Abstract
Protein folding and unfolding are complex phenomena, and it is accepted that multidomain proteins generally follow multiple pathways. Maltose-binding protein (MBP) is a large (a two-domain, 370-amino acid residue) bacterial periplasmic protein involved in maltose uptake. Despite the large size, it has been shown to exhibit an apparent two-state equilibrium unfolding in bulk experiments. Single-molecule studies can uncover rare events that are masked by averaging in bulk studies. Here, we use single-molecule force spectroscopy to study the mechanical unfolding pathways of MBP and its precursor protein (preMBP) in the presence and absence of ligands. Our results show that MBP exhibits kinetic partitioning on mechanical stretching and unfolds via two parallel pathways: one of them involves a mechanically stable intermediate (path I) whereas the other is devoid of it (path II). The apoMBP unfolds via path I in 62% of the mechanical unfolding events, and the remaining 38% follow path II. In the case of maltose-bound MBP, the protein unfolds via the intermediate in 79% of the cases, the remaining 21% via path II. Similarly, on binding to maltotriose, a ligand whose binding strength with the polyprotein is similar to that of maltose, the occurrence of the intermediate is comparable (82% via path I) with that of maltose. The precursor protein preMBP also shows a similar behavior upon mechanical unfolding. The percentages of molecules unfolding via path I are 53% in the apo form and 68% and 72% upon binding to maltose and maltotriose, respectively, for preMBP. These observations demonstrate that ligand binding can modulate the mechanical unfolding pathways of proteins by a kinetic partitioning mechanism. This could be a general mechanism in the unfolding of other large two-domain ligand-binding proteins of the bacterial periplasmic space.
Keywords: Atomic Force Microscopy, Ligand-binding Protein, Protein Conformation, Protein Folding, Single Molecule Biophysics, Force Spectroscopy, Kinetic Partitioning, Maltose-binding Protein, Parallel Pathways, Protein Unfolding
Introduction
Small proteins (with fewer than 100 amino acids) are still under the scrutiny of having smooth versus the rough energy landscape; however, for proteins larger than 100 amino acid residues it is generally believed that they follow multiple pathways involving one or many intermediates during protein unfolding (1–4). Contrary to this notion, there exist few exceptions among large proteins, for example, Borrelia burgdorferi VlsE (5), Alteromonas haloplanctis α-amylase (6), and maltose-binding protein (MBP)2 (7), which are shown to exhibit an apparent two-state equilibrium unfolding pathway in bulk solution. Very recent single-molecule pulling studies showed that out-of-equilibrium mechanical unfolding of MBP is very complex and consists of on-pathway sequential intermediates (8, 9). Our earlier studies on MBP have shown the existence of intermediates during the folding pathway of MBP (10, 11). Traditional bulk biophysical techniques like fluorescence, time-resolved unfolding and folding probe ensembles of molecules and hence suffer many drawbacks compared with methods that probe one molecule at a time. Therefore, single-molecule studies on protein folding/unfolding have proved vital in giving a detailed picture of the protein energy landscape and also measure pathways that are statistically rare and hence averaged out in the ensemble studies (12). Single-molecule force spectroscopy is a technique where individual proteins are investigated under application of an extrinsic mechanical force (13–16). It has been shown earlier that this technique can be used to observe the intermediate states that are difficult to detect in bulk studies (3). Here, we have investigated mechanical unfolding properties of MBP and its precursor protein preMBP using single-molecule force spectroscopy. We discern two different unfolding pathways and show that the associated kinetic partitioning is modulated by ligands maltose and maltotriose.
MBP, a monomeric 370-residue two-domain protein with an α/β structure, is a protein found in the periplasmic space of Gram-negative bacteria and is involved in maltose/maltodextrin uptake and chemotaxis (17). It is synthesized as a precursor protein (preMBP) containing an extra 26 residue N-terminal signal peptide that acts as a translocation signal in the cytosol (18). Previous studies have shown that preMBP is destabilized by approximately 20–40% with respect to MBP and also exhibits a higher unfolding rate (10). MBP consists of two distinct N- and C-globular domains that are discontinuous in a sense that each domain is composed of segments from both the N terminus and C terminus. The two domains rotate with respect to each other by an angle of 35° between the ligand-unbound open conformation and ligand-bound closed conformation. The overall structure forms a “clam”-like shape with a deep spatially buried groove for ligand binding (19, 20). In fact, this is a common structural feature among the class of periplasmic binding proteins, which bind and transport sugars, amino acids, and anions (21). MBP binds to malto-oligosaccharides like maltose and maltotriose with micromolar dissociation constants (KD values) (7, 22, 23). It was shown that preMBP also binds to maltose with affinity similar to that of MBP (10). All these properties make MBP and preMBP an ideal system to understand how ligand-dependent conformational changes in the structure manifest in the folding/unfolding pathway of the two proteins at the single-molecule level.
MATERIALS AND METHODS
Cloning, Expression, and Purification
MBP and preMBP (A14E) genes were amplified from pMAL-p2-MBP vector by a two-step overlap PCR to remove the internal BglII restriction site and introduce the BamHI site at the 5′ end and BglII and KpnI sites at the 3′ end of the full-length amplicons. As BamHI and BglII enzymes generate complementary sequence overhangs, MBP or preMBP gene, digested with BamHI and KpnI, was ligated to pQE80L-(GB1)4 plasmid that was treated with BglII and KpnI enzymes. This resulted in a pQE80L-(GB1)4-MBP or -preMBP construct. pQE80L-(GB1)4 plasmid was treated with BamHI and KpnI to release (GB1)4, and this was then ligated to pQE80L-(GB1)4-MBP or -preMBP that was treated with BglII and KpnI to generate a pQE80L-(GB1)4-(MBP or preMBP)-(GB1)4 construct.
Escherichia coli (DH5α) cells, transformed with pQE80L-(GB1)4-(MBP or preMBP)-(GB1)4 were grown in LB medium in the presence of ampicillin (0.1 mg/ml) at 37 °C and 180 rpm until O.D. at 600 nm ∼0.6. Protein expression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 25 °C, and the culture was allowed to grow overnight. The cells were harvested and lysed by sonication in lysis buffer containing Tris (0.025 m, pH 7.4), NaCl (0.5 m), and PMSF (0.1 mm), and the expressed protein was bound to a Ni-Sepharose 6 Fast Flow (GE Healthcare) column. After binding, the column was washed with lysis buffer containing imidazole (0.04 m), and the elutions were carried out with 0.25 m imidazole. Fractions containing purified proteins were dialyzed extensively against PBS buffer.
