Abstract
In the postnatal vasculature, fully differentiated and quiescent vascular smooth muscle cells (VSMCs) in a “contractile” phenotype are required for the normal regulation of vascular tone. The transforming growth factor-β (TGF-β) superfamily of growth factors (TGF-βs and bone morphogenetic proteins (BMPs)) are potent inducers of contractile phenotype and mediate (i) induction of contractile genes, and (ii) inhibition of VSMC growth and migration. Transcription of contractile genes is positively regulated by a regulatory DNA element called a CArG box. The CArG box is activated by the binding of serum response factor and its coactivators, myocardin (Myocd) or Myocd-related transcription factors (MRTFs). Krüppel-like factor-4 (KLF4) is known to inhibit activation of the CArG box. However, the potential role of KLF4 in the contractile activities of TGF-β or BMP has not been explored. Here, we demonstrate that TGF-β and BMP4 rapidly down-regulate KLF4 through induction of microRNA-143 (miR-143) and miR-145, which leads to a reduction of KLF4 transcripts and decreased KLF4 protein expression. Inhibition of miR-145 prevents down-regulation of KLF4 and activation of contractile genes by TGF-β or BMP4, suggesting that modulation of KLF4 is a prerequisite for induction of contractile genes by TGF-β and BMP4. Interestingly, both TGF-β and BMP4 activate transcription of the miR-143/145 gene cluster through the CArG box, however, TGF-β mediates this effect through induction of Myocd expression, whereas BMP4 utilizes nuclear translocation of MRTF-A. Thus, this study sheds light on both the similarities and the differences of TGF-β and BMP4 signaling in the regulation of KLF4 and contractile genes.
Keywords: Bone Morphogenetic Protein (BMP), Cell Differentiation, Gene Transcription, MicroRNA, RNA Silencing, Smad Transcription Factor, Smooth Muscle, Transcription Factors, Transcription Regulation, Transforming Growth Factor β (TGFbeta)
Introduction
Vascular smooth muscle cells (VSMCs)3 are highly plastic cells that undergo phenotype modulation in response to physiological and pathological cues (1). In response to vascular injury or growth factors, such as platelet-derived growth factor (PDGF) (2), VSMCs dedifferentiate and adopt a highly migratory, proliferative phenotype known as a “synthetic” phenotype that is required for vascular injury repair or during angiogenesis (1). However, prolonged or deregulated dedifferentiation can cause occlusion of the vasculature and contributes to development of vascular proliferative disorders, such as atherosclerosis, restenosis following angioplasty, as well as both systemic and pulmonary hypertension (1). Unlike PDGF, the TGF-β family of growth factors, including TGF-β and BMP4, promote a less migratory and proliferative phenotype known as the “contractile” phenotype (3). Contractile VSMC phenotype is characterized by alterations in the gene expression profile of VSMC. In particular, high expression of VSMC-specific genes, such as smooth muscle α-actin (SMA), calponin1 (CNN), and SM22α (SM22) are associated with the contractile VSMC phenotype. Transcription of contractile genes is regulated by SRF through a DNA sequence motif known as the CArG box (CC(A/T)6GG), which is present in the promoters of VSMC-specific genes (1). A coactivator of SRF, Myocd, interacts with SRF and activates VSMC expression of contractile genes (4–6). Similarly, the Myocd-related transcription factor (MRTF) family of proteins, MRTF-A and MRTF-B, are also involved in the transcriptional regulation of contractile gene markers as coactivators of SRF (7, 8). Myocd is constitutively localized to the nucleus and its activity is regulated primarily at the level of expression. Conversely, MRTFs are sequestered in the cytoplasm through interaction with monomeric G-actin (9, 10). In response to BMP4 or other stimuli, Rho signaling promotes actin polymerization and MRTF translocation into the nucleus where MRTFs associate with SRF (3), resulting in the activation of contractile gene transcription. Unlike BMP4, TGF-β does not activate Rho signaling, and does not induce MRTF nuclear localization. However, it can activate contractile genes in a mechanism distinct from BMP4. For example, in response to TGF-β stimulation, Smad3 can associate with a DNA sequence motif known as the TGF-β control element located in the SM22 gene promoter adjacent to a CArG box (11). In addition to positive regulators, transcription of contractile genes is also controlled by negative regulators, such as KLF4. KLF4 antagonizes contractile gene expression through diverse mechanisms including (i) inhibiting the binding of SRF-Myocd or SRF-MRTFs to the CArG box by direct association with SRF, (ii) altering the chromatin structure surrounding the CArG box, and (iii) suppressing Myocd expression (12). Additionally, a second negative regulator of contractile gene expression, Elk1, is also known to displace Myocd from SRF and suppress contractile gene expression (5).
Recent studies have revealed a critical role for miRNAs in control of the VSMC phenotype switch (13–16). miRNAs are small non-coding RNAs that act as negative regulators of gene expression by associating with partially complementary sequences in the 3′ untranslated regions (UTRs) of mRNAs, which results in mRNA degradation and/or translational inhibition (17). miRNAs are initially transcribed as a long, capped and polyadenylated primary miRNA (pri-miRNA) transcript, which in some cases can encompass multiple mature miRNAs. The RNase III enzyme Drosha processes the pri-miRNA into a ∼72 bp precursor miRNA (pre-miRNA), which is further processed by the RNase Dicer to give rise to the mature, functionally active miRNA. Mature miRNA expression can be regulated transcriptionally, similarly to mRNAs, or post-transcriptionally at either of the processing steps. Both BMP4 and TGF-β induce miR-21 in VSMC, which targets programmed cell death 4 (PDCD4) mRNA, resulting in the induction of contractile genes (14). Conversely, expression of miR-221 is elevated by PDGF-BB signaling and reduces contractile gene expression by down-regulating c-Kit (15). More recently, miR-143 and miR-145, which are encoded as a gene cluster, were found to target KLF4 and play a critical role in regulation of the VSMC phenotype (18–20). miR-143 or miR-145 VSMC knock-out mice exhibit abnormal vascular tone and reduced contractile gene expression (18). The expression of miR-143 and miR-145 has been reported to be repressed during VSMC de-differentiation induced by platelet-derived growth factor (PDGF) and during neointimal formation (19).
Here, we show rapid induction of miR-143/145 primary transcripts (pri-miR143/145) by TGF-β and BMP4 in VSMCs, which in turn down-regulates KLF4. Inhibition of miR-143/145 significantly decreases the induction of the contractile phenotype by TGF-β or BMP4, suggesting that suppression of KLF4 by miR-143/145 is critical for the induction of the contractile phenotype by TGF-β or BMP4. Interestingly, induction of pri-miR-143/145 by TGF-β and BMP4 is mediated by distinct SRF coactivators, Myocd and MRTF-A, respectively. Our results demonstrate a distinct mechanism of regulation of VSMC phenotype by TGF-β and BMP4 via miR-143 and miR-145.
EXPERIMENTAL PROCEDURES
Cell Culture
Human primary pulmonary artery smooth muscle cells (PASMCs) were purchased from Lonza (CC-2581) and maintained in Sm-GM2 media (Lonza) containing 5% FBS. Early passage (passage 4–7) PASMCs were used for this study. Human aortic smooth muscle cell line (AO184), rat PASMC, and Pac1 cells were obtained from ATCC and maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% FBS. Rat aortic smooth muscle cells were a gift from John Castellot (Tufts University) and were maintained in DMEM supplemented with 10% FBS. Phosphoinositide 3-kinase (PI3K) inhibitors wortmanin and LY294002 were purchased from Sigma and used at a final concentration of 10 μm. Recombinant human BMP4, TGF-β, and PDGF-BB were purchased from R&D Systems. Unless otherwise indicated, growth factor concentrations used were: 3 nm BMP4 or 400 pm TGF-β. Cells were cultured under starvation conditions (0.2% FBS) overnight (∼16 h) prior to growth factor stimulation as described (3). Pre-starvation alone does not dramatically alter expression of contractile gene expression or VSMC phenotype.
