Abstract
Ca2+/calmodulin-dependent protein kinase kinase β (CaMKKβ) is a serine/threonine-directed kinase that is activated following increases in intracellular Ca2+. CaMKKβ activates Ca2+/calmodulin-dependent protein kinase I, Ca2+/calmodulin-dependent protein kinase IV, and the AMP-dependent protein kinase in a number of physiological pathways, including learning and memory formation, neuronal differentiation, and regulation of energy balance. Here, we report the novel regulation of CaMKKβ activity by multisite phosphorylation. We identify three phosphorylation sites in the N terminus of CaMKKβ, which regulate its Ca2+/calmodulin-independent autonomous activity. We then identify the kinases responsible for these phosphorylations as cyclin-dependent kinase 5 (CDK5) and glycogen synthase kinase 3 (GSK3). In addition to regulation of autonomous activity, we find that phosphorylation of CaMKKβ regulates its half-life. We find that cellular levels of CaMKKβ correlate with CDK5 activity and are regulated developmentally in neurons. Finally, we demonstrate that appropriate phosphorylation of CaMKKβ is critical for its role in neurite development. These results reveal a novel regulatory mechanism for CaMKKβ-dependent signaling cascades.
Keywords: Calcium/Calmodulin-dependent Protein Kinase (CaMK), Calmodulin, Neurodifferentiation, Protein Phosphorylation, Signal Transduction
Introduction
The Ca2+/calmodulin-dependent protein kinase kinases (CaMKKs)2 are serine/threonine-directed protein kinases that are activated following increases in intracellular Ca2+ concentration. There are two CaMKK isoforms, CaMKKα and CaMKKβ, that are encoded by two separate genes. The CaMKKs have similar domain structures consisting of N- and C-terminal domains of unknown function, a kinase domain, and an autoregulatory domain comprising overlapping autoinhibitory and calmodulin (CaM) binding regions (1). Binding of Ca2+/CaM to the CaM binding domain releases autoinhibition allowing for maximal catalytic activity. Human CaMKKα and CaMKKβ share 65% protein sequence homology, with the highest degree of similarity in their kinase domains, and the biggest differences in the N- and C-terminal domains (2).
The CaMKKs are well characterized activators of the Ca2+/calmodulin-dependent protein kinases I (CaMKI) and IV (CaMKIV) (2–4). In this context, the CaMKKs are upstream activators of Ca2+/CaM-dependent signaling cascades. These cascades are activated following an elevation in intracellular Ca2+, which leads to Ca2+ binding to CaM. Upon Ca2+ binding, CaM assumes an active conformation that allows it to bind target proteins, including the CaMKKs and the CaMKs (5). Binding of Ca2+/CaM to the CaMKs allows the activation loop of the kinase to assume a conformation in which it can then be phosphorylated by the CaMKKs (6, 7). Thus, CaMKK/CaMK signaling cascades are regulated at multiple levels by Ca2+/CaM.
CaMKKα and CaMKKβ are expressed most abundantly in the brain (2), where CaMKK/CaMK signaling cascades play important roles in a number of physiologically important neuronal processes, including learning and memory (8, 9), neuronal differentiation and migration (10), neurite outgrowth (11–13), and synaptogenesis (14). In addition, CaMKKβ, but not CaMKKα, is an upstream activator of the AMP-dependent protein kinase (AMPK) (15–17). The regulation of AMPK by CaMKKβ is important for regulating energy balance in the hypothalamus (18).
Although both CaMKKs activate CaMKI and CaMKIV similarly, there are important differences in how the two are regulated. First, CaMKKα can be phosphorylated by protein kinase A (PKA) leading to the recruitment of 14-3-3 and inactivation of the enzyme (19, 20). Although some PKA sites are conserved in both CaMKKs, the regulation of CaMKKβ by PKA has not been reported. In addition, although CaMKKα activity is almost exclusively dependent on Ca2+/CaM for activity, CaMKKβ has considerable autonomous activity in the absence of Ca2+/CaM binding (2, 21). Despite high autonomous activity, CaMKKβ pathways involving substrates that are not themselves Ca2+/CaM-regulated, such as AMPK, still require a Ca2+/CaM signal for activation (17). In addition, CaMKKβ purified from bacterial and mammalian sources display varying amounts of autonomous activity (15, 21). These observations led us to believe that there are additional Ca2+/CaM-independent methods of CaMKKβ regulation.
In this study, we describe the regulation of CaMKKβ by phosphorylation and identify cyclin-dependent kinase 5 (CDK5) and glycogen synthase kinase 3 (GSK3) as putative upstream kinases. Phosphorylation of CaMKKβ in vitro decreases the Ca2+/CaM-independent, autonomous activity of the enzyme. In addition, we find that CDK5/GSK3 phosphorylation regulates the turnover of CaMKKβ in vivo. Finally, we demonstrate that appropriate phosphorylation of CaMKKβ is critical for its function in neurite development in primary neuronal cultures.
EXPERIMENTAL PROCEDURES
Purification of CaMKKβ
Full-length rat CaMKKβ was expressed as a GST fusion protein in BL21(DE3) Escherichia coli (Stratagene, 200131) from the pGEX-KG-PreS-CaMKKβ vector and purified by glutathione and calmodulin-Sepharose chromatography using a procedure described previously (21). The final purification step involved proteolytic removal of the GST tag. For purification from mammalian cells, full-length rat FLAG-CaMKKβ was expressed from the pSG5 vector transiently overexpressed in HEK 293a cells using Lipofectamine 2000 (Invitrogen, 11668-019). For each transfection, 20 μg of DNA and 40 μl of Lipofectamine 2000 were used per 15-cm dish following the manufacturer's suggested protocol. HEK 293a cells were about 75% confluent at the time of transfection, and cultures were maintained in DMEM (Mediatech, 10-013-CV) with 10% FBS (Gemini Bio-Products, 900-108). Lysates were made from cells 24–36 h after transfection using 1 ml/plate of Nonidet P-40 lysis buffer (25 mm Tris-HCl, pH 7.5, 50 mm NaCl, 0.5% Nonidet P-40, 25 mm NaH2PO4, 2 mm EGTA, 2 mm EDTA, 40 mm NaF, 10 μg/ml leupeptin, 100 μg/ml Pefabloc, and 100 nm okadaic acid). Lysate was clarified by centrifugation at 18,000 × g for 30 min. Supernatant was then incubated for 2 h with constant rocking at 4 °C with 20 μl of anti-FLAG M2-agarose (Sigma, A2220) per ml of lysate. The resin was washed twice with lysis buffer and once with TBS prior to elution for 1 h on ice with 40 μl of TBS containing 300 ng/μl FLAG peptide (Sigma, F3290). For λ-phosphatase-treated CaMKKβ, purification was performed as above up to the washes in lysis buffer. At this point, the resin was washed twice with 1× λ-phosphatase reaction buffer (New England Biolabs, P0753) before being resuspended in 50 μl of λ-phosphatase reaction buffer plus 2.0 μl of λ-phosphatase (New England Biolabs, P0753) per ml of starting lysate. Phosphatase reaction was incubated at 30 °C for 30 min before being washed three times with lysis buffer, once with TBS, and eluted as above. Protein concentrations were measured by fractionating aliquots of each preparation on a 10% SDS-polyacrylamide gel along with BSA standards followed by quantification of Coomassie Blue-stained bands.
Full-length human FLAG-CaMKKβ for mass spectrometry analysis (wild type and individual S129A, S133A, and S137A mutants) was cloned into pcDNA3 and transiently expressed in COS-7 cells. After 28 h, cells were harvested in lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1.0% Triton X-100, 50 mm NaF, 5 mm pyrophosphate, 1 mm EDTA, 1 mm EGTA, and protease inhibitor mixture (Roche Applied Science)) and clarified by centrifugation. Lysates were incubated with 20 μl per 10-cm dish of anti-FLAG M2-agarose overnight at 4 °C with constant rotation. Lysates were washed extensively with PBS. Where required, FLAG-CaMKKβ was eluted in 50-μl fractions using 1 mg/ml FLAG peptide in PBS.
Site-directed Mutagenesis
All mutants used were generated using the QuikChange II XL site-directed mutagenesis kit (Stratagene, 200521). Primers were site-specific and are listed in supplemental Table 1. Mutations were confirmed by sequencing the entire coding region of mutated constructs at the Duke University DNA Analysis Facility.
CaMKKβ Kinase Assays
CaMKKβ catalytic activity was assessed by its ability to phosphorylate a bacterially expressed, catalytically inactive AMPKα1D139Aβ1γ1 heterotrimer. The catalytic aspartic acid (Asp-139) was mutated in the AMPKα1 subunit to eliminate background caused by autophosphorylation. Purification of AMPKα1β1γ1 from bacteria has been described previously (22). For each 25-μl assay, ∼50 ng of partially purified CaMKKβ was incubated in a buffer containing 1.5 μm AMPKα1D139Aβ1γ1, 0.1% Tween 20, 50 mm HEPES, pH 7.0, 150 mm NaCl, 6.25 mm MgCl2, 0.5 mm DTT, 200 μm ATP, and 2 μCi of EasyTides [γ-32P]ATP (PerkinElmer Life Sciences). Assays measuring Ca2+/CaM autonomous activity also contained 1 mm EGTA, and assays measuring Ca2+/CaM-dependent activity contained 1 mm CaCl2 and 1 μm bovine testis CaM. Reactions were terminated at the times indicated in the figure legends by the addition of SDS-loading buffer followed by boiling of the samples. The samples were resolved on 10% SDS-polyacrylamide gels and stained with Coomassie Blue. Gels were dried and subjected to autoradiography. Bands corresponding to the α-subunit of AMPK were cut from the gel, and the amount of 32P incorporated was measured using scintillation counting.