Single-molecule Force Spectroscopy Using Atomic Force Microscope
Single-molecule pulling experiments were done on a custom-built atomic force microscope, which is similar to that described by Fernandez and Li. (24) We have used a top-view optical head with quadrant photodiode for multimode SPM (TVOH-MMAFM) from Veeco Asia Pte. Ltd., Singapore. We replaced the laser in the optical head with a laser diode (51nanoFCM) coupled to a single-mode fiber cable attached to a collimator (60FC-4-M20-10) and microfocusing system (5M-M25-13-S) from Schäfter and Kirchhoff GmbH, Hamburg, Germany. This AFM head was mounted on top of a multiaxis piezoelectric positioning and scanning system with capacitive sensors from Physik Instrumente GmbH, Germany (PicoCube P-363.3.CD) that allows subnanometer resolution. A custom-made electronics module was used to communicate among the quad photodetector, piezoelectic controller, and the data acquisition boards on a personal computer. Data acquisition command modules and further data analysis routines were written using IGOR Pro software from Wavemetrics. The AFM head and piezoelectric positioner were kept on a vibration isolation table (25BM-4 from minus K Technology), which was further kept on a high capacity laboratory table from TMC. We have used a contact mode fluid cell (from Veeco, Singapore) to mount cantilevers and to make measurements in solution. Gold-coated reflective cantilevers with silicon nitride tip (MLCT-AUNM) with spring constants ∼30–50 pN/nm were purchased from Veeco, Singapore. Calibration of cantilevers was done using equipartition theorem (25) prior to each pulling experiment. In a typical pulling experiment, 5 μl of protein (∼1 mg/ml) was added to 50 μl of phosphate-buffered saline (PBS), pH 7.4, kept on a gold-coated glass coverslip, and let sit for 10 min. The cantilever was then calibrated in protein solution and used for force extension (FX) experiments. All experiments were done at ambient temperature ∼25 °C. The pulling speed in all the experiments was 400 nm/s. In pulling experiments containing ligands (maltose and maltotriose), the protein was incubated in ligand-containing buffer for about an hour before doing any pulling. Maltose and maltotriose were obtained from Sigma-Aldrich Co.
Isothermal Titration Calorimetry
ITC measurements were carried out on a VP isothermal titration calorimeter from MicroCal, Inc. (Northampton, MA). Protein (0.02 mm) and maltose or maltotriose (0.85 mm) in CGH buffer (10 mm each, citrate, glycine, and HEPES, pH 7.4) were used for the titrations. The protein and ligand solutions were incubated at 25 °C for at least 1 h and transferred to the cell and syringe, respectively. The reference cell was filled with water. Stirring speed was set at 300 rpm, and after base-line stabilization, 8-μl injections were carried out with an interval of 4 min between each injection until saturation was observed. The dilution of ligand into buffer and buffer into protein was carried out, and these dilution corrections were incorporated before data analysis. Data were analyzed by using the Origin software package and fit to a single binding site model. Results are presented in supplemental Table S1.
RESULTS AND DISCUSSION
Mechanical Properties of MBP
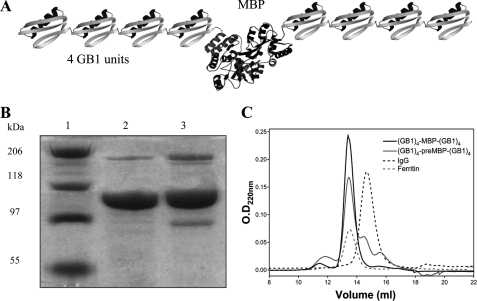
To study the unfolding pathway of MBP and preMBP we have used a single-molecule force spectroscopy approach where we have applied extrinsic mechanical force on polyproteins using a custom-built atomic force microscope. Two chimeric polyproteins, (GB1)4-MBP-(GB1)4 and (GB1)4-preMBP-(GB1)4, consisting of a central MBP (or preMBP) molecule flanked on each side by four GB1 protein domains were constructed by a gene fusion technique (Fig. 1A). A detailed procedure of polyprotein purification is given under “Materials and Methods.” GB1 here acts a molecular marker to fish out the signal resulting from the protein of interest. Li and co-workers (26) have previously characterized mechanical properties of GB1 by single-molecule AFM studies to show that it unfolds at a force of 180 pN unraveling a length of 18 nm. Fig. 1, B and C, shows the SDS-PAGE and gel filtration profiles of the purified polyproteins and that the apparent molecular masses of the proteins were higher than expected on SDS-PAGE. This is likely because the nonglobular shape of the polyprotein causes it to have a higher hydrodynamic radius compared with a globular protein of similar molecular mass. However, the gel filtration clearly shows that the polyproteins are homogenous species and not large aggregates as aggregates would elute at the void volume of the column.
FIGURE 1.
A, schematic of the chimeric protein (GB1)4-MBP-(GB1)4. B, SDS-PAGE (8%) analysis of purified protein. Lane 1, protein marker; lane 2, (GB1)4-MBP-(GB1)4; and lane 3, (GB1)4-preMBP-(GB1)4). C, gel filtration profiles of the proteins on Superdex 200 analytical column. Elution volume for both (GB1)4-MBP-(GB1)4 and (GB1)4-preMBP-(GB1)4 was 13.3 ml. IgG (150 kDa) and ferritin (440 kDa) were used as markers, which eluted at 14.5 ml and 13.4 ml, respectively. The apparent molecular masses from column calibration were 354 and 316 kDa for (GB1)4-MBP-(GB1)4 and (GB1)4-preMBP-(GB1)4, respectively.