Reverse Transcriptase (RT)-Quantitative PCR
Total RNA was extracted by TRIzol (Invitrogen) and 1 μg of RNA was subjected to reverse transcription using the first-strand cDNA synthesis kit (Invitrogen) according to the manufacturer's instructions. SYBR Green-based real time PCR (Bio-Rad) was performed and the quantitative analysis of the change in expression levels was calculated using the comparative Ct method (iQ5, Bio-Rad). PCR cycling conditions were: 94 °C for 3 min and 40 cycles of 94 °C for 15 s, 60 °C for 20 s, and 72 °C for 40 s. For detection of mature miRNAs, the TaqMan MicroRNA assay kit (Applied Biosystems) was used according to the manufacturer's instructions. Data analysis was performed using the comparative Ct method. Average of three experiments each were performed in triplicate with mean ± S.E. presented.
qRT-PCR Primers
The following primers were used: human GAPDH, 5′-AAGGTGAAGGTCGGAGTC-3′ and 5′-GATTTTGGAGGGATCTCG-3′; human SMA, 5′-GCGTGGCTATTCCTTCGTTA-3′ and 5′-ATGAAGGATGGCTGGAACAG-3′; human CNN, 5′-AGCTAAGAGAAGGGCGGAAC-3′ and 5′-CATCTGCAGGCTGACATTGA-3′; human SM22, 5′-AACAGCCTGTACCCTGATGG-3′ and 5′-CGGTAGTGCCCATCATTCTT-3′; human myocardin, 5′-TGCATGCTGCTGTAAAGTCC-3′ and 5′-TAGCTGAATCGGTGTTGCTG-3′; human MRTF-A, 5′-TGTGTCTCAACTTCCGATGG-3′ and 5′-TTCACCTTTGGCTTCAGCTC-3′; human MRTF-B, 5′-GCAACTGCTGCACAAATACC-3′ and 5′-TTGATAAAGGGCTGCTGGAC-3′; human Smad4, 5′-AAGGTGAAGGTGATGTTTG-3′ and 5′-GAGCTATTCCACCTACTGAT-3′; human Id1, 5′-GGCTGTTACTCACGCCTCAAG-3′ and 5′-CCAACTGAAGGTCCCTGATGTAG-3′; human Id3, 5′-GGAGCTTTTGCCACTGACTC-3′ and 5′-TTCAGGCCACAAGTTCACAG-3′; human PAI-1, 5′-GATCGAGGTGAACGAGAGTG-3′ and 5′-AACTTCTCTCCCAGGGTCTC-3′; human KLF4, 5′-CCCAATTACCCATCCTTCCT-3′ and 5′-CGTCCCAGTCACAGTGGTAA-3′; human Pri-miR-143/145, 5′-AACTCCAGCTGGTCCTTAG-3′ and 5′-TCTTGAACCCTCATCCTGT-3′; human Nkx2.5, 5′-TTCTATCCACGTGCCTACAGC-3′ and 5′-CTGTCTTCTCCAGCTCCACC-3′; rat GAPDH, 5′-GGTGTGAACCACGAGAAATATGAC-3′ and 5′-CTCCAGGCGGCATGTCAGATCCAC-3′; rat SMA, 5′-ACTGGGACGACATGGAAAAG-3′ and 5′-CATACATGGCAGGGACATTG-3′; rat KLF4, 5′-TTATCAAGAGCTCATGCCACCGGGA-3′ and 5′-GCGGGCGAATTTCCACCCAC-3′; rat Myocd, 5′-AAACCAGGCCCCCTTCC-3′ and 5′-CGGATTCGAAGCTGTTGTCTT-3′; and rat pri-miR-145, 5′-ACTGCTGAAGGCATCTCTC-3′ and 5′-AGCAACACAAAGGTCAGAAG-3′.
miRNA Mimic and Antisense Oligonucleotides against miRNA
Chemically modified double-stranded RNAs designed to mimic the endogenous mature miR-143, miR-145, and negative control miRNA were purchased from Ambion. miRNA mimics were transfected at a 5 nm concentration using RNAi Max (Invitrogen) according to the manufacturer's instructions. 2′-O-Methyl-modified RNA oligonucleotides complementary to miRNA or GFP (control) sequence were purchased from IDT and transfected at 40 nm using RNAi Max (Invitrogen) according to the manufacturer's instructions using anti-GFP, 5′-AAGGCAAGCUGACCCUGAAGU-3′; anti-miR-143, 5′-GAGCUACAGUGCUUCAUCUCA-3′; and anti-miR-145, 5′-AGGGAUUCCUGGGAAAACUGGAC-3′.
RNA Interference
As a negative control, non-targeting scrambled siRNA (Qiagen) was used. Sequences of other siRNAs used were: KLF4 siRNA, 5′-AUCGUUGAACUCCUCGGUCUCUCUC-3′ (Stealth siRNA, Invitrogen); Myocd siRNA, 5′-AAUGCAACUGCAGAAGCAGAA-3′; and Smad4 siRNA, 5′-CCUGAGUAUUGGUGUUCCAUUGCUU-3′ (Stealth siRNA, Invitrogen). siRNA against MRTF-A and MRTF-B were from Dharmacon using previously described sequences (9). The siRNAs were transfected at 40 nm using RNAi Max (Invitrogen) according to the manufacturer's instructions. FITC-conjugated fluorescent oligonucleotides (Block-it, Invitrogen) were used to evaluate transfection efficiency and confirmed that >90% of cells are transfected by siRNA.
Adenovirus Gene Transduction
To generate a doxycycline-controlled MRTF-A adenoviral construct, a FLAG epitope-tagged mouse MRTF-A cDNA or the MRTF-A cDNA lacking the C-terminal transactivation domain (dominant-negative, Δ630) were cloned into the pTRE-Shuttle2 vector (Clontech) (21, 22). The linearized vectors were ligated into Adeno-X viral DNA and transformed. Correct clones were digested with the PacI restriction enzyme and transfected into AD-293 cells (Stratagene). Recombinant adenovirus were harvested, expanded, and purified with a Vivapure AdenoPack 100 kit (Vivascience). Adenoviral constructs were titered by plaque assay. PASMC were coinfected with adenovirus encoding TRE-MRTF-A(WT or Δ630) and Adeno-XTet-Off encoding rtTA (a tetracycline-controlled transactivator), which binds to the TRE element and activates transcription of MRTF-A. The Adeno-XTet-Off virus alone was used as a control.
Immunoblot Assay
Cells were lysed in TNE buffer (1% Nonidet P-40, 10 mm Tris-HCl (pH 7.5), 1 mm EDTA, 150 mm NaCl). Total cell lysates were separated on a SDS-PAGE, transferred to PVDF membranes (Millipore), immunoblotted with antibodies, and visualized using an enhanced chemiluminescence detection system (Amersham Biosciences). Antibodies used for immunoblotting are: anti-GAPDH antibody (2E3–2E10, Abnova), anti-β-Actin (clone AC-15, Sigma), anti-KLF4 (4038, Cell Signaling), anti-Myocd (Clone 355521, R&D Systems), anti-Smad4 (Clone H-552, Santa Cruz), anti-CNN (Clone C2687, Sigma), and anti-SMA (clone 1A4, Sigma).