CaMKKβ Phosphopeptide Sequencing
FLAG-CaMKKβ (100 ng) was heated for 5 min at 100 °C, then reduced and alkylated with iodoacetamide, and digested in solution with trypsin (Promega) overnight at 37 °C. In solution, digestion was required for identification of the 121–139 and 121–142 peptides. Digests were acidified with formic acid and analyzed by LC-MS/MS. Chromatography was performed on a PepMap300 C18 column running at 500 nl/min with buffer A, 0.1% formic acid, and buffer B, 80% acetonitrile, 0.1% formic acid, using a Dionex 3000 rapid separation liquid chromatography-nano liquid chromatography. Peptides were resolved with a 40-min 4–60% B solvent gradient. Mass spectrometry was performed on a 5600 TripleTOF MS (AB-Sciex) using an MS/MS program that specifically targeted CaMKKβ(121–139) and -(121–142) peptides as well as a standard information-dependent acquisition MS/MS method.
CaMKKβ TOF Analysis
FLAG-CaMKKβ was eluted from anti-FLAG M2-agarose beads by washing with 100 mm glycine, pH 2.7, precipitated with methanol, and resuspended in 30% acetic acid before injection onto the liquid chromatography-MS. Chromatography was performed on a Dionex 3000 liquid chromatography using a 2.1 × 100-mm 300SB-C8 column (Agilent) at a flow rate of 250 μl/min. Buffer A is 0.1% formic acid; Buffer B is acetonitrile, 0.1% formic acid. Proteins were resolved on a 3–55% B gradient over 30 min. Mass spectrometry was performed on a QSTAR Pulsar i Q-TOF (Applied Biosystems) using a TurboIonSpray source (Applied Biosystems). Source conditions were as follows: gas = 16 p.s.i., gas temperature = 120 °C, and source voltage = 5000 V.
Phosphorylation of CaMKKβ by p35/CDK5 and GSK3β
GST-CaMKKβ was purified from E. coli through the glutathione-Sepharose step (21). Five μg of GST-CaMKKβ D311A or GST-CaMKKβ D311A, S137A were then incubated with 0.234 μg of active CDK5/p35 (Millipore, 14-477) in a 25-μl reaction containing 0.1% Tween 20, 50 mm HEPES, pH 7.0, 150 mm NaCl, 6.25 mm MgCl2, 0.5 mm DTT, 200 μm ATP, and 5 μCi of EasyTides [γ-32P]ATP (PerkinElmer Life Sciences). Reactions were terminated at the indicated times by adding SDS-PAGE loading dye and boiling. The samples were resolved on 10% SDS-polyacrylamide gels and stained with Coomassie Blue. Gels were dried and subjected to autoradiography.
To examine the requirement for CDK5/p35 priming of CaMKKβ, 0.5 μg of phosphatase-treated FLAG-CaMKKβ purified from HEK 293a cells was preincubated with 0.234 μg of active CDK5/p35 (Millipore, 14-477) in a 25-μl reaction containing 0.1% Tween 20, 50 mm HEPES, pH 7.0, 150 mm NaCl, 6.25 mm MgCl2, 0.5 mm DTT, and 200 μm ATP for an hour. Samples were then treated with 1 μm roscovitine (Santa Cruz Biotechnology, sc-24002) to inhibit CDK5 before addition of 0.234 μg of active GSK-3β (Millipore, 14-538). Reactions were terminated at the indicated times by addition of SDS-PAGE loading dye and boiling. Samples were then fractionated on 10% SDS-polyacrylamide gels and immunoblotted with the indicated antibodies.
Immunoblotting
Extracts were resolved on 8 or 10% SDS-polyacrylamide gels and transferred to Immobilon-FL PVDF membranes (Millipore, IPFL00010). Membranes were blocked in a buffer containing 0.2× PBS, 0.1% casein, 0.5% fish gelatin, and 0.04% sodium azide for 1 h at room temperature. Primary antibodies were diluted in blocking buffer with 0.1% Tween 20 at the following concentrations: panCaMKK (BD Biosciences, 610545) 1:1000; phosphoserine (BD Biosciences, 612546) 1:1000; FLAG (Sigma, F1804) 1:1000; FLAG (Santa Cruz Biotechnology, sc-807) 1:500; CaMKI 1:1000; CDK5 (Santa Cruz Biotechnology, sc-173) 1:500; p35 (Santa Cruz Biotechnology, sc-820) 1:500; β-actin (Sigma, A5441) 1:10,000; and pan-14-3-3 (Abcam, ab14112) 1:1000. After incubation with primary antibody, membranes were washed with 1× PBS containing 0.1% Tween 20. Membranes were then incubated with appropriate fluorescently conjugated secondary antibodies, either IRDye800 goat anti-mouse IgG (Rockland, 610-132–121) or Alexa Fluor 680 goat anti-rabbit IgG (Molecular Probes, A21109), at a 1:5000 dilution in blocking buffer containing 0.1% Tween 20 and 0.02% SDS. Membranes were washed with 1× PBS containing 0.1% Tween 20. Membranes were then scanned with an Odyssey Infrared Imager (Li-Cor Biosciences), and quantification of bands was performed with the Odyssey software version 3.0.
Two-dimensional Gel Electrophoresis
Isoelectric focusing step was performed in the Protean IEF cell (Bio Rad) using ReadyStrip IPG strips (Bio-Rad, 163-2015) with a pH range of 4–7 according to the manufacturer's instructions. For the second dimension, the IPG strip was resolved on a Criterion 12.5% Tris-HCl precast gel (Bio-Rad, 345-0102). Two-dimensional gels were then analyzed by immunoblot as above.
FLAG Immunoprecipitations
For immunoprecipitation of CaMKKβ from HEK 293a cells, 10 μg of FLAG-CaMKKβ were transiently transfected using 20 μl of Lipofectamine 2000 per 100 mm dish according to the manufacturer's protocol. Cells were treated with 10 mm roscovitine (Santa Cruz Biotechnology, sc-24002) or 100 nm GSK3 inhibitor XV (Calbiochem, 361558) as indicated. Lysates were made from cells 36–48 h after transfection in Nonidet P-40 lysis buffer. Lysates were clarified by centrifugation at 18,000 × g for 30 min and then incubated with 10 μl of anti-FLAG M2-agarose (Sigma, A2220) for 2 h at 4 °C with constant rocking. Beads were then washed twice in Nonidet P-40 lysis buffer and once in TBS and eluted with 1× TBS containing 300 ng/μl FLAG peptide (Sigma, F3290). Immunoprecipitates were then subjected to immunoblot analysis.
Pulse-Chase Experiments
FLAG-tagged CaMKKβ lentivirus was constructed by inserting two annealed primers containing the FLAG sequence and NheI overhangs into an NheI cut pRRLsin.PPT.hPGK.IRES.EGFP-mouse full-length CaMKKβ construct that has been described previously (10). CaMKKβ(S129A,S133A,S137A) was made by site-directed mutagenesis as described above. Primary mouse embryonic fibroblast (MEFs) were infected with concentrated lentivirus stocks expressing wild type FLAG-CaMKKβ or FLAG-CaMKKβ(S129A,S133A,S137A) following previously described procedures (10). MEFs were trypsinized and sorted by flow cytometry for GFP-positive cells 48 h after infection. Gates were set to collect populations of MEFs from both infections that had the same amount of GFP expression. Between 2 and 4 × 105 cells were plated in 60-mm culture dishes in DMEM (Mediatech, 10-013-CV) containing 10% FBS, 1 mm HEPES (Invitrogen, 13630), and penicillin/streptomycin solution (Mediatech, 30-002-CI) for 24 h after sort. Before labeling, cells were switched to DMEM lacking cysteine and methionine (Invitrogen, 21013-024) containing 10% FBS, 1 mm HEPES, and penicillin/streptomycin solution for 15 min. Cells were then labeled with 0.44 mCi/ml EasyTag EXPRE35S35S protein labeling mix (PerkinElmer Life Sciences, NEG772) for 10 min. After the 10-min pulse, cells were chased in regular DMEM supplemented with 10% FBS, 5 mm cysteine, 5 mm methionine, 1 mm HEPES, and penicillin/streptomycin solution. At the indicated times, cells were lysed in Nonidet P-40 lysis buffer, and FLAG-CaMKKβ was immunoprecipitated using anti-FLAG M2-agarose as described above. Immunoprecipitates were fractionated on an SDS-polyacrylamide gel and silver-stained to visualize protein. 35S incorporation into CaMKKβ bands was determined using a Storm 840 phosphorimager (GE Healthcare), and the signal was quantified using ImageQuant version 5.2 software (GE Healthcare).
Animals
Camkk2−/− mice were generated in our laboratory as described previously (18) and backcrossed into a C57BL/6J background for 10 generations. Control mice were generated from F1 heterozygous matings and backed-crossed into the C57BL/6J background for 10 generations. All animals were bred in the Duke University Medical Sciences Research Building animal facility under a 12-h light (6:00 a.m. to 6:00 p.m.), 12-h dark (6:00 p.m. to 6:00 a.m.) cycle. Food and water were provided ad libitum, and all animal care was in compliance with the National Institutes of Health and Duke University institutional guidelines governing the use of laboratory and experimental animals. CDK5 conditional knock-out brains and matching controls were a kind gift from the L-H. Tsai laboratory and are described elsewhere (23).