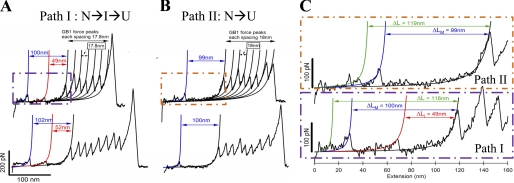
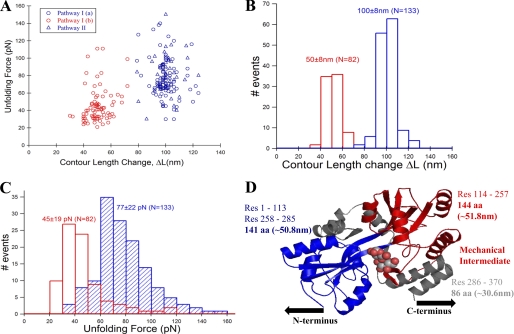
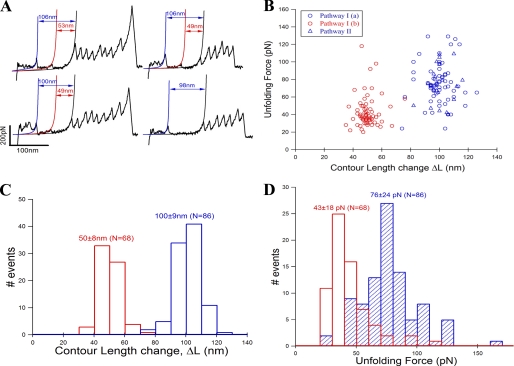
Single-molecule pulling experiments were performed on a custom-built atomic force microscope, and a detailed description of the apparatus is given under “Materials and Methods.” Fig. 2A shows the experimental results from pulling single (GB1)4-MBP-(GB1)4 molecules at a velocity of 400 nm/s. The single-molecule FX traces show characteristic regularly spaced saw-tooth pattern force peaks with an unfolding force of ∼200 pN. These peaks have been fitted with a worm-like chain (WLC) model of polymer elasticity (27) (shown as black curves), and they measure an average contour length change of 17.6 nm. The observed mechanical properties (i.e. contour length and the unfolding force) match those expected from GB1 protein allowing GB1 to act as a fingerprint molecule for the chimeric construct (26). In addition to the GB1 saw-tooth pattern, there are two distinct force peaks in the beginning of the trace occurring at much lower force than GB1. These two peaks have also been fitted to the WLC model as shown in red and blue curves. We measure the change in contour length of these peaks from the first GB1 unfolding event in the FX trace similar to the way in yellow fluorescent protein studied earlier (28). The measured contour length changes for the two force peaks are ∼100 nm (ΔLM) and ∼50 nm (ΔLI) (see also Table 1). Based on the design of the chimeric polyprotein construct (GB1)4-MBP-(GB1)4, there should be at least five GB1 peaks in a single-molecule FX trace to be sure that MBP has been subjected to the stretching force. Hence, we can unambiguously assign the two force peaks prior to the GB1 saw-tooth pattern to the mechanical signature arising from MBP. If MBP unfolds in a two-state or all-or-none manner there would be only a single force peak within a contour length change of ∼128 nm (370aa * 0.36 nm/aa − 4.35 nm; aa = amino acid residue) from the first GB1 force peak (29). Here, 4.35 nm is the length of the folded MBP protein, i.e. the distance between the N and C termini in the MBP crystal structure (Protein Data Bank ID code 1OMP). The contour length change of 128 nm would correspond to a single transition state during unfolding. However, we instead observe the first force peak at a shorter distance of 100 ± 8 nm from the first GB1 unfolding, in addition to encountering an extra peak at 50 ± 8 nm. This indicates that the unfolding pathway involves two transition states that are separated by an intermediate occurring at approximately half the length of the first force peak. We call this pathway of mechanical unfolding via an intermediate as path I, where I(a) corresponds to protein transition from native to intermediate state (N→I) and I(b) corresponds to protein transition from intermediate to the fully unfolded state (I→U). The unfolding forces and the corresponding contour length changes measured from many single-molecule measurements have been plotted as a scatter plot (Fig. 3A), and it can be clearly seen that the intermediate unfolding occurred at a lower force than that of the initial unfolding. From the unfolding forces histogram of the intermediate (45 ± 19 pN) and initial peak (77 ± 22 pN), it is clear that the intermediate is mechanically less stable than the initial state (Fig. 3, B and C).
FIGURE 2.
Representative single-molecule FX traces of (GB1)4-MBP-(GB1)4. A, FX traces of the chimeric protein from constant velocity (400 nm/s) pulling experiments are shown. These FX traces contain two force peaks in the beginning followed by a characteristic saw-tooth pattern of GB1. The GB1 peaks have the expected average values of 17.6 nm for the unfolding contour length change and 200 pN for the unfolding force. The two force peaks in the beginning are due to MBP unfolding, and their unfolding contour lengths are measured from the first GB1 unfolding peak using the WLC model of polymer elasticity (fits are shown in color). Mechanical stretching of MBP results in partial unfolding leading to an intermediate state, which is the initial unfolding event (fitted by blue WLC curve) with a peak force of ∼77 pN and an unfolding contour length (ΔLM) of ∼100 nm. Further stretching of the intermediate results in a completely unraveled MBP, which is the intermediate unfolding event (fitted by red WLC curve) with peak force ∼45 pN and the unfolding contour length ΔLI ∼ 50 nm. We call this mechanical unfolding pathway path I, which contains an intermediate (N→I→U). In 62% of all the observed FX traces, MBP unfolds via path I. B, in the remaining 38% of the FX traces, stretching of MBP results in a single force peak with peak force ∼77 pN and the unfolding contour length (ΔLM ∼ 100 nm), which is similar to that of path I; however, the intermediate peak is absent. We call this mechanical unfolding pathway path II (N→U), which is devoid of the intermediate of path I. C, FX trace shows the MBP unfolding region (from A and B) in greater detail for path I and path II. Force peaks with <30 pN at contour lengths ∼115–120 nm preceding the initial unfolding event can be seen in these representative traces (fitted by green WLC curves; see “Results” for more details). However, distinction of these low force peaks was not clearly possible in all of the FX traces of different pulling events due to their very weak mechanical nature.
TABLE 1.
Mechanical unfolding properties of MBP and preMBP
| Sample | Contour length change of initial peak ΔLM | Contour length change of intermediate ΔLI | Unfolding force of initial peak | Unfolding force of intermediate | % Traces with intermediatea | |
|---|---|---|---|---|---|---|
| nm | nm | pN | pN | |||
| MBP | No ligand | 100 ± 8b (n = 133)c | 50 ± 8 (n = 82) | 77 ± 22 | 45 ± 19 | 61.7 ± 4.3 |
| Maltose (0.2–10 mm) | 100 ± 9 (n = 86) | 50 ± 8 (n = 68) | 76 ± 24 | 43 ± 18 | 79.1 ± 4.5 | |
| Maltotriose (0.5–10 mm) | 102 ± 11(n = 57) | 50 ± 9 (n = 47) | 79 ± 29 | 45 ± 21 | 82.5 ± 5.2 | |
| PreMBP | No ligand | 100 ± 9 (n = 46) | 51 ± 8 (n = 24) | 75 ± 20 | 34 ± 10 | 52.2 ± 7.4 |
| Maltose (0.2–10 mm) | 100 ± 8 (n = 78) | 53 ± 10 (n = 53) | 77 ± 30 | 52 ± 30 | 67.9 ± 5.4 | |
| Maltotriose (0.5–10 mm) | 102 ± 14 (n = 53) | 52 ± 10 (n = 38) | 79 ± 30 | 55 ± 37 | 71.7 ± 6.2 | |
a Percent traces with Intermediate = (No. of traces with the intermediate/total number of traces) × 100. The errors are S.E., calculated using the bootstrap method (49, 50) (see supplemental text).
b All reported values are average ± S.D.
c The number of measurements per experiment is given in parentheses.