Immunofluorescence Staining
PASMCs were fixed and permeabilized in a 50% acetone, 50% methanol solution and subjected to staining using anti-SMA antibody (clone 1A4, Sigma) conjugated with fluorescein isothiocyanate (FITC) and nuclear staining with 4′,6-diamidino-2-phenylindole (DAPI, Invitrogen). For quantiation, highly stained cells were counted from 4 randomly chosen fields relative to the total number of DAPI-marked nuclei. Approximately 600 total cells were counted in each condition.
Collagen Matrix Contraction Assay
Collagen matrix contraction assay was performed as described (23). Briefly, PASMCs transfected with anti-miR-145 or control oligonucleotides were embedded in attached collagen matrices, followed by 3 nm BMP4 for 24 h. Upon releasing the collagen lattice from the culture dish, the embedded cells become free to contract the deformable collagen lattice and the surface area of collagen lattice is reduced. Twenty-four h after detachment of the gel from the dish, the gel surface area was measured and demonstrated as the percentage of the gel surface area at the time of detachment.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed as described (28). Briefly, soluble chromatin was prepared from PASMC following cross-linking with 1% formaldehyde for 10 min, sonication, and centrifugation. Soluble chromatin was incubated with anti-RNA polymerase II antibody (Clone Ctd4h8, Millipore) for positive control, anti-Smad4 antibody (Clone H-552, Santa Cruz), anti-Smad2/3 antibody (Upstate), or an IgG control antibody. PCR primers for the human Myocd gene promoter were: primer A forward, 5′-AGAGGTCCCAGATAATCCAT-3′ and reverse, 5′-TTTTGGACCCTTCTTTTTAC-3′; primer B forward, 5′-AGAAACTGCTTCCCTGCAAA-3′ and reverse, 5′-TCAGTGCAAGTTGGGATGAA-3′; and primer C forward, 5′-CCCACCAATCTCTTCCCTGT-3′ and reverse, 5′-TCAACCCCCAGCCCCC-3′.
Luciferase Reporter Assay
Myocardin promoter luciferase reporter constructs were generated by PCR amplification of DNA fragments from human genomic DNAs and incorporation of HindIII and XbaI restriction endonuclease sites, followed by cloning into pGL2-vector (Invitrogen) containing a TATA box. The sequences used were: F1, 5′-AACAAGCTTAGAAACTGCTTCCCTGCAAA-3′; F2, AACAAGCTTGCCAGCTCATCTAGGTCAT-3′; F3, 5′-AACAAGCTTTCTCTGCTCAGATAATTTTGC-3′; R1, 5′AAGTCTAGATCAGTGCAAGTTGGGATGAA-3′; and R2, 5′-AAGTCTAGAGGTCTTCAATGAGAAGAAGG-3′. The SBE1–7-luc construct was generated with primers F1 and R2, SBE1–4-luc with primers F1 and R1, SBE3/4-luc with primers F2 and R1, and SBE6/7-luc with primers F3 and R2. For miR-145 binding site luciferase constructs, sequences corresponding to the human miR-145 binding site in KLF4, or a mutated version (see supplemental Fig. S4 for sequences), were synthesized with SacI and XbaI overhangs and inserted into pIS0 vector. All constructs were verified by sequencing. For the luciferase assay, the indicated reporter construct was transfected together with a β-galactosidase (β-gal) plasmid as an internal transfection control. Luciferase assays were carried out as previously described (24).
Statistical Analysis
The results presented are average of at least three experiments each performed in triplicate with mean ± S.E. Statistical analyses were performed by analysis of variance, followed by Tukey's multiple comparison test or by Student's t test as appropriate, using Prism 4 (GraphPad Software Inc.). p values of <0.05 were considered significant and are indicated with asterisks.
RESULTS
Down-regulation of KLF4 by TGF-β and BMP4
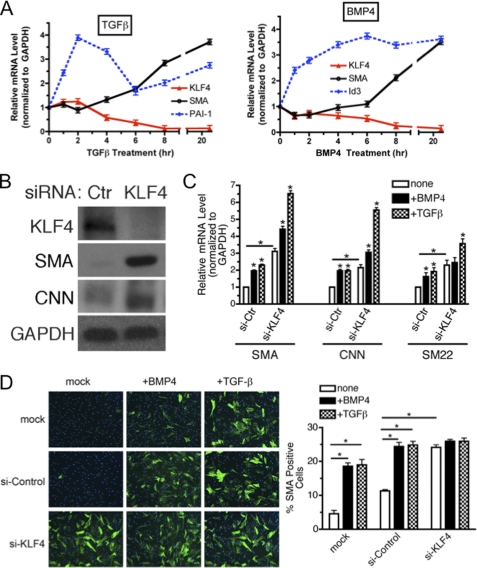
TGF-β and BMP4 induce contractile gene expression through various mechanisms, including transcriptional activation of contractile genes through a CArG box element located in the promoters of contractile genes, to which the critical transcription activator SRF binds as a complex with Myocd, or MRTFs (3). Association of the SRF-Myocd/MRTF complex with the CArG box can be inhibited by KLF4. Thus, we hypothesized that in addition to positive regulation of MRTF and Myocd activity, BMP4 and TGF-β-mediated induction of contractile genes may also involve inhibition of the negative regulator, KLF4. To examine this possibility, PASMCs were treated with TGF-β or BMP4 for various lengths of time (2–24 h). Activation of the TGF-β or BMP4 signaling pathways was confirmed by induction of target genes; plasminogen activator inhibitor-1 (PAI-1) for TGF-β (25), and inhibitor of DNA binding 3 (Id3) for BMP4 (26) (Fig. 1A). As expected, SMA was gradually elevated during 24 h using TGF-β or BMP4 treatments (Fig. 1A). Other contractile markers, such as CNN and SM22 were also induced similarly to SMA (data not shown). Interestingly, KLF4 mRNA was gradually decreased during the first 8 h after treatment and continued to be repressed 24 h after treatment (Fig. 1A). Unlike contractile markers or KLF4, no change in GAPDH mRNA or protein level was observed following BMP4 or TGF-β treatments (supplemental Fig. S1), which justifies the use of GAPDH as a loading control. Similar results were obtained in human aortic SMC Ao184 cells (supplemental Fig. S2A). These results demonstrate that both TGF-β and BMP repress KLF4 prior to induction of contractile genes in various VSMCs.
FIGURE 1.