Primary Cerebellar Granule Cell (CGC) Isolation
CGCs were isolated from 6-day-old mice using a previously published procedure (10). Cells were cultured in Neurobasal media (NB, Invitrogen, 21103) supplemented with B27 (Invitrogen, 17504-044), sodium pyruvate, l-glutamine, 0.06 mg/ml glucose, and 25 mm KCl. CGCs were plated at a density of 2–4 × 106 cell/well in 6-well dishes that had been coated overnight with 10 μg/ml poly-d-lysine. CGCs were harvested for either protein or RNA analysis after 24 h in culture.
Cerebellar Granule Cell Transfection and Axon Growth Measurement
CGCs were plated on 12-mm round poly-d-lysine-coated coverslips placed in 24-well dishes at a density of 250,000 cells per well in NB media supplemented with B27, sodium pyruvate, l-glutamine, 0.06 mg/ml glucose, 25 mm KCl, and 10% Donor Equine Serum. After allowing the cells to rest for 4 h, medium was removed and replaced with NB medium with no supplements. Cells were transfected in this medium with Lipofectamine 2000. For each well, 1 μg of total DNA and 1 μl of Lipofectamine 2000 were used. After a 45-min incubation with the DNA-Lipofectamine 2000 complexes, the medium was replaced with the original culture medium. CGCs were then cultured for an additional 3 days at 37 °C with 5% CO2. After 3 days, cells were fixed in 4% formaldehyde in 1× PBS for 15 min at room temperature. Cells were permeabilized and blocked for 45 min at room temperature in a solution containing 0.2% Triton X-100, 1% Normal Donkey Serum, and 1× PBS. Primary antibodies were diluted as follows in 1× PBS, anti-GFP (Molecular Probes, A11122) 1:200, and anti-Tau (Sigma, T9450) 1:500. Coverslips were incubated with primary antibody overnight at 4 °C in a humidified chamber. The next day, coverslips were washed with 1× PBS before incubation with appropriate secondary antibodies, Alexa Fluor 488 anti-mouse IgG (Molecular Probes), or Cy3 anti-rabbit IgG (Jackson ImmunoResearch), diluted 1:500 in 1× PBS. After application of secondary antibody, coverslips were washed with 1× PBS with the last wash also containing 0.25 μg/ml DAPI. Coverslips were mounted on slides using ProLong Gold antifade mounting media (Invitrogen, P36934).
Stained CGCs were visualized on a Zeiss LSM 510 inverted confocal microscope using a 63× oil immersion objective. GFP staining was used to indicate transfected neurons, and care was taken to find GFP-positive neurons that did not overlap allowing for accurate measurement of neurite length. Length measurements were made with the line trace function of MetaMorph Premier software version 7.6.5. Neurons were characterized as polarized when the longest neurite was more than twice as long as the next longest neurite, as has been described previously (24). In all experiments, there were very few dead cells as determined by nuclei condensation, and only live cells were considered for analysis. Transfection efficiency was estimated by counting the number of GFP-positive cells as a percentage of total cells. For CGCs transfected with GFP alone, transfection efficiency was about 10%. All GFP-CaMKKβ constructs used had similar transfection efficiencies around 7%.
RNA Isolation and Quantitative Real Time PCR
Total RNA was extracted from ∼2 × 106 fresh or 1-day in vitro cerebellar granule neurons using the RNeasy mini kit (Qiagen, 74104), including the optional on-column DNase digestion (Qiagen, 79254). RNA yield was determined spectrophotometrically. Single-stranded cDNA was synthesized using SuperScript II RNase H− reverse transcriptase (Invitrogen, 18064-022) according to the manufacturer's protocol. Real time PCR was carried out on a CFX 96 real time system (Bio-Rad) using iQ SYBR Green Supermix (Bio-Rad, 170-8882) and the following primers: CaMKK2, 5′-CATGAAGGACGCTGC-3′ (forward) and 5′-TGACAACGCCATAGGAGCC-3′ (reverse); 18 S ribosomal protein, 5′-AGGGTTCGATTCCGGAGAGG-3′ (forward) and 5′-CAACTTTAATATACGCTATTGG-3′ (reverse). Relative levels of CaMKKβ mRNA were calculated using the (ΔΔC(t)) method with 18 S ribosomal protein as a reference gene.
RESULTS
N-terminal Phosphorylation of CaMKKβ Results in Altered CaM Dependence
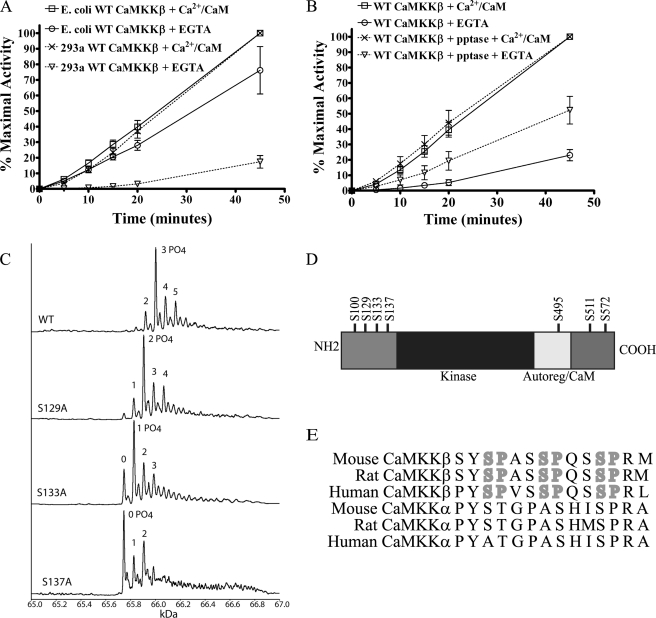
To evaluate the differences in CaMKKβ autonomous activity between enzyme preparations isolated from bacterial and mammalian sources, we purified CaMKKβ from E. coli and the human HEK 293a cell line. We tested the Ca2+/CaM-dependent and -independent activities of these enzyme preparations using bacterially expressed AMPKα1D139Aβ1γ1 as a substrate. AMPK was chosen as a substrate because it does not require Ca2+/CaM binding to be phosphorylated by CaMKKβ, as is the case with CaMKI and CaMKIV (7, 25, 26, 28). Thus, the use of AMPK makes it possible to measure the Ca2+/CaM dependence of CaMKKβ without concern for the Ca2+/CaM dependence of its substrate. When we tested the activity of our preparations of CaMKKβ, we confirmed the observation that bacterially expressed CaMKKβ has higher levels of Ca2+/CaM autonomous activity than that prepared from mammalian cells (Fig. 1A). This led us to hypothesize that CaMKKβ is modified in mammalian cells in a manner that keeps autonomous activity low and increases Ca2+/CaM dependence. As CaMKKβ has many putative phosphorylation sites, we questioned if this modification could be phosphorylation. To examine this possibility, we isolated CaMKKβ from cells that had been labeled with 32P-orthophosphate, treated it with λ-phosphatase, and found that phosphatase treatment decreased the total phosphate incorporation by 80% (supplemental Fig. 1). This result reveals that CaMKKβ is phosphorylated in vivo and that we can successfully remove the majority of this phosphorylation with λ-phosphatase treatment. Next, the Ca2+/CaM-dependent and -independent activities of phosphatase-treated HEK 293a-produced CaMKKβ were assayed using the same bacterially expressed AMPKα1D139Aβ1γ1 as a substrate. As shown in Fig. 1B, the phosphatase-treated CaMKKβ had increased Ca2+/CaM-independent activity compared with untreated CaMKKβ. Using TOF mass spectrometry analysis of FLAG-CaMKKβ produced in the mammalian COS-7 cell line, we observed multiple peaks corresponding to multiple phosphorylated forms of CaMKKβ (Fig. 1C). Phosphopeptide sequencing was then used to identify seven phosphorylation sites on CaMKKβ (Fig. 1D and supplemental Fig. 2, and data not shown). To determine which of these sites contribute to the Ca2+/CaM dependence of CaMKKβ, a series of serine to alanine mutants was constructed. Ser-100, Ser-495, and Ser-511 are homologous to PKA sites in CaMKKα (29, 30), and phosphorylation of CaMKKα by PKA on Ser-100 and Ser-511 leads to the recruitment of 14-3-3 and inactivation of the enzyme (19, 20). Interestingly, although we were able to confirm the observation that CaMKKα associates with 14-3-3 after stimulation of PKA activity, CaMKKβ was unable to recruit 14-3-3 under the same conditions (supplemental Fig. 3). Furthermore, a CaMKKβ S100A,S495A,S511A triple mutant had activity similar to the wild type protein in our in vitro kinase assays (data not shown). From these data, we conclude that the phosphorylated residues that alter CaMKKβ autonomous activity are not homologous to the PKA sites in CaMKKα.
FIGURE 1.