FIGURE 3.
The intermediate on path I of MBP unfolding is mechanically weaker than the native protein. A, unfolding force and contour length change values of MBP obtained from many single-molecule FX traces (see Fig. 2, A and B) were used in making unfolding force versus contour length change scatter plot. The plot clearly shows two clusters (or populations) of force peaks: one for the initial unfolding event (blue circles from path I and blue triangles from path II) and another for the intermediate unfolding event (red circles, only from path I). B, contour length change histograms for the initial (ΔLM = 100 ± 8 nm, blue bars) and intermediate states (ΔLI = 50 ± 8 nm, red bars). C, unfolding force histograms of the initial (77 ± 22 pN, blue bars) and intermediate unfolding (45 ± 19 pN, red bars). Unfolding force of initial peak from both path I (blue circles) and II (blue triangles) are combined, indicating that they are mechanically indistinct. The intermediate is mechanically weaker than the native state of MBP. D, proposed contour length mapping onto MBP structure (maltose-bound MBP structure Protein Data Bank ID code 1ANF). Protein region labeled gray likely gives the ∼28-nm low unfolding force peaks preceding the initial force peak. Path I: N-terminal domain (colored blue) manifests in the initial peak (N→I), and C-terminal domain (colored red) gives the intermediate unfolding (I→U). However, in path II, both N- and C-terminal domains unfold together giving a single peak (N→U) without any on-pathway intermediate.
As mentioned earlier, the largest measured contour length change (100 nm) in our FX traces is less than the expected contour length change, thus giving an unaccountable difference in length of ∼28 nm. Very rarely, we do observe peaks in the beginning of the FX traces at forces <30 pN (i.e. force peaks before the 100-nm-long initial force peak as shown by the green WLC curves in Fig. 2C). Although a detailed characterization of these low force peaks has not been possible because of their rare occurrence and the presence within the noise floor of the instrumental setup (∼20 pN), it is quite possible that the difference in contour length change (∼28 nm) could be accounted by these very low force peaks arising due to the unraveling of MBP from the termini before the 100-nm force peak. Indeed, this was the case as shown by recent single-molecule studies on mechanical unfolding of MBP by Bertz and Rief (8) and Bechtluft et al. (9). In both of these studies, the low force unfolding event was noticed, which corroborates our observations. While Bechtluft et al. (9) used optical tweezers, comprising a very soft spring constant to assess the subpiconewton force regime to study MBP unfolding; Bertz and Rief (8) have been unable to observe this low force event in all of their FX traces (Fig. 2 in Ref. 8), indicating that this unfolding event is indeed mechanically too weak to be captured by AFM pulling studies clearly. Hence, we did not try to further assign these peaks in our analysis, although the molecular extension prior to the 100-nm peak accounts for the difference in the measured and expected contour lengths.
Unfolding Pathway of MBP
Information obtained from the single-molecule FX profiles during the N- and C-terminal unfolding can assist in predicting the unfolding pathway of MBP based on its x-ray crystallographic structure (Fig. 3D) (17). The C terminus consists of α-helices, which are known to be mechanically very weak (30) and can account for the initial loss of ∼25–30-nm extension in the FX profiles as observed in our measurements and previous studies (8, 9). The same has been shown by steered molecular dynamics simulations where the C-terminal helices are the first to unfold, followed by the unfolding of the remaining 278-residue core protein structure (8). However, unlike the optical tweezers experiment and the steered molecular dynamics simulations (9), we observed the presence of an intermediate en route to unfolding. On the basis of our studies and Bertz and Rief. (8), we can map the observed contour length changes onto the structure to assign the initial higher force peak at 100 nm possibly resulting from the unfolding of the N-terminal domain. This is primarily due to the richness of mechanically stronger β-sheets as well as the direction of force-vector lying along the N terminus and the unraveled C-terminal helices (both transmit the force into N-terminal domain). This leads to an extension of 50 nm to produce an intermediate (I) possibly consisting of an intact C-terminal domain. Further stretching of the intermediate could unfold this second domain, leading to complete unraveling of the protein structure. Regions of terminal helices, N-domain, and C-domain unfolding are depicted in gray, blue, and red in x-ray crystallographic structure in Fig. 3D. The higher unfolding force (77 ± 22 pN) of the initial peak prior to the comparatively weaker C-domain region supports the structural mapping based on contour length changes and direction of force application. This scenario is similar to what has been shown by Bertz and Rief (8) except that we do not observe the hierarchical unfolding of MBP but instead observe a single intermediate.
MBP Unfolds via Two Different (Parallel) Pathways
Apart from the similarities of C-terminal helices unfolding before the major force peak, there is a discrepancy between the two previous studies on the mechanical unfolding of MBP (8, 9). A mechanical intermediate (50 nm), similar to that shown in Fig. 2A, has been observed in AFM studies by Bertz and Rief (8), whereas it was completely absent in the optical tweezers study by Bechtluft et al. (9). To understand this discrepancy, we performed experiments on many single molecules. All observed FX traces can be categorized into two classes where 62% of the MBP unfolding traces have the intermediate force peak as shown in Fig. 2A, but the remaining FX traces show different behavior (Fig. 2B). In these remaining 38% traces, we observe only one force peak at 100 nm for MBP, and the intermediate force peak at 50 nm is not observed (see Table 1). This indicates that part of the time MBP is unfolding via a simpler pathway devoid of the 50-nm intermediate (path II: N→U). This result is similar to that observed by Bechtluft et al. (9) in their optical tweezers experiment. Because the resolution of our instrument is ∼20 pN, any unfolding event occurring above 20 pN should have been clearly observed as a force peak as in Fig. 2A. Hence, the absence of an intermediate force peak in Fig. 2B indicates the absence of a mechanically stable intermediate during unfolding via path II.