KLF4 is a critical regulator of the VSMC phenotype switch upon TGF-β/BMP treatment. A, PASMCs were starved overnight and then treated with 400 pm TGF-β (left) or 3 nm BMP4 (right) for various lengths of time as indicated. Relative expression of SMA or KLF4 mRNA was examined by qRT-PCR and presented after normalizing to GAPDH. As controls for TGF-β or BMP4 treatments, transcriptional targets of TGF-β, PAI-1, and BMP4, ID3, were examined. The relative mRNA levels were presented as mean ± S.E., with each experiment conducted in triplicate (n = 3). B, hPASMCs were transfected with a non-targeting siRNA (si-Ctr) or siRNA targeting KLF4 (si-KLF4). 24 h after transfection, cells were switched to starvation media for an additional 24 h. Cells were then harvested and KLF4, SMA, CNN, or GAPDH (loading control) proteins were examined by immunoblotting. C, hPASMC were treated as in B. Relative expression of SMA, CNN, or SM22, normalized to GAPDH, was determined by qRT-PCR. Relative mRNA levels were presented as mean ± S.E. (*, p < 0.05, n = 3). D, PASMCs were mock transfected or transfected with si-Control or si-KLF4. 24 h following transfection, cells were starved overnight prior to stimulation with 3 nm BMP4 or 400 pm TGF-β for an additional 48 h before fixation and immunostaining with SMA antibody. Left panel, SMA protein and nuclei were visualized by FITC-labeled antibodies (green) and DAPI (blue), respectively. Right panel, percent of SMA-positive cells was calculated by a number of SMA-positive cells divided by a total number of cells. Approximately 600 cells were counted from four randomly chosen fields under each condition (*, p < 0.05).
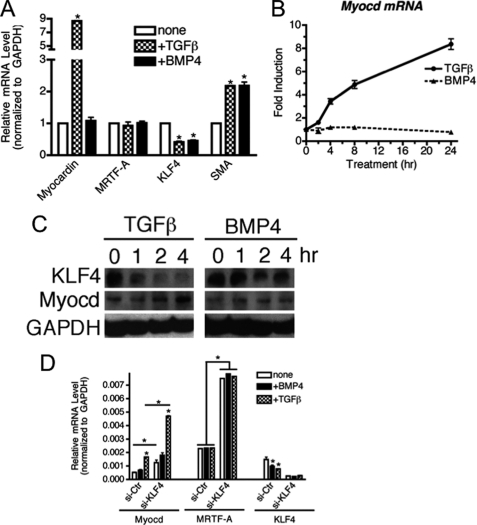
To examine a role of KLF4 in the control of contractile gene expression, we used siRNA to knockdown KLF4, thus mimicking the effect of BMP4 and TGF-β. Endogenous KLF4 protein was down-regulated more than 90% using siRNA for KLF4 (si-KLF4) in PASMC (Fig. 1B). Under this condition, levels of contractile proteins, SMA and CNN, were dramatically elevated (Fig. 1B), indicating that the endogenous KLF4 constitutively represses the expression of contractile genes. Similarly to proteins, transcripts of the contractile genes (SMA, CNN, and SM22) were ∼3-fold increased at the basal state (Fig. 1C). Upon treatment with TGF-β or BMP4, these mRNAs were further elevated (Fig. 1C). Immunofluorescence staining of SMA (Fig. 1D) and immunoblotting (supplemental Fig. S3) further confirmed an inhibitory role of KLF4 in the induction of contractile genes by TGF-β and BMP4.
Induction of Pri-miR-143/145 by TGF-β or BMP4
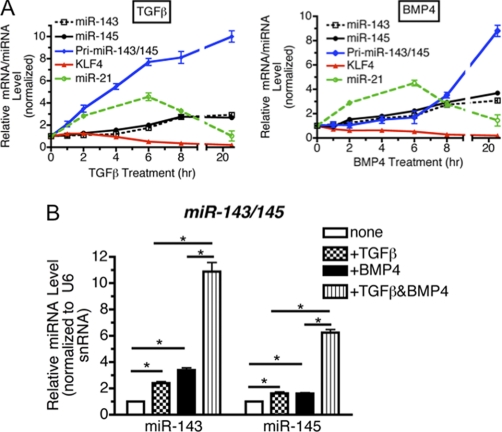
miRNAs are known to be involved in the repression of target mRNAs through imperfect binding to regions in the 3′ UTR, additionally, we have previously identified miRNAs as important players in the VSMC phenotype switch (13–16). The rapid reduction of KLF4 expression in response to TGF-β and BMP4 suggested that a miRNA could be involved. KLF4 is known to be targeted by several miRNAs, including miR-1 (27), miR-10b (28), miR-25 (29), miR-143 and miR-145 (18, 20). We previously performed a miRNA array to analyze the expression of miRNAs 24 h following BMP4 and TGF-β treatments in PASMC (16). Of the miRNAs identified to target KLF4, only miR-143 and miR-145 were induced at least 1.5-fold by both BMP4 and TGF-β, suggesting that they may be critical for BMP4- and TGF-β-mediated reduction of KLF4. To verify the array data, we utilized qRT-PCR to follow the expression of mature miR-143 and miR-145 1–24 h after BMP4 or TGF-β stimulation of PASMC (Fig. 2A). Induction of mature miR-21, which is known to be induced by BMP4 and TGF-β in PASMC (14, 16), served as a control for BMP4 or TGF-β treatments (Fig. 2A, green line for miR-21). Mature miR-143 and miR-145 were induced 6–8 h after TGF-β or BMP4 treatments (Fig. 2A, black dotted line for miR-143 and black solid line for miR-145). Importantly, a decrease in KLF4 mRNA was observed concomitant with miR-143 and miR-145 up-regulation with BMP4/TGF-β stimulation.
FIGURE 2.
TGF-β and BMP4 activate transcription miR-143/145, which down-regulates KLF4. A, PASMCs were starved overnight followed by stimulation with 400 pm TGF-β (left panel) or 3 nm BMP4 (right panel) for various lengths of time and subjected to qRT-PCR analysis. Relative expression of KLF4 mRNA, primary miR-143/145 transcripts (pri-miR-143/145), normalized to GAPDH was plotted. Mature miR-21, miR-143, or miR-145 expression normalized to U6 snRNA expression was also plotted. B, following overnight starvation, PASMCs were treated with 100 pm TGF-β, 1 nm BMP4, or a combination of both for 8 h, followed by qRT-PCR analysis of miR-143 or miR-145. Results were plotted as mean ± S.E. (*, p < 0.05).
We next sought to examine the mechanism of miR-143/145 induction by BMP4 and TGF-β. miRNAs can be regulated either at the level of transcription of the pri-miRNA, or at the Drosha or Dicer processing steps. miR-143 and miR-145 are encoded as a bicistronic miRNA gene cluster, which is transcribed as a common long primary transcript (pri-miR143/145) and processed into two distinct mature miRNAs: miR-143 and miR-145 (18). The promoter region of the miR-143/145 gene cluster contains a CArG box, which is found in many VSMC-specific gene promoters and serves as a binding site for SRF in complex with coactivators Myocd and MRTFs (18). As it was previously implicated that TGF-β and BMP4 induce contractile genes through activating the CArG box, we examined whether the level of pri-miR-143/145 is regulated by TGF-β or BMP4 in PASMC. One h after treatment with TGF-β, primary transcripts of miR-143 or miR-145 were elevated ∼3-fold relative to untreated cells (Fig. 2A, TGF-β, blue line). Unlike TGF-β treatment, BMP4 treatment induced the pri-miR143/145 level more gradually and only after a 8-h treatment was pri-miR-143/145 elevated over 3-fold (Fig. 2A, blue lines). As the induction of mature miR-143 and miR-145 occurs following induction of the pri-miR-143/145 transcript, these results suggest that the primary regulatory point of miR-143/145 expression is at the level of transcription. Interestingly, the time course of pri-miR-143/145 induction varied in response to BMP4 and TGF-β, thus, we hypothesized that different signaling pathways may be involved in inducing pri-miR-143/145 transcription. Treatment with a lower concentration of TGF-β (100 pm) or BMP4 (1 nm) alone for 8 h induced miR-143 and miR-145 only ∼2–3-fold, however, cotreatment with TGF-β and BMP4 synergistically elevated both miR-145 and miR-143 for ∼6–10-fold (Fig. 2B). Altogether, these results indicate that TGF-β and BMP4 are able to potentiate transcription of the miR-143/145 gene cluster.