CaMKKβ activity is modified by phosphorylation. A, in vitro kinase assay measuring the ability of mammalian 293a CaMKKβ or E. coli CaMKKβ to phosphorylate AMPK. CaMKKβ preparations were incubated with bacterially produced AMPKα1D139Aβ1γ1 in the presence of [γ-32P]ATP and Ca2+/CaM or EGTA for the indicated amounts of time. Reactions were then subjected to SDS-PAGE and staining with Coomassie Blue. Bands corresponding to the AMPKα subunit were excised, and the amount of 32P incorporated was measured by scintillation counting. Activities are presented as a percent of the maximal activity (CaM-dependent activity at 45 min) to correct for variation in protein yields. Data shown are the average of at least three independent experiments. Error bars represent standard error. B, in vitro kinase assay comparing the activity of wild type and λ-phosphatase-treated CaMKKβ purified from mammalian cells. Assays were performed as in A. Data shown are the average of at least three independent experiments. Error bars represent standard error. C, TOF-MS analysis of wild type (WT) CaMKKβ. The major peak corresponds to a triple-phosphorylated form of CaMKKβ. Shift of the dominant peak in TOF-MS analysis of CaMKKβ S129A, S133A, and S137A single mutants implies sequential phosphorylation. D, schematic of CaMKKβ domain structure showing phosphorylated residues identified by mass spectrometry. Autoreg/CaM, autoregulatory domain and calmodulin binding domains that overlap in CaMKKβ. E, alignment of region surrounding N-terminal phosphorylation sites Ser-129, Ser-133, and Ser-137. Phosphorylation sites with neighboring prolines are highlighted in boldface type. Motif is conserved in mouse, rat, and human CaMKKβ but not in CaMKKα.
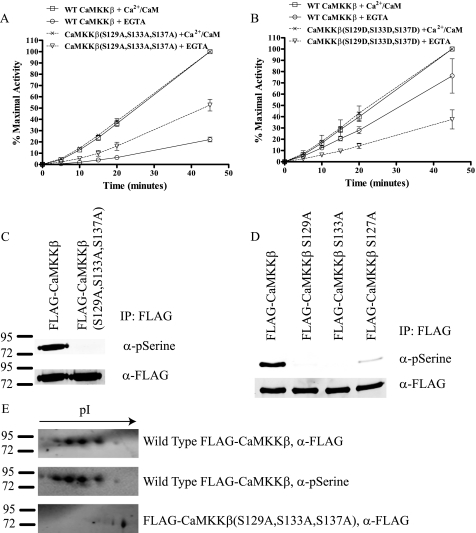
Previous studies involving truncation mutants of CaMKKβ suggest that a region of the N terminus spanning residues 129–151 is responsible for generating autonomous activity of the enzyme (21). Our mass spectrometry phosphopeptide sequencing data revealed three CaMKKβ phosphorylation sites, Ser-129, Ser-133, and Ser-137, which fall within this region (supplemental Fig. 2). These sites are conserved in mouse, rat, and human isoforms of CaMKKβ, but the region shares little homology with CaMKKα (Fig. 1E). Thus, we made a CaMKKβ S129A,S133A,S137A triple mutant (CaMKKβ(S129A,S133A,S137A)) and compared its activity with the wild type protein. We found that this phospho-deficient mutant did indeed exhibit increased autonomous activity with no change in Ca2+/CaM-dependent activity (Fig. 2A). In addition, activity of CaMKKβ(S129A,S133A,S137A) was unaltered by λ-phosphatase treatment indicating that these are the primary residues responsible for altering Ca2+/CaM autonomous activity (data not shown).
FIGURE 2.
Phosphorylation of CaMKKβ on Ser-129, Ser-133, and Ser-137 in the N terminus is responsible for decreased autonomous activity. A, in vitro kinase assay measuring the ability of wild type CaMKKβ and CaMKKβ(S129A,S133A,S137A) produced in mammalian cells to phosphorylate AMPK. Assays were performed as in Fig. 1A. Data shown are the average of at least three independent experiments. Error bars represent standard error. B, in vitro kinase assay measuring the activity of bacterially expressed wild type CaMKKβ or CaMKKβ(S129D,S133D,S137D). Assays were performed as in Fig. 1A. Data shown are the average of at least three independent experiments. Error bars represent standard error. C, FLAG-tagged wild type CaMKKβ or CaMKKβ(S129A,S133A,S137A) were partially purified from HEK 293a cells and immunoblotted with a commercially available phosphoserine (pSerine) antibody. The membrane was also blotted for FLAG as a loading control. D, FLAG-tagged wild type CaMKKβ or CaMKKβ S129A, S133A, or S137A single point mutants were partially purified from HEK 293a cells and immunoblotted with a commercially available phosphoserine antibody. The membrane was also blotted for FLAG as a loading control. E, two-dimensional gel analysis of wild type CaMKKβ and CaMKK(S129A,S133A,S137A) purified from mammalian cells. Top and middle panels show wild type CaMKKβ blotted with FLAG and phosphoserine antibodies, respectively. Bottom panel shows CaMKKβ(S129A,S133A,S137A) blotted with FLAG antibody.
To further validate importance of Ser-129, Ser-133, and Ser-137 in generation of CaMKKβ autonomous activity, we mutated them to aspartic acid in a bacterially produced version of CaMKKβ (CaMKKβ(S129D,S133D,S137D)) in an attempt to mimic the phosphorylated state. Comparison of the activity of this phospho-mimetic to the wild type protein revealed that CaMKKβ(S129D,S133D,S137D) had decreased autonomous activity (Fig. 2B). Thus, the bacterially produced CaMKKβ(S129D,S133D,S137D) mimics the phosphorylated form of the wild type protein purified from mammalian cells. Taken together, these data indicate that phosphorylation of CaMKKβ at Ser-129, Ser-133, and Ser-137 in the N terminus of the protein leads to decreased Ca2+/CaM autonomous activity and that phosphorylation of these sites can be mimicked in the bacterially expressed form of the enzyme by mutating the serines to aspartic acids.
The dominant peak from our TOF mass spectrometry corresponds to a triple phosphorylated form of CaMKKβ. We then questioned if this peak corresponded to CaMKKβ phosphorylated on Ser-129, Ser-133, and Ser-137. To test this, we performed TOF mass spectrometry on the corresponding serine to alanine mutants of CaMKKβ expressed in COS-7 cells. The S137A mutant resulted in the dominant CaMKKβ peak having no phosphates, whereas the S133A mutant major peak had a single phosphate and the S129A mutant had two phosphates (Fig. 1C). These data indicate that Ser-129, Ser-133, and Ser-137 contribute to the dominant phosphorylated form of CaMKKβ. In addition, these data indicate that phosphorylation of Ser-129, Ser-133, and Ser-137 are coordinately regulated with priming of Ser-137 being required before subsequent phosphorylation of Ser-133 and then Ser-129. To further validate our TOF mass spectrometry data, we performed a two-dimensional gel analysis on wild type CaMKKβ and CaMKKβ(S129A,S133A,S137A). To detect phosphorylated protein, we used a commercially available phosphoserine antibody, which we found recognized wild type CaMKKβ but not CaMKKβ(S129A,S133A,S137A) (Fig. 2C). To determine which sites the phosphoserine antibody recognized, we tested its reactivity with CaMKKβ S129A, S133A, and S137A single point mutants. As seen in Fig. 2D, mutating any one of these three sites individually leads to loss of reactivity with the phosphoserine antibody. Because our TOF mass spectrometry data with the CaMKKβ single mutants indicate mutation of Ser-133 blocks phosphorylation of Ser-129 and mutation of Ser-137 blocks phosphorylation of Ser-133 and Ser-129, we suggest that the phosphoserine antibody is most likely recognizing Ser-129. In the two-dimensional gel, wild type CaMKKβ resolved into a series of four spots, which all reacted with the phosphoserine antibody (Fig. 2E). CaMKKβ(S129A,S133A,S137A) resolved into a single spot that was right-shifted when compared with the wild type protein and failed to show any reactivity with the phosphoserine antibody (Fig. 2E and data not shown). From this experiment, we conclude that in cells CaMKKβ is phosphorylated constitutively on Ser-129, Ser-133, and Ser-137.
N Terminus of CaMKKβ Is Sequentially Phosphorylated by CDK5 and GSK3
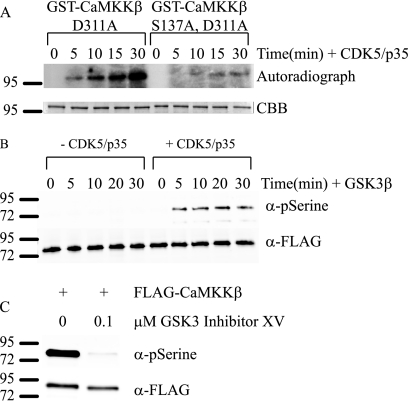
Because CaMKKβ is phosphorylated in mammalian cells, we wanted to determine the protein kinase or kinases responsible. From our TOF mass spectrometry data, we suspected that phosphorylation of Ser-129, Ser-133, and Ser-137 is coordinately regulated. Supporting this idea, the spacing of the residues resembles a consensus glycogen synthase kinase 3 (GSK3) phosphorylation motif, and it is common for GSK3 to require a priming phosphorylation event (31). If this were the case for CaMKKβ, the prediction would be that Ser-137 was phosphorylated first, followed by GSK3-dependent phosphorylation of Ser-133 and Ser-129. To help direct the search for the putative CaMKKβ priming kinase, we used the Scansite program to search kinases that might recognize the region around the Ser-137 site and identified cyclin-dependent kinase 5 (CDK5) (32). To test this idea, we incubated purified active CDK5 with bacterially produced and catalytically inactive GST-CaMKKβ D311A in the presence of [γ-32P]ATP and found CaMKKβ to be phosphorylated with 32P (Fig. 3A). Because there was a greatly diminished phosphorylation of a GST-CaMKKβ D311A,S137A mutant, it is likely that Ser-137 is the primary site of phosphorylation by CDK5 (Fig. 3A). We then examined the phosphorylation of CaMKKβ by GSK3β. For these experiments, we incubated phosphatase-treated HEK 293a-purified CaMKKβ in the presence or absence of CDK5 for an hour before addition of purified, active GSK3β. We used the phosphoserine antibody to detect phosphorylation of CaMKKβ. We found that incubation with either CDK5 or GSK3β alone was insufficient to produce reactivity with the phosphoserine antibody. However, incubation with CDK5 followed by GSK3β produced a robust phosphoserine signal. These data demonstrate that CDK5 and GSK3β can phosphorylate CaMKKβ in vitro and that the phosphorylation events catalyzed by these protein kinases are sequential, with phosphorylation of Ser-137 by CDK5 being required for the subsequent phosphorylation of CaMKKβ by GSK3β on Ser-133 and Ser-129.