In the scatter plot (Fig. 3A), it can be clearly seen that the force of unfolding of the initial peak (77 pN) and the contour length (100 nm) are independent of the pathway, i.e. whether MBP is unfolding via path I or path II. Hence, it can be concluded that the mechanical properties of the transition states of the initial peaks on paths I and II are similar. It is also important to note that we have never observed the intermediate force peak alone without the 100-nm initial peak in any of the FX traces, which indicates that the intermediate is not off-pathway of the initial transition state. These two different types of traces, one with a 50-nm mechanically stable intermediate (in 62% cases) and the remaining traces without any intermediate at 50 nm (in 38% cases), indicate that the unfolding pathway of MBP is complex, and it follows two independent routes (paths I and II). It can be concluded from our results that MBP unfolding via path I in AFM experiments was observed by Bertz and Rief (8) and via path II was observed by Bechtluft et al. (9).
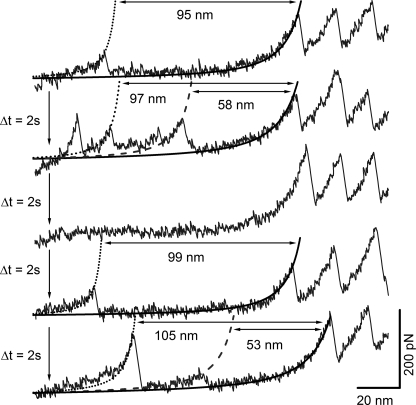
Furthermore, we have performed repeated stretch-relax cycles on the same MBP molecule similar to ones conducted on I27 polyprotein (13, 31). Here, repeated cycles of pulling are performed without detaching the polyprotein molecule, with each pulling event followed by relaxation of the force to allow refolding to take place. As shown in Fig. 4, the same MBP molecule was observed to unfold via both path I and path II in different cycles of repeated pulling. The choice of MBP between path I or path II was stochastic and independent of the path chosen in the very first pulling event. From this experiment, it can be concluded that the parallel pathways we observe on unfolding single molecules is intrinsic to MBP and not due to heterogeneous population in the sample. We have also performed experiments on apo MBP at varying pulling speeds (supplemental Fig. S1). We observe that the flux through the intermediate remains largely unchanged in the pulling speed range (400–4000 nm/s).
FIGURE 4.
FX traces of (GB1)4-MBP-(GB1)4 from repeated stretch-relax cycles performed on a single molecule. The molecule was relaxed for 2s before stretching in each cycle. Each stretching event was length-clamped at 170 nm such that the molecule could not detach from either the cantilever tip or the Au coverslip. WLC curves are also shown: in black dotted for the initial state and in gray dashed for the intermediate state of MBP and black solid for the first GB1 unfolding. Cycle 2 and 5 (FX traces 2 and 5 from top): MBP unfolds via path I (i.e. through an intermediate) in these cycles, and the contour length changes measured by the WLC curves for the initial (ΔLM) and intermediate states (ΔLI) are ∼100 nm and ∼50 nm, respectively. Cycles 1 and 4 (FX traces 1 and 4 from top): MBP unfolds via path II in these cycles, resulting in a single force peak with a contour length change of ∼100 nm for the initial state (ΔLM) as measured by the WLC curves. Cycle 3 (FX trace 3): MBP failed to refold within the delay time of 2 s and hence did not show any force peak. However, the same MBP molecule refolded in the next cycles 4 and 5. In all the cycles, GB1 unfolding force peaks with (∼18-nm contour length change) served as the molecular fingerprint.
The division of unfolding into distinct parallel pathways, as observed here for MBP, is called “kinetic partitioning” in the literature. This mechanism was previously observed in ensemble solution studies in the case of barstar (32) and immunoglobin (Ig) domain of the human cardiac muscle protein titin (I27) (33). It was recently demonstrated that T4 lysozyme also exhibits kinetic partitioning during mechanical unfolding as shown by single-molecule AFM pulling experiments (3). MBP, to the best of our knowledge, becomes the largest protein and the second such example after T4 lysozyme where mechanical unfolding occurs via kinetic partitioning. However, unlike T4 lysozyme, the partitioning observed in the case of MBP is limited to two specific pathways, and additionally, the relative flux down each pathway is shown to be modulated by ligand binding, as described in detail in the next section.
Maltose and Maltotriose Modulate the Kinetic Partitioning in MBP Unfolding
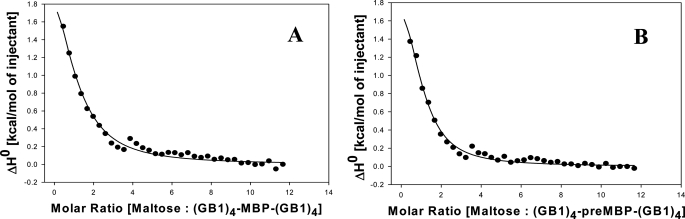
MBP binds maltose (disaccharide) and maltotriose (trisaccharide), and the KD of the bound complexes are 1–3 μm and 0.3 μm, respectively (7, 22). We have carried out ITC studies and a thermal shift assay which clearly show that maltose and maltotriose bind to MBP in the (GB1)4-MBP-(GB1)4 polyprotein construct (Fig. 5 and supplemental text). We determined the dissociation constant (KD) of the (GB1)4-MBP-(GB1)4-maltose complex from ITC experiments to be 17 μm. The binding is somewhat weaker in the polyprotein compared with MBP alone. To determine the effect of maltose on the mechanical unfolding pathway of MBP, we have performed the single-molecule pulling experiments on MBP in the presence of 0.2–10 mm maltose (traces shown in Fig. 6A). The mechanical properties (contour lengths and peak forces) of maltose-bound MBP are similar to that of apoMBP (Fig. 6). Maltose-bound MBP also unfolds via two parallel pathways I and II (Table 1). The contour length increments show no change in the ligand-bound compared with the unbound protein, and the unfolding force for both the initial peak and the intermediate remains the same as the free protein. Similar observations were made by Bertz and Rief (8) in their N- and C-terminal pulling geometry of MBP. However, when they performed the stretching along the direction that perturbs the ligand binding groove directly, a ligand-induced mechanical stabilization of protein by 15 pN was reported (34). In our study, which probes the protein along the N- to C-terminal direction, we also do not observe any additional mechanical stabilization of the MBP upon ligand binding. However, the protein responds to the ligand binding by altering its choice for unfolding pathway taken. Maltose-bound MBP unfolded via path I in 79% of the cases whereas in 21% cases there was no intermediate (path II). Although the pathways (and hence the energy landscape) are the same between the bound and unbound form, the flux through different pathways is modified upon maltose binding, thus causing an enhancement in the number of instances or the probability of the bound protein to undergo unfolding via path I involving an intermediate. Similar enhancement is observed for maltotriose, where maltotriose-bound MBP unfolded via path I in 83% cases (see supplemental Fig. S2 and Table 1), and the remaining 17% follow unfolding through path II. Because the enthalpy of binding (ΔH°) of maltotriose to the proteins at 25 °C was low, the KD values could not be determined by ITC for this ligand. However, the binding of maltotriose to the polyprotein constructs was confirmed by using a thermal shift assay (35) that measures the increase in thermal stability of (GB1)4-MBP-(GB1)4 as a function of added ligand concentration. The thermal stability (Tm) of the protein increased progressively with increase in the molar ratio of ligand to protein, and the data suggests that both maltose and maltotriose ligands bind with similar affinity to (GB1)4-MBP-(GB1)4 (supplemental Table S2). Binding to maltotriose skews the flux toward path I to a similar extent as that of maltose. This could be expected as the polyprotein constructs do not show any drastic differences in binding strength to both the ligands. However, in either case, the flux through path I did not increase further even at very high (10 mm) ligand concentrations. To our knowledge, such modulation of unfolding pathways upon ligand binding has not been reported until now, and this study is the first to show that ligands can modulate the kinetic partitioning of mechanical unfolding of a protein without apparently changing the pathways themselves.