Induction of miR-145 Is Critical for Down-regulation of KLF4 by BMP4 and TGF-β
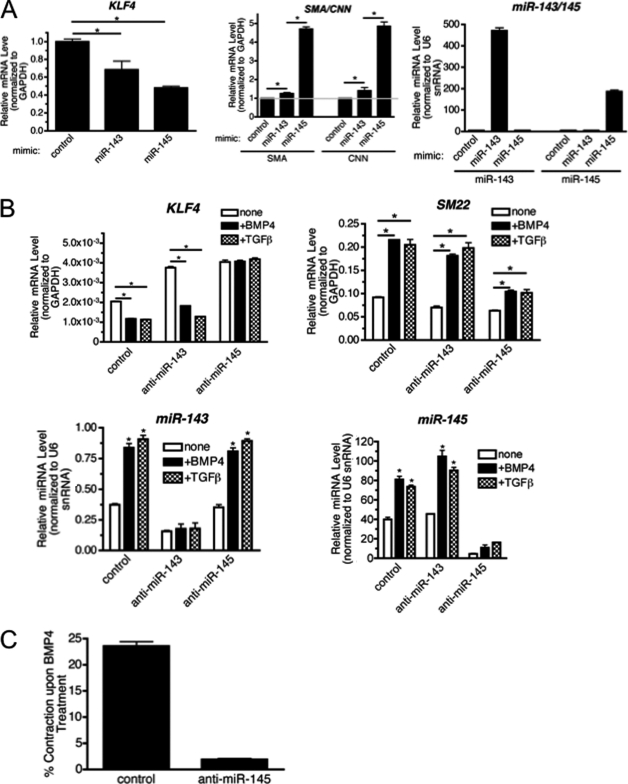
It is reported that both miR-143 and miR-145 mediate silencing of KLF4 gene expression (18). To examine whether miR-143 or miR-145 are able to down-regulate KLF4 mRNA in PASMC, modified short RNA designed to mimic mature miR-143 or miR-145 (miRNA mimic) were transfected into PASMCs. miR-143 mimic and miR-145 mimic expressed ∼94- and ∼37-fold over endogenous levels, respectively (Fig. 3A). Although the total levels of miRNA obtained following transfection with miRNA mimics is much higher that that obtained by BMP4 or TGF-β, only a small percentage of the exogenous miRNA is likely to be properly loaded into RISC. Thus, we speculate that the amount of functionally active miRNA is not dramatically different than that obtained with BMP4 or TGF-β treatments. Under this condition, miR-143 mimic and miR-145 mimic reduced KLF4 mRNA to ∼68 and ∼48% of the endogenous level, respectively (Fig. 3A), suggesting that miR-145 is a more potent regulator of KLF4 in comparison with miR-143. This observation is consistent with the result obtained using a KLF4-luciferase reporter assay in which the full-length 3′ UTR of KLF4 is cloned downstream of the luciferase gene (18). To further confirm the direct association of miR-145 with the 3′ UTR of KLF4, the predicted the miR-145 binding site in the human KLF4 3′ UTR was cloned into a luciferase reporter vector. Overexpression of miR-145, but not a control miRNA mimic, repressed expression of the WT KLF4 binding site reporter (supplemental Fig. S4). Importantly, miR-145 did not repress a vector containing a mutated miR-145 binding site (supplemental Fig. S4). Overexpression of miR-145 potently induced contractile gene expression (Fig. 3A). However, despite down-regulation of KLF4 with high levels of miR-143 mimic, SMC markers were only modestly elevated. We hypothesize that miR-143 and miR-145 may have additional divergent targets that may explain the differential effects on SMC contractile gene expression. These results demonstrate that the level of KLF4 expression is negatively regulated by miR-145/143 in PASMC and plays a critical role in contractile gene expression levels.
FIGURE 3.
Repression of KLF4 is required for TGF-β/BMP4-mediated induction of the contractile phenotype in VSMCs. A, miR-143 mimic or miR-145 mimic were transfected in hPASMCs, 24 h after transfection, cells were incubated for an additional 24 h in starvation media. Cells were then harvested and qRT-PCR analysis of KLF4 and contractile genes (SMA and CNN), normalized to GAPDH, was performed. The expression of miR-143 or miR-145 was measured by qRT-PCR analysis normalized to U6 snRNA (*, p < 0.05). B, anti-miR-143 or anti-miR-145 were transfected in PASMCs, the cells were then starved overnight prior to stimulation with TGF-β or BMP4 for an additional 24 h. qRT-PCR analysis was used to determine levels of KLF4, and the contractile gene SM22, normalized to GAPDH. The level of expression of miR-143 or miR-145 was measured by qRT-PCR analysis normalized to U6 snRNA (*, p < 0.05). C, PASMCs were transfected with control oligonucleotides, or anti-miR-145, followed by treatment with 3 nm BMP4 in starvation media for 24 h. Cells were then embedded into collagen gel lattices on the 12-well plate. Twenty-four h after the collagen lattices were dissociated from the well, gel lattices were photographed. Relative lattice area was quantitated by dividing the area of gel lattice by the initial area of the well. Experiments were performed three times. Data represent mean ± S.E. (*, p < 0.01).
To examine the role of miR-143 and miR-145 in BMP4- and TGF-β-induced contractile gene expression, endogenous miR-143 or miR-145 was down-regulated by transfecting 2′-O-methyl-modified RNA oligonucleotides, which are complementary to the miR-143 (anti-miR-143) or miR-145 sequences (anti-miR145). Both anti-miR-143 and anti-miR-145 reduced >80% of basal and TGF-β or BMP4 induced expression of miR-143 and miR-145, respectively (Fig. 3B). Anti-miR-143 or anti-miR-145 elevated the basal expression of KLF4 (Fig. 3B) presumably because KLF4 is released from constitutive down-regulation by endogenous miR-143 or miR-145. When miR-145 was down-regulated by anti-miR-145, the effect of TGF-β or BMP4 on KLF4 was abolished, whereas in anti-miR-143-transfected cells, KLF4 was reduced about 50% upon TGF-β or BMP4 treatments (Fig. 3B). As anti-miR-143 and anti-miR-145 reduce the endogenous miR-143 and miR-145 similarly (Fig. 3B), these results suggest a dominant role of miR-145 in the TGF-β or BMP4-mediated control of KLF4. Importantly, knockdown of miR-145 dramatically reduced TGF-β- and BMP4-mediated induction of SMC markers: SM22 (Fig. 3B), SMA, and CNN (data not shown). Transcriptional induction of non-contractile genes by TGF-β or BMP was intact in cells transfected with anti-miR-145 (supplemental Fig. S5), thus, the lack of induction of SM22 is not due to a general inhibition of the TGF-β signaling pathway but more specifically due to elevated expression of KLF4.
Increased expression of contractile genes is associated with an increase in VSMC contractility, which can be quantitated by a collagen gel matrix contraction assay. Consistent with the up-regulation of contractile genes, BMP4 treatment of VSMCs increases the ability of VSMC to deform a collagen gel matrix and reduced gel size ∼23% (Fig. 3C, control) (13, 23). However, knockdown of miR-145 abolished BMP4-mediated contraction (Fig. 3C). This result further supports a critical role of miR-145 in BMP4-mediated contractile gene expression (Fig. 3B).