FIGURE 3.
CDK5 and GSK3 phosphorylate CaMKKβ in vivo. A, in vitro kinase assay in which 5 μg of bacterially purified GST-CaMKKβ D311A or GST-CaMKKβ D311A, S137A was incubated with 0.234 μg of active p35/CDK5 in the presence of [γ-32P]ATP for the indicated times. Top panel is autoradiograph, and bottom panel is Coomassie-stained gel to show loading. CBB, Coomassie Blue. B, in vitro kinase assay in which 0.5 μg of HEK 293a-expressed FLAG-CaMKKβ was incubated with active GSK3β either with (right) or without (left) preincubation with CDK5/p35. C, FLAG-CaMKKβ was transiently overexpressed in HEK 293a cells. The cells were then treated overnight with 0.1 μm GSK3 inhibitor XV before being lysed. CaMKKβ was immunoprecipitated from the lysate and immunoblotted using the phosphoserine (pSerine) antibody described in Fig. 2C and FLAG antibody as a loading control.
We next questioned if CaMKKβ could be phosphorylated by CDK5 and GSK3 in our HEK 293a cell system. To test if CaMKKβ is phosphorylated by GSK3, we inhibited GSK3 activity using the GSK3 inhibitor XV compound (33). After an overnight treatment of the cells with 0.1 μm of the GSK3 inhibitor, all reactivity with the phosphoserine antibody was lost (Fig. 3C). This indicates that CaMKKβ fails to be phosphorylated in the absence of GSK3 activity. We also treated HEK 293a cells with 10 μm roscovitine, an inhibitor of CDC2, CDK2, and CDK5. Treatment of cells with roscovitine was limited to 1 h as previous reports have demonstrated that CDK5/p35 is turned over rapidly, and newly synthesized kinase will be free of inhibitor (34). Following this short treatment, we were unable to detect any change in phosphoserine levels on CaMKKβ (data not shown). This is most likely due to the short duration of treatment, and it does not exclude CDK5 as the kinase responsible for CaMKKβ phosphorylation.
CDK5-dependent Phosphorylation Regulates the Turnover of CaMKKβ
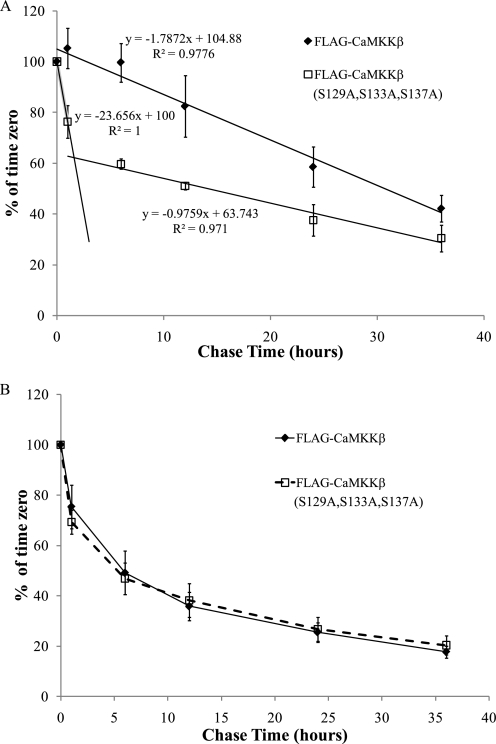
While working with the CaMKKβ mutants, we noted that expression of CaMKKβ(S129D,S133D,S137D) was higher than wild type CaMKKβ (data not shown). This suggested to us that phosphorylation of CaMKKβ may influence its turnover. To test this theory, we used a pulse-chase experiment to measure the rate of turnover of wild type CaMKKβ and CaMKKβ(S129A,S133A,S137A). MEFs, a readily available primary cell line, were infected with lentiviruses expressing either wild type CaMKKβ or CaMKKβ(S129A,S133A,S137A). The MEFs were then pulsed with media containing 35S-labeled methionine and -cysteine. After a 10-min pulse, the cells were chased in label-free media for the indicated times. We found that wild type CaMKKβ decays linearly with a half-life of 30.7 h (Fig. 4A). Interestingly, decay of CaMKKβ(S129A,S133A,S137A) is bi-phasic. During the first phase, the mutant protein is rapidly degraded with a half-life of 2.1 h. However, by the 6-h chase time point, ∼60% of the labeled protein remains, and the decay curve levels off giving a half-life of 51.2 h. As a control, we measured the rate of 35S turnover in crude lysates, and we found no difference between those derived from the CaMKKβ(S129A,S133A,S137A) or wild type lysates (Fig. 4B). These data indicate that phosphorylation of newly synthesized CaMKKβ is critical for its stability, as protein that cannot be phosphorylated is rapidly degraded. Thus, one function of phosphorylation of CaMKKβ is to regulate stability of newly synthesized protein. Because CDK5 phosphorylation is required prior to GSK3β, we believe that regulation of CDK5 activity is probably most influential in determining CaMKKβ phosphorylation status. Thus, we predicted that the absence of CDK5 activity would lead to decreased levels of CaMKKβ protein.
FIGURE 4.
Phosphorylation of CaMKKβ regulates its turnover. A, pulse-chase experiment examining the half-life of CaMKKβ in MEFs. MEFS were infected with a lentivirus containing either wild type FLAG-CaMKKβ or FLAG-CaMKKβ(S129A,S133A,S137A). Cells were pulsed with 35S-labeled Cys/Met mixture and after that pulse cells were chased in normal medium supplemented with 5 mm methionine and cysteine. Cells were harvested at the indicated times, and CaMKKβ was immunoprecipitated with a-FLAG M2-agarose. The immunoprecipitates were subjected to SDS-PAGE and silver-stained, and 35S incorporated into CaMKKβ was quantified using a PhosphoImager. These values were expressed as a percent of signal at time 0. Data are average of four experiments. Error bars represent standard error. B, to ensure that 35S labeling was properly quenched during the pulse-chase experiments in A, 5–10 μg of total protein from each time point was precipitated with TCA, and total counts present were quantified by scintillation counting. As expected, amount of 35S radioactivity decreases rapidly throughout the experiment. Data are average of four experiments. Error bars represent standard error.
CDK5/p35 Activity Regulates the Level of CaMKKβ in Primary Brain Tissues and Cultured Neurons
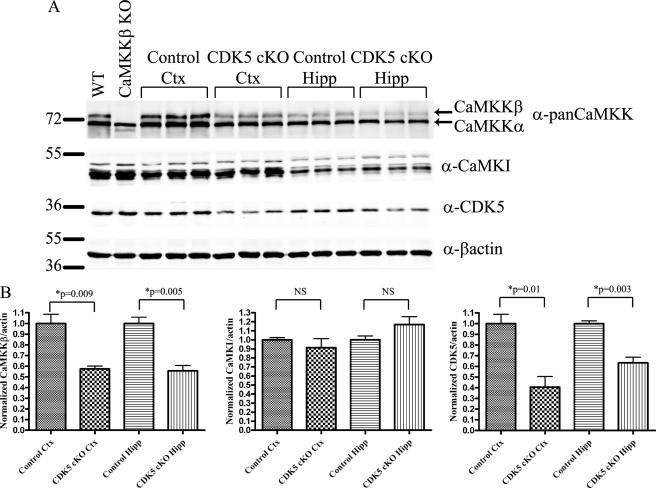
To examine the consequence of loss of CDK5 activity on CaMKKβ, we examined CaMKKβ levels in a CDK5 knock-out mouse model. The CDK5 global knock-out mouse displays embryonic and perinatal mortality (35), so we used CDK5 conditional knock-outs that lack CDK5 in CaMKIIα-positive neurons (23). We examined CaMKKβ in cortex and hippocampus from these animals, as these are regions of the brain where both CaMKKβ and CDK5 are expressed (2, 36). In accordance with our prediction that CDK5 phosphorylation regulates CaMKKβ levels, we found a 43% reduction of CaMKKβ in cortex and a 45% reduction in hippocampus in the CDK5 conditional knock-outs (Fig. 5, A and B). Levels of the CaMKKβ substrate CaMKI were unchanged, indicating specificity for CaMKKβ (Fig. 5, A and B). From this, we concluded that absence of CDK5 activity leads to a reduction in CaMKKβ, and this effect is specific as CaMKI levels are not altered.