FIGURE 5.
ITC measurements of maltose binding to (GB1)4-MBP-(GB1)4 (A) and (GB1)4-preMBP-(GB1)4 (B) at 25 °C in CGH buffer (10 mm each citrate, glycine, and HEPES, pH 7. 4). Data points are shown as solid circles, and the fit is shown as a solid line.
FIGURE 6.
Representative single-molecule FX traces of (GB1)4-MBP-(GB1)4 in the presence of 0. 2–10 mm maltose. A, the unfolding features of bound MBP are very similar to those of apoMBP. Ligand-bound MBP also unfolds via two parallel pathways I and II. Three of the traces show the intermediate peak whereas one does not. B, force versus contour length change scatter plot. The plot clearly shows two clusters (or populations) of force peaks: one for the initial unfolding event (blue circles from path I and blue triangles from path II) and another for the intermediate unfolding event (red circles, only from path I). C, contour length change histograms for the initial (ΔLM = 100 ± 9 nm, blue bars) and intermediate states (ΔLI = 50 ± 8 nm, red bars). However, the flux through path I is higher (79%) compared with that of apoMBP (62%). D, unfolding force histograms of the initial (76 ± 24 pN, blue bars) and intermediate unfolding (43 ± 18 pN, red bars).
Structural comparison between apoMBP and ligand-bound MBP have shown a ligand-induced conformational change in the protein structure (open to close), resulting in the two N- and C-terminal clefts coming closer to each other and thus enclosing the ligand molecule in the spatially buried binding groove (36–38). The bound ligand locally stabilizes both the N- and C-domains around its binding site through hydrogen bonding (20). Moreover, the ligand binding site is far from the termini. In mechanical unfolding of MBP, the force is transmitted through the termini, and thus the initial events of unfolding (i.e. unraveling of C-terminal helices and N terminus) would not be affected as the ligand binding site does not interact with them directly. A similar observation has been made earlier both in mechanical unfolding studies (39–41) and proteasomal degradation of proteins (where mechanical unfolding precedes peptide lysis) (42). However, the intermediate manifested from unraveling the C-terminal domain of MBP should have been affected by ligand binding, and the unfolding force should have been influenced. Surprisingly, this is not the case, and the unfolding force of the intermediate remains unchanged. While the intermediate occurred more frequently in the presence of ligand, still its occurrence could not be seen in every MBP pulling event. This could be explained by considering the scenario of apoMBP in equilibrium between both open and close conformations, where ligand binding shifts the equilibrium to the closed conformation for the protein. Tang et al. (38) have shown that apoMBP exists largely in the open state, but a small amount of a closed-like conformation is also present. This is also true in the case of enzymes, for example, in dihydrofolate reductase (43). It is hence possible that these two conformations of MBP follow different pathways in mechanical unfolding: closed conformation via path I with an intermediate and open conformation via path II without an intermediate. Now, on ligand binding the equilibrium shifts more toward the closed conformation, and hence the unfolding flux also increases through path I causing increased events of intermediate occurrence. If this is true, then under saturating conditions of ligand the entire unfolding flux should go through path I. But, the intermediate unfolding flux is not observed to increase on increasing the ligand concentration. This can be explained as follows: as ligand binding is an equilibrium process between binding and unbinding, the ligand might be falling off from MBP during the single-molecule mechanical unfolding. In that case the protein shuttles through open conformation during the unfolding and eventually follows path II. This explains why the flux through path I is not 100% even under saturating ligand concentrations.
Unfolding Pathways of MBP and PreMBP Are the Same
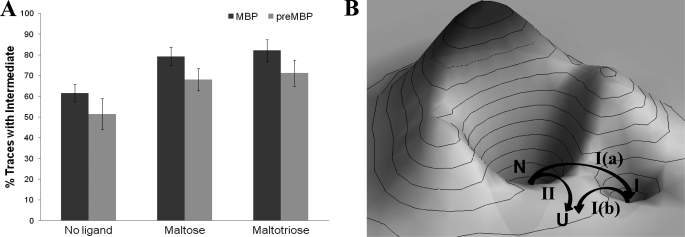
We have also made a chimeric polyprotein, (GB1)4-preMBP-(GB1)4. The FX traces for this polyprotein are shown in supplemental Fig. S3, and the results are summarized in Table 1. Although preMBP contains an extra 26-residue N-terminal signal (10), we did not find any difference in the contour lengths and unfolding forces from those of MBP. This clearly indicates that the signal peptide has been unfolded at very low force before even reaching the 100-nm force peak and does not affect the mechanical stability of the initial and intermediate force peaks. This observation is consistent with earlier proteolysis studies (10) that demonstrate that the signal peptide does not interact with the rest of the mature protein in the native state, but rather stabilizes the unfolded state of the protein. The similar unfolding forces for MBP and preMBP therefore also imply that the transition state for mechanical unfolding is closer to the native state than to the unfolded state and hence, the transition states for the 100-nm peak and 50-nm peak are similar to those of MBP. In the case of preMBP unfolding, the intermediate was observed in 53% cases only. We do not know the reason for the discrepancy in the flux through path I between MBP (62%) and preMBP (53%) because both have similar contour length changes upon unfolding and exhibit similar unfolding forces. However, we have found the trend of ligand-modulated mechanical properties to be the same for preMBP as for MBP (supplemental Figs. S4 and S5). Maltose-bound (GB1)4-preMBP-(GB1)4 has a dissociation constant of 11 μm, and maltotriose also binds with similar affinity as shown by ITC and thermal shift assay, respectively (Fig. 5 and supplemental Table S2). The mechanical properties of preMBP in the presence and absence of ligands are given in Table 1. Similar to MBP, both the ligands modulated the unfolding pathways by channeling preMBP more through the pathway containing the intermediate (path I), 68% cases by maltose and 72% cases by maltotriose. The results from the preMBP study further support the assertion that bound ligands can modulate the kinetic partitioning in the mechanical unfolding pathways of MBP (Fig. 7A).