Differential Mechanism of Activation of miR-143/145 by TGF-β and BMP4
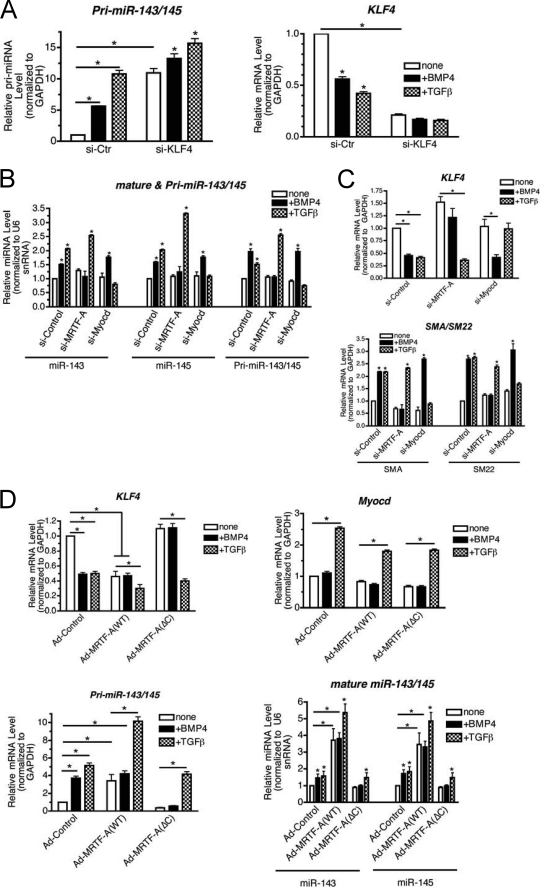
The promoter region of the miR-143/145 gene cluster contains a CArG box similarly to the contractile gene promoters. In vivo observations demonstrate a physiological significance of the CArG box in the spatial and temporal expression of miR-143/145 during cardiac development (18). Thus, we hypothesized that transcriptional regulation of the miR-143/145 gene may also be negatively regulated by KLF4 similarly to contractile genes. About 80% of endogenous KLF4 was down-regulated by siRNA in PASMCs (Fig. 4A, right panel). In comparison with si-Ctr cells, the basal pri-miR-143 level was ∼10-fold augmented in si-KLF4 cells, and TGF-β or BMP4 further elevated pri-miR-143/145 (Fig. 4A, pri-miR-143/145). This result indicates that, similarly to contractile genes, pri-miR-143/145 is negatively regulated by KLF4. Next, we examined whether down-regulation of CArG box binding factors, such as MRTFs and Myocd, abrogate the induction of miR-143/145 gene transcription. Primary transcripts of miR-143/145 (pri-miR-143/145), mature miR-143, or miR-145 were examined in PASMC transfected with siRNA against MRTF-A (si-MRTF-A), Myocd (si-Myocd), or a control, scrambled siRNA (si-Control), followed by BMP4 or TGF-β treatments. si-Myocd and si-MRTF-A down-regulated the endogenous Myocd and MRTF-A levels to ∼10 and ∼40% of si-Control-treated cells, respectively (supplemental Fig. S6). BMP4 was able to induce both pri-miR-143/145 and mature miR-143/145 in the cells down-regulated in Myocd (Fig. 4B). However, no induction of pri-miR-143/145 or mature miR-143/145 was observed in cells down-regulated in MRTF-A (Fig. 4B). This result suggests that MRTF-A, but not Myocd, is essential for BMP4-dependent induction of miR-143/145 gene transcription. Another member of the MRTF family, MRTF-B, does not play a significant role in the regulation of miR-143/145 nor KLF4 as knockdown of MRTF-B did not affect the regulation of these genes (supplemental Fig. S7). Unlike BMP4, TGF-β-mediated induction of pri-miR-143/145 or mature miR-143/145 was abolished upon Myocd knockdown, but was unaltered upon MRTF-A knockdown (Fig. 4B). These results demonstrate that MRTF-A is essential for the BMP4-mediated induction of pri-miR-143/145 and mature miR-143/145, whereas TGF-β-mediated induction of miR-143/145 requires Myocd (Fig. 4B). Consistent with the result of miR-143/145, BMP4 was able to repress KLF4 and induce SMC marker expression in the si-Myocd-treated cells but not in si-MRTF-A-treated cells (Fig. 4C), indicating that Myocd is not essential for BMP-mediated regulation of KLF4 via miR-143/145. Conversely, TGF-β-mediated repression of KLF4 and SMC marker induction was abolished in si-Myocd cells, but not in si-MRTF-A cells (Fig. 4C). Altogether, these results indicate that BMP4 and TGF-β utilize divergent signaling mechanisms to induce the expression of miR-143/145, down-regulate KLF4, and subsequently elevate VSMC marker expression.
FIGURE 4.
Differential mechanism of activation of miR-143/145 transcription by TGF-β and BMP4. A, hPASMCs were transfected with si-Ctr or siRNA against KLF4. 24 h after transfection, cells were switched to starvation media overnight, followed by BMP4 or TGF-β treatments for an additional 24 h. qRT-PCR analysis of pri-miR-143/145 and KLF4, normalized to GAPDH, was performed (*, p < 0.01). B, hPASMCs were transfected with si-Ctr or siRNA against Myocd (si-Myocd) or MRTF-A (si-MRTF-A), 24 h after transfection cells were starved overnight followed by BMP4 or TGF-β treatments for 8 h. RNA was harvested and qRT-PCR analysis of pri-miR-143/145 was normalized to GAPDH, or mature miR-143 or miR-145 normalized to U6 snRNA was performed (*, p < 0.01). C, PASMCs transfected with si-Ctr, si-MRTF-A, or si-Myocd were starved overnight and then stimulated with BMP4 or TGF-β for an additional 24 h, followed by qRT-PCR analysis of KLF4 (top panel) or contractile genes (SMA and SM22) (bottom panel) normalized to GAPDH (*, p < 0.01). D, wild type MRTF-A(WT) or a mutant of MRTF-A deleted in C terminus transactivation domain (ΔC) were transduced to hPASMCs by adenoviral vector, followed by overnight starvation and treatment with BMP4 or TGF-β for 24 h. KLF4, Myocd mRNA (top panel), pri-miR-143/145 (bottom left) normalized to GAPDH, and mature miR-143/145 (bottom right panel) normalized to U6 snRNA were examined by qRT-PCR analysis. Data represent mean ± S.E. (*, p < 0.01).