FIGURE 5.
CDK5 activity correlates with CaMKKβ levels in mouse brain tissues. A, lysates made from control or CDK5 conditional knock-out (CDK5 cKO) cortex (Ctx) or hippocampus (Hipp) were blotted with antibodies to pan-CaMKK, CaMKI, β-actin, and CDK5. To help in the identification of CaMKKβ versus CaMKKα, brain extracts from wild type and CaMKKβ knock-out mice were run in the left two lanes. B, quantification of A. Error bars represent standard error, and p values were calculated using an unpaired Student's t test.
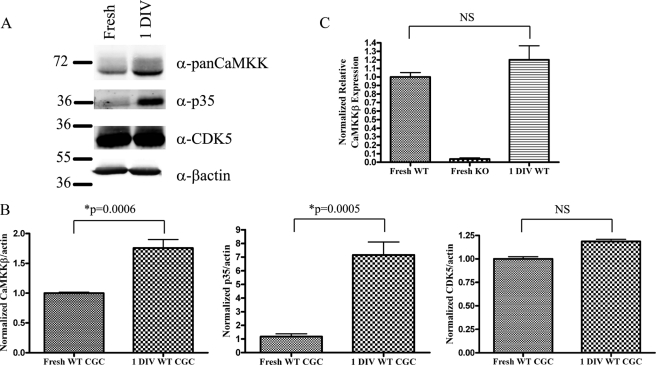
Interestingly, levels of CDK5 activity are developmentally regulated in the central nervous system and correlate with the cessation of neuronal proliferation and the start of migration and neurite outgrowth (36). Much like the cyclin-dependent kinases, CDK5 activity is dependent on expression and binding of a small protein activator (37). Two CDK5 activators, p35 and p39, have been identified with p35 being the better studied of the two. A good in vitro model of neuronal differentiation is the culture of cerebellar granule cells (CGCs). Cerebellar granule precursors can be isolated from neonatal mouse cerebellum and differentiate in culture. Previous work from our laboratory has shown that lack of CaMKKβ impairs the ability of CGCs to cease proliferation in the external granule cell layer, and migrate into the internal granule cell layer of the cerebellum (10). Thus, we decided to examine the relationship between CDK5 and CaMKKβ in cultured primary CGCs. We compared the protein levels of CDK5, p35, and CaMKKβ in lysates from freshly isolated, mitotically active CGC precursors and CGCs that had been cultured for 1 day in vitro allowing them to differentiate. Although levels of CDK5 were unchanged, we observed a large increase in p35 levels in differentiated cells, which correlated with an increase in total CaMKKβ (Fig. 6, A and B). This increase in CaMKKβ was not due to a change in the level of CaMKKβ transcription, as levels of mRNA remained constant (Fig. 6C). Thus, levels of CaMKKβ increase during neuronal development, and this increase correlates with increased CDK5 activity.
FIGURE 6.
CDK5 activity correlates with CaMKKβ levels in primary CGC cultures. A, 75 μg of lysate from freshly isolated WT or 1 day in vitro (1DIV) WT CGCs were immunoblotted with pan-CaMKK, p35, CDK5, and β-actin antibodies. Representative blot is from four independent experiments. B, quantification of A. Combined analysis is from four independent experiments. Error bars represent standard error, and p values were calculated using an unpaired Student's t test. C, real time PCR analysis of CaMKKβ mRNA from freshly isolated WT or 1 day in vitro (1DIV) WT CGCs. Combined analysis is from three independent experiments. Error bars represent standard error, and changes in CaMKKβ mRNA were determined to be nonsignificant (NS) based on a p value >0.05 as calculated by an unpaired Student's t test.
Phosphorylation of CaMKKβ by CDK5 Is Important in Neurite Outgrowth
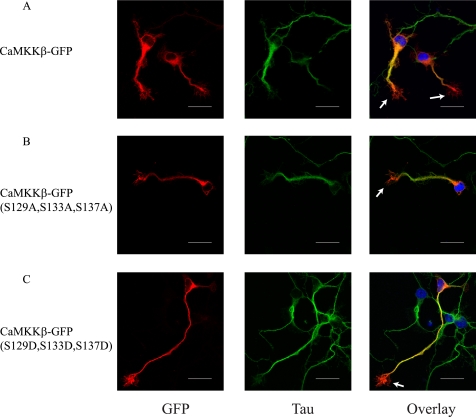
We next questioned why induction of CaMKKβ is important during development of CGC. To address this question, we first examined the subcellular localization of CaMKKβ in developing CGCs after transfection of CaMKKβ null CGCs with GFP fusions of wild type CaMKKβ, CaMKKβ(S129A,S133A,S137A), or CaMKKβ(S129D,S133D,S137D). CaMKKβ was visualized by staining for GFP in fixed neurons, and axons were visualized with Tau-1 staining. We found that all three CaMKKβ constructs shared a similar distribution pattern with signal in the cell body and along the axon. In addition, we found CaMKKβ to be enriched in growth cones (indicated with arrows, Fig. 7, A–C). Thus, CaMKKβ is widely distributed in neurons, and localization is not grossly disrupted by changing phosphorylation status.
FIGURE 7.
Localization of CaMKKβ in developing CGCs. A–C, CaMKKβ null CGCs were transfected with GFP-tagged wild type CaMKKβ (A), CaMKKβ(S129A,S133A,S137A) (B), or CaMKKβ(S129D,S133D,S137D) (C) constructs as indicated on the left. Cells were then stained for GFP (red) to visualize CaMKKβ and Tau-1 (green) to visualize axons. Panel on right shows overlay of signals along with DAPI (blue) to stain nucleus. Arrows point to growth cones. Scale bar, 20 μm. Representative images are from four independent experiments.
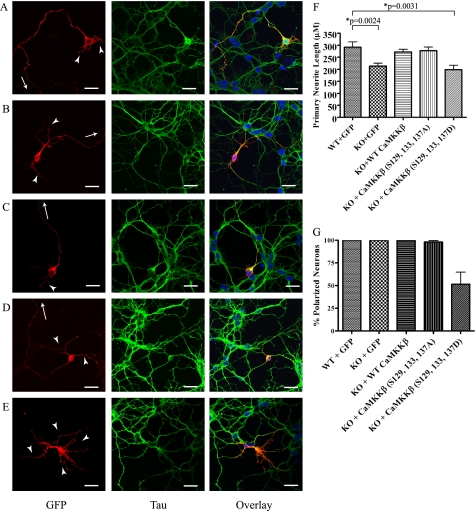
The growth cone is a specialized structure that controls outgrowth of neurites by responding to extracellular cues, some of which trigger Ca2+ signaling (38). Because CaMKKβ localizes to growth cones, we hypothesized that it may play a role in neurite outgrowth. In cerebellar granule neurons, as in most other neurons, neurite outgrowth is polar leading to a typical morphology characterized by a single long axon and several shorter dendrites. To examine the role of CaMKKβ in neurite outgrowth, we examined the morphology of CGCs isolated from wild type and CaMKKβ null mice transfected with GFP. After 3 days in vitro, neurons isolated from both wild type and CaMKKβ null mice displayed a consistent polar morphology, including a single long process (axon) and multiple shorter processes (dendrites) (Fig. 8, A, B, and G). However, when we measured the length of the longest primary neurite, we found a significant decrease in the CaMKKβ null CGCs (Fig. 8F). This indicated a defect in neurite extension and is consistent with previous reports (11, 13). We then questioned what effect wild type CaMKKβ, CaMKKβ(S129A,S133A,S137A), or CaMKKβ(S129D,S133D,S137D) would have on CGC morphology and neurite outgrowth. Thus, we transfected CaMKKβ null CGCs with GFP fusions of these constructs and analyzed them as we did the wild type and CaMKKβ null CGCs. We found that the neurons expressing both the wild type and CaMKKβ(S129A,S133A,S137A) constructs had normal polar morphology (Fig. 8, C, D, and G). However, the neurons transfected with CaMKKβ(S129D,S133D,S137D) displayed a striking phenotype whereby they failed to establish appropriate axon-dendrite polarity and instead grew several long neurites (Fig. 8, E and G). This defect is similar to that seen in rat hippocampal neurons overexpressing active CaMKI (12) and indicates a defect in establishment of appropriate polarity during neurite outgrowth. We next measured the length of the longest neurite in the CaMKKβ null CGC expressing wild type CaMKKβ, CaMKKβ(S129A,S133A,S137A), and CaMKKβ(S129D,S133D,S137D) constructs. We found that both wild type CaMKKβ and CaMKKβ(S129A,S133A,S137A) were able to restore the decrease in primary neurite length observed in the CaMKKβ null neurons (Fig. 8F). However, CaMKKβ(S129D,S133D,S137D) was unable to rescue the defect in primary neurite length (Fig. 8F). This is consistent with the observation that these neurons are nonpolar and fail to grow a single long process (Fig. 8, E and G). Together, these data indicate that appropriate regulation of CaMKKβ by phosphorylation is critical for neurite outgrowth during neuronal development.
FIGURE 8.
