Roth A. F., Drisdel R. C., Mastro R., Green W. N., Yates J. R., 3rd, Davis N. G., El-Husseini A. (2008) Nature 456, 904–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Linder M. E., Deschenes R. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 74–84 [DOI] [PubMed] [Google Scholar]
- 11. Hemsley P. A., Grierson C. S. (2008) Trends Plant Sci. 13, 295–302 [DOI] [PubMed] [Google Scholar]
- 12. Sorek N., Bloch D., Yalovsky S. (2009) Curr. Opin. Plant Biol. 12, 714–720 [DOI] [PubMed] [Google Scholar]
- 13. Sanders S. L., Portoso M., Mata J., Bähler J., Allshire R. C., Kouzarides T. (2004) Cell 119, 603–614 [DOI] [PubMed] [Google Scholar]
- 14. Kawasaki H., Schiltz L., Chiu R., Itakura K., Taira K., Nakatani Y., Yokoyama K. K. (2000) Nature 405, 195–200 [DOI] [PubMed] [Google Scholar]
- 15. Wang Y., Zhang W., Jin Y., Johansen J., Johansen K. M. (2001) Cell 105, 433–443 [DOI] [PubMed] [Google Scholar]
- 16. Robzyk K., Recht J., Osley M. A. (2000) Science 287, 501–504 [DOI] [PubMed] [Google Scholar]
- 17. Shin J. A., Choi E. S., Kim H. S., Ho J. C., Watts F. Z., Park S. D., Jang Y. K. (2005) Mol. Cell 19, 817–828 [DOI] [PubMed] [Google Scholar]
- 18. Zempleni J., Chew Y. C., Hassan Y. I., Wijeratne S. S. (2008) Nutr. Rev. 66, Suppl. 1, S46–S48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kotova E., Lodhi N., Jarnik M., Pinnola A. D., Ji Y., Tulin A. V. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 6205–6210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilson J. P., Raghavan A. S., Yang Y. Y., Charron G., Hang H. C. (2011) Mol. Cell Proteomics, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen B. B., Coon T. A., Glasser J. R., Mallampalli R. K. (2011) Mol. Cell Biol. 31, 1905–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zou C., Butler P. L., Coon T. A., Smith R. M., Hammen G., Zhao Y., Chen B. B., Mallampalli R. K. (2011) J. Biol. Chem. 286, 2719–2727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Agassandian M., Chen B. B., Schuster C. C., Houtman J. C., Mallampalli R. K. (2010) FASEB J. 24, 1271–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cooley J., McDonald B., Accurso F. J., Crouch E. C., Remold-O'Donnell E. (2008) J. Leukoc. Biol. 83, 946–955 [DOI] [PubMed] [Google Scholar]
- 25. Brown E. M., Shoback D. M. (1984) Prog. Clin. Biol. Res. 168, 139–144 [PubMed] [Google Scholar]
- 26. Pszczolkowski M. A., Lee W. S., Liu H. P., Chiang A. S. (1999) Mol. Cell Endocrinol. 158, 163–171 [DOI] [PubMed] [Google Scholar]
- 27. Ryan A. J., Chen B. B., Vennalaganti P. R., Henderson F. C., Tephly L. A., Carter A. B., Mallampalli R. K. (2008) J. Biol. Chem. 283, 24628–24640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Favale N. O., Fernández-Tome M. C., Pescio L. G., Sterin-Speziale N. B. (2010) Biochim. Biophys. Acta 1801, 1184–1194 [DOI] [PubMed] [Google Scholar]
- 29. Mazzarelli P., Pucci S., Bonanno E., Sesti F., Calvani M., Spagnoli L. G. (2007) Cancer Biol. Ther. 6, 1606–1613 [DOI] [PubMed] [Google Scholar]
- 30. Mansilla F., da Costa K. A., Wang S., Kruhøffer M., Lewin T. M., Orntoft T. F., Coleman R. A., Birkenkamp-Demtröder K. (2009) J. Mol. Med. 87, 85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]