MRTF-A is primarily localized in the cytoplasm in unstimulated cells; upon stimulation with BMP4, MRTF-A translocates into the nucleus to promote changes in gene expression. Exogenous expression of MRTF-A results in forced nuclear accumulation and subverts the requirement of BMP4 for MRTF-A activation (3). To examine a critical role of MRTF-A in miR-143/145 induction and KLF4 repression by BMP4, adenovirus encoding wild type MRTF-A (MRTF-A(WT)) or a dominant-negative mutant of MRTF-A(ΔC), which is deleted in the carboxyl terminus transactivation domain (amino acid 630 to end) (21) was transduced in PASMCs. As a negative control, adenovirus encoding rtTA was transduced (Ad-Control). MRTF-A(WT) and MRTF-A(ΔC) were expressed at a similar level and unchanged by BMP4 or TGF-β treatments (supplemental Fig. S8). MRTF-A(WT) augmented the basal levels of pri-miR-143/145, as well as miR-143 and miR-145 (Fig. 4D). Consistent with high levels of miR-143 and miR-145, the KLF4 level was reduced in MRTF-A(WT) transduced cells compared with control virus-transduced cells (Fig. 4D, KLF4). In the presence of MRTF-A(WT), BMP4 did not further decrease the KLF4 level, whereas TGF-β reduced KLF4. This is consistent with further induction of pri-miR-143/145 and mature miR-143/145 by TGF-β through a MRTF-A independent mechanism (Fig. 4D, Myocd). The dominant-negative MRTF-A(ΔC) reduced the basal levels of pri-miR/143/145 and miR-143/145, which resulted in an increase in KLF4 (Fig. 4D). BMP4 was unable to repress KLF4 in the presence of MRTF-A(ΔC) (Fig. 4D, KLF4). This is consistent with the result that pri-miR-143/145 and miR-143/145 were not induced by BMP4 in the presence of MRTF-A(ΔC) (Fig. 4D and supplemental Fig. S8). In the presence of MRTF-A(ΔC), TGF-β retained the ability to induce pri-miR-143/145 and miR-143/145 and reduce KLF4 (Fig. 4D). In summary, these results further confirm that BMP4 requires MRTF-A, whereas TGF-β requires Myocd for the induction of pri-miR-143/145 and down-regulation of KLF4.
Induction of Myocardin by TGF-β
We next turned our attention to the mechanism of miR-143/145 induction by TGF-β via Myocd. The observation that Myocd is required for TGF-β-mediated induction of miR-143/145 suggested that the level or activity of Myocd could be altered by TGF-β. Although Myocd is well recognized as a critical regulator of VSMC marker expression, the mechanisms that regulate Myocd expression in VSMC are poorly understood. We tested if TGF-β may regulate the expression of Myocd in VSMC. Following treatment of PASMC with TGF-β or BMP4 for varying times, the expression of Myocd and MRTF-A was analyzed by qRT-PCR. Both TGF-β and BMP4 mediated down-regulation of KLF4 and induction of SMA, indicating active signaling by BMP4 or TGF-β (Fig. 5A, SMA). Interestingly, only TGF-β was able to induce Myocd (Fig. 5A, myocardin). Time course analysis indicated that Myocd mRNA is induced as early as 1 h after TGF-β stimulation, but not by BMP4 stimulation throughout the times examined (Fig. 5B). Similarly to mRNA, Myocd protein was elevated upon TGF-β, but not BMP4 treatment, whereas KLF4 protein was decreased (Fig. 5C). Similar regulation of Myocd by TGF-β was also observed in aortic SMC Ao184, confirming that induction of Myocd by TGF-β is conserved among VSMCs (supplemental Fig. S2). Consistent with a previous report showing that overexpression of KLF4 represses Myocd mRNA (12), down-regulation of endogenous KLF4 by siRNA >70% strongly elevated the Myocd mRNA level, which was further elevated by TGF-β treatment (Fig. 5D). Interestingly, the MRTF-A mRNA level was also elevated upon si-KLF4 transfection similarly to Myocd (Fig. 5D). These results indicate that (i) TGF-β and KLF4 regulate Myocd transcription positively and negatively, respectively, and (ii) KLF4 represses both Myocd and MRTF-A.
FIGURE 5.
TGF-β induces myocardin. A, hPASMCs were starved overnight and then treated with 3 nm BMP4 or 400 pm TGF-β for 24 h. Relative expression of Myocd, MRTFA, KLF4, or SMA mRNAs normalized to GAPDH was examined by qRT-PCR and plotted as mean ± S.E., n = 3 (*, p < 0.001). B, hPASMC were starved overnight prior to treatment with TGF-β or BMP4 for the indicated times. The expression of Myocd mRNA, normalized to GAPDH, was determined by qRT-PCR. C, hPASMCs were treated as in A for 1, 2, or 4 h. KLF4 or Myocd protein expression levels were examined by immunoblot analysis. GAPDH serves as a loading control. D, hPASMCs were transfected with si-Ctr or siRNA against KLF4, 24 h after transfection cells were starved overnight, followed by BMP4 or TGF-β treatments for 24 h and qRT-PCR analysis of Myocd, MRTF-A, and KLF4 normalized to GAPDH (*, p < 0.01).
Smad-dependent Transcriptional Activation of Myocd by TGF-β
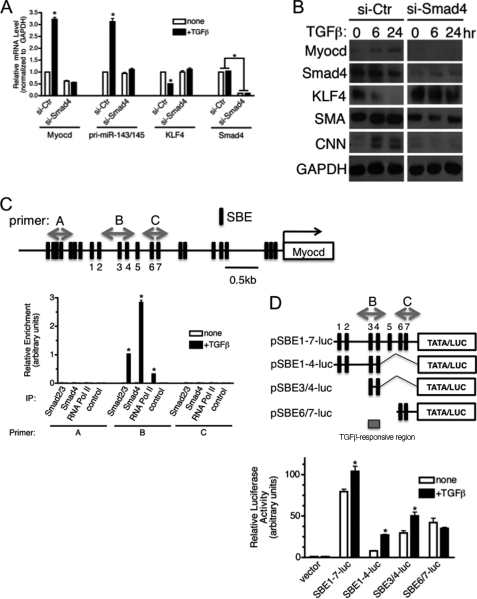
To examine a mechanism of induction of Myocd mRNA by TGF-β, Smad4, a signal transducer that is critical for transcriptional regulation by TGF-β, was down-regulated by siRNA (si-Smad4), followed by TGF-β treatment. When ∼90% of Smad4 was down-regulated (Fig. 6, A for mRNA, and B for protein), Myocd mRNA induction by TGF-β was abolished, suggesting that Smad4 plays a critical role in transcriptional activation of the MYOCD gene (Fig. 6A, Myocd). Similarly to Myocd mRNAs, induction of primary transcripts of miR-143/145 by TGF-β was abolished in si-Smad4 cells (Fig. 6A, pri-miR-143/145). Furthermore, KLF4 was no longer down-regulated upon TGF-β stimulation in si-Smad4 cells (Fig. 6A, KLF4). Consistent with the results of mRNAs in Fig. 6A, both the basal and TGF-β-induced Myocd protein levels were reduced in si-Smad4 cells in comparison with si-Control cells (Fig. 6B). Knockdown of the BMP4-regulated Smad 1 and 5 did not alter the basal or levels of Myocd (data not shown), further confirming a TGF-β-specific effect on Myocd expression.
FIGURE 6.
Induction of Myocd by TGF-β is Smad-dependent. A, hPASMCs were transfected with si-Ctr or siRNA against Smad4 (si-Smad4). After 24 h, cells were starved overnight followed by treatment with TGF-β for an additional 24 h. qRT-PCR analysis of Myocd, pri-miR-143/145, KLF4, or Smad4 mRNA, normalized to GAPDH, was performed (*, p < 0.05). B, hPASMCs were transfected with si-Ctr or si-Smad4. After 24 h, cells were starved overnight and then treated with TGF-β for 6 or 24 h, followed by immunoblot analysis of Myocd, Smad4, KLF4, or contractile markers (SMA and CNN). GAPDH serves as a loading control. C, upper, schematic of the Myocd promoter and location of SBE sites. Lower, PASMC were starved overnight and then stimulated with 400 pm TGF-β for 2 h. Cells were harvested, and ChIP assay was performed using anti-Smad2/3 or anti-Smad4 antibody. ChIP with nonspecific control IgG and anti-RNA polymerase II (RNA pol II) antibody were used as negative and positive controls, respectively. qRT-PCR was performed using different primer sets (A–C) to measure enrichment of the DNA fragment containing different regions of the Myocd promoter. Results were plotted as relative enrichment to input, mean ± S.E., n = 3. D, upper, schematic of Myocd promoter luciferase constructs. The B and C regions correspond to ChIP primers in C and SBE sites are numbered as in C. Lower, PAC1 cells were transfected with empty luciferase vector or the indicated Myocd promoter luciferase: pSBE1–7-luc, pSBE1–4-luc, pSBE3/4-luc, or pSBE6/7-luc. 24 h after transfection, cells were treated with TGF-β in starvation media for 24 h and subjected to luciferase assay. Results were plotted as relative luciferase activity normalized to β-galactosidase activity mean ± S.E., n = 3. *, p < 0.001. IP, immunoprecipitation.