Phosphorylation of CaMKKβ is important in neurite outgrowth. A–E, representative images showing morphology of wild type neurons transfected with GFP (A), CaMKKβ null neurons transfected with GFP (B), or CaMKKβ null neurons rescued with wild type CaMKKβ-GFP (C), CaMKKβ(S129A,S133A,S137A)-GFP (D), or CaMKKβ(S129D,S133D,S137D)-GFP (E). Neurons were stained with GFP (red) to visualize CaMKKβ and Tau (green) to visualize axons. Panel on right shows overlay of signals along with DAPI (blue) to stain nucleus. Arrows point to primary neurite (axon) that continues out of view, and arrowheads indicate shorter secondary neurites (dendrites). Scale bar, 20 μm. F, length of axons as measured in WT or CaMKKβ null (KO) CGCs transfected with the indicated GFP fusion constructs after 3 days in vitro. At least 40 neurons were measured for each condition from three independent experiments. Error bars represent standard error, and p values were calculated using an unpaired Student's t test. G, polarity of neurons was determined for WT and CaMKKβ null (KO) CGCs transfected with the indicated GFP fusion constructs after 3 days in vitro. At least 40 neurons were measured for each condition from three independent experiments. Error bars represent standard error.
DISCUSSION
In this study, we report the novel regulation of CaMKKβ activity by multisite phosphorylation. We find that CaMKKβ autonomous activity is decreased following phosphorylation on three sites in the N terminus (Ser-129, Ser-133, and Ser-137), and we identify CDK5 and GSK3 as the kinases responsible for these phosphorylation events and show they act sequentially with CDK5 priming for subsequent phosphorylation by GSK3. In addition, we find that CaMKKβ isolated from mammalian cells is constitutively phosphorylated, leading us to believe that this is a mechanism by which cells keep CaMKKβ autonomous activity low allowing for tight control of activity by Ca2+/CaM.
In addition to regulation of autonomous activity, we find that phosphorylation of CaMKKβ by CDK5 and GSK3 regulates its half-life. From the results of pulse-chase experiments, we conclude that phosphorylation of Ser-129, Ser-133, and Ser-137 is critical for stability of newly synthesized protein. Because CaMKKβ needs to be phosphorylated by CDK5 before it can be phosphorylated by GSK3β, we predicted that regulation of CDK5 activity would correlate with CaMKKβ phosphorylation and stability. Our data from CDK5 conditional knock-out mice support this as the cortex and hippocampus from these animals have a significant decrease in CaMKKβ protein. In addition to CaMKKβ, the CDK5 substrate tyrosine hydroxylase is regulated in a similar fashion as phosphorylation also regulates its stability (39). Thus, one important effect of CaMKKβ phosphorylation is to stabilize and thereby increase cellular levels of CaMKKβ.
CDK5 activity is developmentally regulated and correlates with a switch from a proliferative state to a migratory state in neurons (36). Although CDK5 is ubiquitously expressed, its activity is regulated by co-expression of two small protein activators p35 and p39 (37, 40, 41). Studies in CDK5, p35, and p39 knock-out models demonstrate the importance of CDK5 activity in neuronal development, especially in the cortex (35, 42). In addition, CDK5 activity is important in the migration of granule cells in the developing cerebellum (43). Previous data from our laboratory demonstrated that the absence of CaMKKβ causes a defect in the developing cerebellum whereby granule cells fail to cease proliferation and migrate properly (10). This led us to believe that induction of CDK5 activity during development would increase CaMKKβ levels allowing for its proper function in developing neurons. Our data from cultured CGCs support this theory. In CGCs, we reveal that CaMKKβ levels increase in concert with p35 levels as the cells differentiate in culture. Thus, CaMKKβ levels are regulated during development.
We then questioned the role up-regulation of CaMKKβ may play during CGC development. When we examined CaMKKβ localization in CGC, we noticed it was enriched in growth cones. Enrichment of CDK5 and its activator p35 has also been reported in growth cones, and work in cultured cortical neurons demonstrated a requirement for CDK5 during neurite outgrowth (44). Previous work has demonstrated that a CaMKK/CaMKI pathway contributes to axon outgrowth through the regulation of growth cone motility (11). This led us to believe that phosporylation of CaMKKβ by CDK5 in the growth cone could be important in neurite outgrowth, so we examined neurite outgrowth in our CGC model. We found that CaMKKβ null CGCs grow shorter primary neurites than wild types, and we were able to rescue this defect with wild type CaMKKβ and CaMKKβ(S129A,S133A,S137A). This indicates that phosphorylation of CaMKKβ is not critical for its role in neurite extension. Surprisingly, CaMKKβ(S129D,S133D,S137D) was unable to recover the defect in primary neurite outgrowth and caused an additional defect in the establishment of appropriate neuronal polarity. This indicates CaMKKβ has an additional role in the early stages of neurite outgrowth during which appropriate axon-dendrite polarity is established from the polarized emergence of axons and dendrites (45). This is consistent with previous work showing that constitutive activation of CaMKK/CaMKI pathways during the early stages of hippocampal neuron differentiation leads to the development of multiple axons (12). Thus, we conclude from our data that appropriate phosphorylation of CaMKKβ is most critical during the early stages of neurite outgrowth in which neuronal polarity is established.
During the establishment of neuronal polarity, signaling molecules acquire a polarized distribution because of processes such as local protein translation and degradation in the emerging processes (45). One intriguing explanation for the phenotype generated by CaMKKβ(S129D,S133D,S137D) is that CaMKKβ is locally synthesized and phosphorylated in the neurite that will become the axon leading to its enhanced stability. Local synthesis of polarity proteins such as Par-3 is known to occur in axons and be essential for axon specification and growth (46). In addition, the CaMKKβ downstream target cAMP-response element-binding protein is also known to be locally translated in axons where it is postulated to be more susceptible to modification before being transported to the nucleus where it influences transcription of target genes (47). If CaMKKβ was also locally modified by CDK5, in the case of CaMKKβ(S129D,S133D,S137D) this localized regulation would be lost and could lead to the inappropriate initiation and growth of the multiple nonpolarized neurites seen in our experiments. Additional work will be required to confirm this theory.
There are several possible CaMKKβ downstream targets that could mediate its role in neurite outgrowth. In cortical neurons, CaMKK-CaMKIα and CaMKK-CaMKIγ pathways have been implicated in the growth of axons and dendrites, respectively (13, 48). In addition, regulation of CaMKIγ by CaMKK has also been implicated in the early stages of axon formation (12). In cortical and hippocampal neurons, both CaMKKα and CaMKKβ are expressed, and it is not clear in these model systems which isoform is most important for CaMKI activation. Because CGCs express higher levels of CaMKKβ than CaMKKα (49), it is reasonable to assume CaMKKβ may be the more important kinase isoform in this type of neuron.
In addition to regulating CaMKI, CaMKKβ also regulates CaMKIV. Our previous work on the cerebellum of CaMKIV and CaMKKβ null animals implicates a CaMKKβ/CaMKIV/cAMP-response element-binding protein signaling pathway in the regulation of brain-derived neurotrophic factor (BDNF) transcription. BDNF is an important neurotrophin in CGC development influencing survival, migration, neurite outgrowth, and synapse development (50–52). Recent work also demonstrates that the BDNF receptor TrkB adopts a polarized distribution in CGCs with the highest levels present in the leading process (53). This polarization is critical for migration of CGCs along BDNF gradients in the developing cerebellum. Because CaMKKβ regulates BDNF levels, disruption of this signaling pathway may adversely influence the establishment of BDNF gradients critical for appropriate CGC polarization and migration.
Recent work has also implicated the CaMKKβ target AMPK in neuronal polarization and neurite growth (27). In these studies, hippocampal neurons were treated with the AMPK agonists metformin or 5-aminoimidazole-4-carboxamide-1-β-riboside, and Amato et al. (27) noticed a failure to establish appropriate polarity as well as a decrease in total neurite length. This is similar to what we observe when we overexpress CaMKKβ(S129D,S133D,S137D). Further work is required to determine whether the phenotype we observe is due to increased activation of AMPK.
Thus, we have described herein a novel CDK5/GSK3-dependent mechanism responsible for the regulation of CaMKKβ activity. In the future, it will be necessary to consider the phosphorylation status of CaMKKβ in addition to Ca2+/CaM binding to better understand regulation of CaMKKβ-dependent signaling pathways.
Supplementary Material
Acknowledgments
We greatly appreciate the kind gift of the CMV-p35 construct as well as CDK5 conditional knock-out brain samples from Susan Su and Dr. Li-Huei Tsai (Picower Institute for Learning and Memory, Massachusetts Institute of Technology, Cambridge, MA). We also thank Josep Colomer, Kristin Anderson, and Libby MacDougall for their helpful comments on the manuscript and Tom Ribar for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants DK074701 and GM033976.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3 and Table 1.