FIGURE 7.
Energy landscape of the ligand-modulated parallel mechanical unfolding pathways of MBP and preMBP. A, bar chart showing the ligand-modulated flux (percent traces with intermediate via path I) for MBP and preMBP. The error bars are the S.E. calculated using the bootstrap method (see Table 1). B, schematic showing the conceptual energy landscape of MBP and its mechanical unfolding pathways. On mechanical stretching, the native protein takes one of the two paths, I and II. Path I is followed by 62% of cases, and the remaining 38% of cases take path II. On path I, the protein transits to an intermediate state (as shown by the path I(a), which then takes path I(b) to the completely stretched unfolded state). On the other hand, the native protein transits a single barrier to reach the unfolded state on path II. Similar energy landscape can be envisaged for the mechanical unfolding of preMBP.
Energy Landscape
Based on our findings we can construct a schematic of the energy landscape of MBP as shown in Fig. 7B. The mechanical stretching of MBP unravels in two different pathways under the kinetic partitioning mechanism. In path I, the protein goes from its native state (N) to the intermediate state (I) via transition state (TS1). The intermediate (I) then passes the transition state (TS2) to a completely unfolded state (U). In path II, the protein goes from its native state (N) to the unfolded state (U) via a single barrier (TS3) without a detour. We found that the transition states TS1 and TS3 have similar mechanical properties. It is highly possible that they differ enough to give parallel pathways I and II and yet are energetically similar so as to be populated simultaneously, which is the case for titin I27 (33). It is very interesting that ligand binding modulates kinetic partitioning: the preference to path I increased from 62% to 79% upon binding to maltose and it further enhances to 82% upon binding to maltotriose. Similar energy landscapes can be envisaged for preMBP from the kinetic partitioning results shown in Fig. 6A.
CONCLUSION
Our study conclusively demonstrates that MBP unfolds via parallel unfolding pathways and that bound ligands (maltose and maltotriose) modulate the relative flux through each of the parallel unfolding pathways. In addition, ligand binding to the precursor protein of MBP (preMBP) also shows the same effect as in the case of MBP. The “new view” of protein folding can be described using an energy landscape where there are multiple pathways for folding and unfolding (44). Because of the roughness of the energy landscape, unfolded molecules follow both direct and indirect pathways, and the folding flux undergoes kinetic partitioning (45). Most studies have focused on relatively small proteins such as lysozyme, barstar, and titin I27 (3, 32, 33). It is only in the case of T4 lysozyme that parallel pathways have been revealed using mechanical unfolding. None of these studies has examined the effects of ligands on modulating the relative flux through different pathways. Our single-molecule experiments on the mechanical unfolding of MBP and preMBP give further evidence to this kinetic partitioning mechanism. In previous studies, ligand binding has been shown to affect the mechanical properties of proteins, for example, dihydrofolate reductase (46) and protein G (47), and acylphosphatase (48). In all of them, the force required to unfold a protein is enhanced upon ligand binding, and this could be explained by an increase in the barrier height. However, in the current study we show that ligands can modulate the kinetic pathways of the protein unfolding without modifying the mechanical properties such as unfolding force and contour length. Furthermore, many periplasmic binding proteins that bind and transport sugars, amino acids, and anions have structures similar to MBP consisting of two large domains that undergo open-to-close conformational change upon ligand binding (21). Hence, the ligand-modulated kinetic partitioning studies on MBP that we report here can help in forming a general prediction about the unfolding properties and energy landscape of the other periplasmic proteins. In addition, our results underscore the importance of single-molecule AFM studies along with ensemble experiments to understand the complex behavior of even large biomolecules that get obscured in averaging effects of the bulk.
Supplementary Material
Acknowledgments
We thank Prof. Hongbin Li for providing the GB1 gene for constructing the polyprotein. We thank K. Hemachandra for comments and critical reading of the manuscript.
This work was supported by the Tata Institute of Fundamental Research (to S. R. K. A.).

The on-line version of this article (available at http://www.jbc.org) contains Tables S1 and S2, Figs. S1–S5, text, and additional references.
- MBP
- maltose-binding protein
- AFM
- atomic force microscopy
- ΔLM and ΔLI
- contour length changes for initial and intermediate states, respectively
- FX
- force extension
- ITC
- isothermal titration calorimetry
- pN
- piconewton
- preMBP
- precursor protein of MBP
- WLC
- worm-like chain.