- CaMKK
- Ca2+/calmodulin-dependent protein kinase kinase
- AMPK
- AMP-dependent protein kinase
- CaM
- calmodulin
- CaMK
- Ca2+/calmodulin-dependent protein kinase
- CGC
- cerebellar granule cell
- BDNF
- brain-derived neurotrophic factor
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1. Tokumitsu H., Wayman G. A., Muramatsu M., Soderling T. R. (1997) Biochemistry 36, 12823–12827 [DOI] [PubMed] [Google Scholar]
- 2. Anderson K. A., Means R. L., Huang Q. H., Kemp B. E., Goldstein E. G., Selbert M. A., Edelman A. M., Fremeau R. T., Means A. R. (1998) J. Biol. Chem. 273, 31880–31889 [DOI] [PubMed] [Google Scholar]
- 3. Lee J. C., Edelman A. M. (1994) J. Biol. Chem. 269, 2158–2164 [PubMed] [Google Scholar]
- 4. Tokumitsu H., Enslen H., Soderling T. R. (1995) J. Biol. Chem. 270, 19320–19324 [DOI] [PubMed] [Google Scholar]
- 5. Means A. R. (2000) Mol. Endocrinol. 14, 4–13 [DOI] [PubMed] [Google Scholar]
- 6. Chatila T., Anderson K. A., Ho N., Means A. R. (1996) J. Biol. Chem. 271, 21542–21548 [DOI] [PubMed] [Google Scholar]
- 7. Tokumitsu H., Soderling T. R. (1996) J. Biol. Chem. 271, 5617–5622 [DOI] [PubMed] [Google Scholar]
- 8. Mizuno K., Antunes-Martins A., Ris L., Peters M., Godaux E., Giese K. P. (2007) Neuroscience 145, 393–402 [DOI] [PubMed] [Google Scholar]
- 9. Mizuno K., Ris L., Sánchez-Capelo A., Godaux E., Giese K. P. (2006) Mol. Cell. Biol. 26, 9094–9104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kokubo M., Nishio M., Ribar T. J., Anderson K. A., West A. E., Means A. R. (2009) J. Neurosci. 29, 8901–8913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wayman G. A., Kaech S., Grant W. F., Davare M., Impey S., Tokumitsu H., Nozaki N., Banker G., Soderling T. R. (2004) J. Neurosci. 24, 3786–3794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davare M. A., Fortin D. A., Saneyoshi T., Nygaard S., Kaech S., Banker G., Soderling T. R., Wayman G. A. (2009) J. Neurosci. 29, 9794–9808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ageta-Ishihara N., Takemoto-Kimura S., Nonaka M., Adachi-Morishima A., Suzuki K., Kamijo S., Fujii H., Mano T., Blaeser F., Chatila T. A., Mizuno H., Hirano T., Tagawa Y., Okuno H., Bito H. (2009) J. Neurosci. 29, 13720–13729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Saneyoshi T., Wayman G., Fortin D., Davare M., Hoshi N., Nozaki N., Natsume T., Soderling T. R. (2008) Neuron 57, 94–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hawley S. A., Pan D. A., Mustard K. J., Ross L., Bain J., Edelman A. M., Frenguelli B. G., Hardie D. G. (2005) Cell Metab. 2, 9–19 [DOI] [PubMed] [Google Scholar]
- 16. Woods A., Dickerson K., Heath R., Hong S. P., Momcilovic M., Johnstone S. R., Carlson M., Carling D. (2005) Cell Metab. 2, 21–33 [DOI] [PubMed] [Google Scholar]
- 17. Hurley R. L., Anderson K. A., Franzone J. M., Kemp B. E., Means A. R., Witters L. A. (2005) J. Biol. Chem. 280, 29060–29066 [DOI] [PubMed] [Google Scholar]
- 18. Anderson K. A., Ribar T. J., Lin F., Noeldner P. K., Green M. F., Muehlbauer M. J., Witters L. A., Kemp B. E., Means A. R. (2008) Cell Metab. 7, 377–388 [DOI] [PubMed] [Google Scholar]
- 19. Davare M. A., Saneyoshi T., Guire E. S., Nygaard S. C., Soderling T. R. (2004) J. Biol. Chem. 279, 52191–52199 [DOI] [PubMed] [Google Scholar]
- 20. Ichimura T., Taoka M., Hozumi Y., Goto K., Tokumitsu H. (2008) FEBS Lett. 582, 661–665 [DOI] [PubMed] [Google Scholar]
- 21. Tokumitsu H., Iwabu M., Ishikawa Y., Kobayashi R. (2001) Biochemistry 40, 13925–13932 [DOI] [PubMed] [Google Scholar]
- 22. Neumann D., Woods A., Carling D., Wallimann T., Schlattner U. (2003) Protein Expr. Purif. 30, 230–237 [DOI] [PubMed] [Google Scholar]
- 23. Samuels B. A., Hsueh Y. P., Shu T., Liang H., Tseng H. C., Hong C. J., Su S. C., Volker J., Neve R. L., Yue D. T., Tsai L. H. (2007) Neuron 56, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. de la Torre-Ubieta L., Gaudillière B., Yang Y., Ikeuchi Y., Yamada T., DiBacco S., Stegmüller J., Schüller U., Salih D. A., Rowitch D., Brunet A., Bonni A. (2010) Genes Dev. 24, 799–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Matsushita M., Nairn A. C. (1998) J. Biol. Chem. 273, 21473–21481 [DOI] [PubMed] [Google Scholar]
- 26. Haribabu B., Hook S. S., Selbert M. A., Goldstein E. G., Tomhave E. D., Edelman A. M., Snyderman R., Means A. R. (1995) EMBO J. 14, 3679–3686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amato S., Liu X., Zheng B., Cantley L., Rakic P., Man H. Y. (2011) Science 332, 247–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hawley S. A., Selbert M. A., Goldstein E. G., Edelman A. M., Carling D., Hardie D. G. (1995) J. Biol. Chem. 270, 27186–27191 [DOI] [PubMed] [Google Scholar]
- 29. Okuno S., Kitani T., Fujisawa H. (2001) J. Biochem. 130, 503–513 [DOI] [PubMed] [Google Scholar]
- 30. Kitani T., Okuno S., Fujisawa H. (2001) J. Biochem. 130, 515–525 [DOI] [PubMed] [Google Scholar]
- 31. Doble B. W., Woodgett J. R. (2003) J. Cell Sci. 116, 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Obenauer J. C., Cantley L. C., Yaffe M. B. (2003) Nucleic Acids Res. 31, 3635–3641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Atilla-Gokcumen G. E., Williams D. S., Bregman H., Pagano N., Meggers E. (2006) ChemBioChem. 7, 1443–1450 [DOI] [PubMed] [Google Scholar]
- 34. Nikolic M., Tsai L. H. (2000) in Methods in Enzymology (Balch W. E., Der C. J., Hall A. eds) pp. 200–213, Academic Press; [DOI] [PubMed] [Google Scholar]
- 35. Ohshima T., Ward J. M., Huh C. G., Longenecker G., Veeranna, Pant H. C., Brady R. O., Martin L. J., Kulkarni A. B. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 11173–11178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsai L. H., Takahashi T., Caviness V. S., Jr., Harlow E. (1993) Development 119, 1029–1040 [DOI] [PubMed] [Google Scholar]
- 37. Tsai L. H., Delalle I., Caviness V. S., Jr., Chae T., Harlow E. (1994) Nature 371, 419–423 [DOI] [PubMed] [Google Scholar]
- 38. Gomez T. M., Zheng J. Q. (2006) Nat. Rev. Neurosci. 7, 115–125 [DOI] [PubMed] [Google Scholar]
- 39. Moy L. Y., Tsai L. H. (2004) J. Biol. Chem. 279, 54487–54493 [DOI] [PubMed] [Google Scholar]
- 40. Tang D., Yeung J., Lee K. Y., Matsushita M., Matsui H., Tomizawa K., Hatase O., Wang J. H. (1995) J. Biol. Chem. 270, 26897–26903 [DOI] [PubMed] [Google Scholar]
- 41. Ko J., Humbert S., Bronson R. T., Takahashi S., Kulkarni A. B., Li E., Tsai L. H. (2001) J. Neurosci. 21, 6758–6771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chae T., Kwon Y. T., Bronson R., Dikkes P., Li E., Tsai L. H. (1997) Neuron 18, 29–42 [DOI] [PubMed] [Google Scholar]
- 43. Ohshima T., Gilmore E. C., Longenecker G., Jacobowitz D. M., Brady R. O., Herrup K., Kulkarni A. B. (1999) J. Neurosci. 19, 6017–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nikolic M., Dudek H., Kwon Y. T., Ramos Y. F., Tsai L. H. (1996) Genes Dev. 10, 816–825 [DOI] [PubMed] [Google Scholar]
- 45. Polleux F., Snider W. (2010) Cold Spring Harbor Perspect. Biol. 2, a001925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hengst U., Deglincerti A., Kim H. J., Jeon N. L., Jaffrey S. R. (2009) Nat. Cell Biol. 11, 1024–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cox L. J., Hengst U., Gurskaya N. G., Lukyanov K. A., Jaffrey S. R. (2008) Nat. Cell Biol. 10, 149–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Takemoto-Kimura S., Ageta-Ishihara N., Nonaka M., Adachi-Morishima A., Mano T., Okamura M., Fujii H., Fuse T., Hoshino M., Suzuki S., Kojima M., Mishina M., Okuno H., Bito H. (2007) Neuron 54, 755–770 [DOI] [PubMed] [Google Scholar]
- 49. Vinet J., Carra S., Blom J. M., Harvey M., Brunello N., Barden N., Tascedda F. (2003) Mol. Brain Res. 111, 216–221 [DOI] [PubMed] [Google Scholar]
- 50. Jin X., Hu H., Mathers P. H., Agmon A. (2003) J. Neurosci. 23, 5662–5673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Segal R. A., Pomeroy S. L., Stiles C. D. (1995) J. Neurosci. 15, 4970–4981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Hong E. J., McCord A. E., Greenberg M. E. (2008) Neuron 60, 610–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou P., Porcionatto M., Pilapil M., Chen Y., Choi Y., Tolias K. F., Bikoff J. B., Hong E. J., Greenberg M. E., Segal R. A. (2007) Neuron 55, 53–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.