REFERENCES
- 1. Brockwell D. J., Radford S. E. (2007) Curr. Opin. Struct. Biol. 17, 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Radford S. E., Dobson C. M., Evans P. A. (1992) Nature 358, 302–307 [DOI] [PubMed] [Google Scholar]
- 3. Peng Q., Li H. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Udgaonkar J. B. (2008) Annu. Rev. Biophys. 37, 489–510 [DOI] [PubMed] [Google Scholar]
- 5. Jones K., Wittung-Stafshede P. (2003) J. Am. Chem. Soc. 125, 9606–9607 [DOI] [PubMed] [Google Scholar]
- 6. Feller G., d'Amico D., Gerday C. (1999) Biochemistry 38, 4613–4619 [DOI] [PubMed] [Google Scholar]
- 7. Ganesh C., Shah A. N., Swaminathan C. P., Surolia A., Varadarajan R. (1997) Biochemistry 36, 5020–5028 [DOI] [PubMed] [Google Scholar]
- 8. Bertz M., Rief M. (2008) J. Mol. Biol. 378, 447–458 [DOI] [PubMed] [Google Scholar]
- 9. Bechtluft P., van Leeuwen R. G., Tyreman M., Tomkiewicz D., Nouwen N., Tepper H. L., Driessen A. J., Tans S. J. (2007) Science 318, 1458–1461 [DOI] [PubMed] [Google Scholar]
- 10. Beena K., Udgaonkar J. B., Varadarajan R. (2004) Biochemistry 43, 3608–3619 [DOI] [PubMed] [Google Scholar]
- 11. Krishnan B., Kulothungan S. R., Patra A. K., Udgaonkar J. B., Varadarajan R. (2009) J. Mol. Biol. 385, 1243–1256 [DOI] [PubMed] [Google Scholar]
- 12. Michalet X., Weiss S., Jäger M. (2006) Chem. Rev. 106, 1785–1813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carrion-Vazquez M., Oberhauser A. F., Fowler S. B., Marszalek P. E., Broedel S. E., Clarke J., Fernandez J. M. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 3694–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Borgia A., Williams P. M., Clarke J. (2008) Annu. Rev. Biochem. 77, 101–125 [DOI] [PubMed] [Google Scholar]
- 15. Puchner E. M., Gaub H. E. (2009) Curr. Opin. Struct. Biol. 19, 605–614 [DOI] [PubMed] [Google Scholar]
- 16. Galera-Prat A., Gómez-Sicilia A., Oberhauser A. F., Cieplak M., Carrión-Vázquez M. (2010) Curr. Opin. Struct. Biol. 20, 63–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spurlino J. C., Lu G. Y., Quiocho F. A. (1991) J. Biol. Chem. 266, 5202–5219 [DOI] [PubMed] [Google Scholar]
- 18. Bassford P. J., Jr. (1990) J. Bioenerg. Biomembr. 22, 401–439 [DOI] [PubMed] [Google Scholar]
- 19. Millet O., Hudson R. P., Kay L. E. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 12700–12705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quiocho F. A., Spurlino J. C., Rodseth L. E. (1997) Structure 5, 997–1015 [DOI] [PubMed] [Google Scholar]
- 21. Quiocho F. A., Ledvina P. S. (1996) Mol. Microbiol. 20, 17–25 [DOI] [PubMed] [Google Scholar]
- 22. Thomson J., Liu Y., Sturtevant J. M., Quiocho F. A. (1998) Biophys. Chem. 70, 101–108 [DOI] [PubMed] [Google Scholar]
- 23. Schwartz M., Kellermann O., Szmelcman S., Hazelbauer G. L. (1976) Eur. J. Biochem./ FEBS 71, 167–170 [DOI] [PubMed] [Google Scholar]
- 24. Fernandez J. M., Li H. (2004) Science 303, 1674–1678 [DOI] [PubMed] [Google Scholar]
- 25. Florin E. L., Rief M., Lehmann H., Ludwig M., Dornmair K., Moy V. T., Gaub H. E. (1995) Biosens. Bioelectron. 10, 895–901 [Google Scholar]
- 26. Cao Y., Lam C., Wang M., Li H. (2006) Angew Chem. Int. Ed Engl. 45, 642–645 [DOI] [PubMed] [Google Scholar]
- 27. Bustamante C., Marko J. F., Siggia E. D., Smith S. (1994) Science 265, 1599–1600 [DOI] [PubMed] [Google Scholar]
- 28. Perez-Jimenez R., Garcia-Manyes S., Ainavarapu S. R., Fernandez J. M. (2006) J. Biol. Chem. 281, 40010–40014 [DOI] [PubMed] [Google Scholar]
- 29. Ainavarapu S. R., Brujic J., Huang H. H., Wiita A. P., Lu H., Li L., Walther K. A., Carrion-Vazquez M., Li H., Fernandez J. M. (2007) Biophys. J. 92, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rief M., Pascual J., Saraste M., Gaub H. E. (1999) J. Mol. Biol. 286, 553–561 [DOI] [PubMed] [Google Scholar]
- 31. Ainavarapu S. R., Wiita A. P., Huang H. H., Fernandez J. M. (2008) J. Am. Chem. Soc. 130, 436–437 [DOI] [PubMed] [Google Scholar]
- 32. Zaidi F. N., Nath U., Udgaonkar J. B. (1997) Nat. Struct. Biol. 4, 1016–1024 [DOI] [PubMed] [Google Scholar]
- 33. Wright C. F., Lindorff-Larsen K., Randles L. G., Clarke J. (2003) Nat. Struct. Biol. 10, 658–662 [DOI] [PubMed] [Google Scholar]
- 34. Bertz M., Rief M. (2009) J. Mol. Biol. 393, 1097–1105 [DOI] [PubMed] [Google Scholar]
- 35. Niesen F. H., Berglund H., Vedadi M. (2007) Nat. Protoc. 2, 2212–2221 [DOI] [PubMed] [Google Scholar]
- 36. Duan X., Hall J. A., Nikaido H., Quiocho F. A. (2001) J. Mol. Biol. 306, 1115–1126 [DOI] [PubMed] [Google Scholar]
- 37. Sharff A. J., Rodseth L. E., Spurlino J. C., Quiocho F. A. (1992) Biochemistry 31, 10657–10663 [DOI] [PubMed] [Google Scholar]
- 38. Tang C., Schwieters C. D., Clore G. M. (2007) Nature 449, 1078–1082 [DOI] [PubMed] [Google Scholar]
- 39. Hann E., Kirkpatrick N., Kleanthous C., Smith D. A., Radford S. E., Brockwell D. J. (2007) Biophys. J. 92, L79–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Junker J. P., Ziegler F., Rief M. (2009) Science 323, 633–637 [DOI] [PubMed] [Google Scholar]
- 41. Kedrov A., Krieg M., Ziegler C., Kuhlbrandt W., Muller D. J. (2005) EMBO Rep. 6, 668–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee C., Schwartz M. P., Prakash S., Iwakura M., Matouschek A. (2001) Mol. Cell 7, 627–637 [DOI] [PubMed] [Google Scholar]
- 43. Zhang Z., Rajagopalan P. T., Selzer T., Benkovic S. J., Hammes G. G. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 2764–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Onuchic J. N., Luthey-Schulten Z., Wolynes P. G. (1997) Annu. Rev. Phys. Chem. 48, 545–600 [DOI] [PubMed] [Google Scholar]
- 45. Thirumalai D., Klimov D. K., Woodson S. A. (1997) Theor. Chem. Acc. 96, 14–22 [Google Scholar]
- 46. Ainavarapu S. R., Li L., Badilla C. L., Fernandez J. M. (2005) Biophys. J. 89, 3337–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cao Y., Balamurali M. M., Sharma D., Li H. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15677–15681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Arad-Haase G., Chuartzman S. G., Dagan S., Nevo R., Kouza M., Mai B. K., Nguyen H. T., Li M. S., Reich Z. (2010) Biophys. J. 99, 238–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Efron B. (1982) The Jackknife, the Bootstrap, and Other Resampling Plans, Society for Industrial and Applied Mathematics, Philadelphia, PA [Google Scholar]
- 50. Wiita A. P., Perez-Jimenez R., Walther K. A., Gräter F., Berne B. J., Holmgren A., Sanchez-Ruiz J. M., Fernandez J. M. (2007) Nature 450, 124–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.