In silico analysis of the MYOCD gene promoter identified >20 putative Smad-binding elements (SBEs) (CAGAC/G) within ∼2 kb of the transcription initiation site, including several that are highly evolutionarily conserved (Fig. 6C and supplemental Fig. S9). To map the region of the MYOCD promoter that is critical for the TGF-β-mediated transcriptional activation, chromatin immunoprecipitation (ChIP) analysis was performed by immunoprecipitating genomic fragments associated with the TGF-β signal transducers: Smad2/3 or Smad4, followed by PCR amplification using different primer sets (Fig. 6C, A–C). Robust increase in the recruitment of Smad2/3 and Smad4 to the region amplified with primer B was observed 2 h after stimulation with TGF-β (Fig. 6C, primer B). Neither the upstream (Fig. 6C, primer A) nor the downstream regions (Fig. 6C, primer C) relative to the primer B region showed recruitment of Smad2/3 or Smad4, indicating that TGF-β-dependent association of Smad proteins occurs proximal to the primer B region. To further examine the region of the MYOCD promoter required for TGF-β-mediated induction, primers spanning 7 SBE sites within ChIP primers B and C were used to amplify the MYOCD promoter from genomic DNA. The product was inserted into a minimal, TATA-box containing luciferase construct (Fig. 6D, pSBE1–7). Sequential deletions were then generated to delineate the regions required for TGF-β response (pSBE1–4, pSBE3/4, and pSBE6/7). The luciferase constructs were transfected into rat Pac1 cells and luciferase production was measured following a 24-h TGF-β treatment. Consistent with the ChIP assay result, all three reporter constructs spanning the primer B region (Fig. 6D, pSBE1–7-luc, pSBE1–4-luc, and pSBE3/4-luc) conferred TGF-β responsiveness, whereas a construct containing only the primer C region did not respond to TGF-β (Fig. 6D, pSBE6/7-luc). These results demonstrate that association of Smad proteins with SBE3/4 is essential for transcriptional activation of the MYOCD gene and subsequent induction miR-143/145 by TGF-β.
DISCUSSION
In this study we demonstrate a critical role and mechanism of regulation of KLF4 by the TGF-β family of growth factor signaling pathways in VSMCs phenotype control. The importance of the signaling pathway mediated by the TGF-β family of growth factors in promoting the VSMC contractile phenotype has been demonstrated in a number of pathological models, including vascular injury models and pulmonary arterial hypertension animal models (30). It is clear that one of the activities of the TGF-β family of growth factor signaling cascades is to induce VSMC-specific contractile gene expression both transcriptionally (3) and post-transcriptionally through the modulation of miRNA expression, such as miR-21 (14). Induction of miR-21 by TGF-β or BMP4 leads to down-regulation of the mRNA translation initiation inhibitor PDCD4 (14). Although the precise mechanism utilized by PDCD4 to repress expression of contractile genes is unknown, it is plausible to speculate that miR-21-mediated repression of PDCD4 could facilitate protein synthesis of different contractile gene products as well as Myocd. As transcription complexes SRF-Myocd or SRF-MRTFs potently activate contractile genes through the CArG box, it is believed that activation of the CArG box is sufficient for induction of contractile genes by TGF-β or BMP4. In this study, we demonstrate that induction of contractile genes by TGF-β and BMP4 is greatly reduced when down-regulation of KLF4 by miR-143/145 is abrogated. We speculate that KLF4 might be responsible for setting a threshold so that low levels of contractile signals are unable to activate contractile gene expression.
KLF4 is a critical factor in the regulation of cellular differentiation and development in a number of systems, including VSMCs, and its expression is tightly regulated. In some cellular contexts, such as skin epithelial cells, high expression of KLF4 is associated with enhanced differentiation (31). Conversely, in VSMC, both a previous study (32) and this study demonstrate that KLF4 expression is linked to dedifferentiation. Similarly, KLF4 is important to maintain an undifferentiated cellular state in stem cells as exogenous expression of KLF4 together with three additional factors (c-Myc, Oct3/4, and Sox2) is able to reprogram non-pluripotent adult somatic cells into pluripotent stem cells known as induced pluripotent stem (iPS) cells (33). The level of KLF4 in embryonic stem cells (ESCs) is controlled, at least in part, through regulation of miR-143/145 expression. In human ESCs, miR-145 is expressed at a low level in self-renewing cells but is highly up-regulated during differentiation (34). Interestingly, miR-145 targets not only KLF4 but other pluripotency factors, OCT4 and SOX2 (34, 35), suggesting that it is a key regulator of ESC phenotype. We show here that members of the TGF-β family rapidly induce miR-143/145. As TGF-β and BMP play essential roles in the differentiation of ESC (36), we speculate that induction of miR-143/145 in response to TGF-β or BMP may be important to trigger down-regulation of multiple pluripotency factors simultaneously and promote differentiation in ESCs.
In addition to the regulation of differentiation status, KLF4 plays an important, context dependent, role in the regulation of cellular proliferation. This function of KLF4 is particularly relevant to tumorigenesis as aberrant expression of KLF4 is linked to development of various human cancers, including colorectal, gastrointestinal, breast, prostate, and lung cancer, as well as adenomatous polyposis coli (37–44). KLF4 was initially identified as a tumor suppressor as loss of KLF4 is often observed in various tumors and transgenic mice lacking KLF4 in the gastric compartment develop stomach polyps and hyperplasia (45). More recently it was demonstrated that KLF4 is highly expressed in more than 70% of breast cancers and plays a role in maintenance of cancer stem cells, as well as tumor cell migration and invasion (46). Thus, KLF4 has both pro- and anti-oncogenic properties depending on the specific cell type. An additional role for KLF4 in the repression of cardiac hypertrophy has been uncovered (47). KLF4 is induced in cultured cardiomyocytes upon hypertrophic stimuli, and in mice following transverse aortic constriction. In this system, induction of KLF4 reduces the development of pathologic cardiac hypertrophy (47).
In SMCs and ESCs, repression of KLF4 by miR-143/145 promotes differentiation. Consistently, down-regulation of miR-143 or miR-145 has been reported in a number of cancers (48–50). Thus, it is plausible to speculate that some tumors may develop due to reduced expression of miR-143/145, and elevated KLF4, as a result of reduced TGF-β activities or Myocd expression. A recent study shows that transcription of the miR-143/145 gene cluster is repressed by KRAS and the Ras-responsive element-binding protein (RREB1) in pancreatic carcinoma with KRAS mutation. This is consistent with the observation that loss of miR-143/145 expression is frequently observed in KRAS mutant pancreatic cancers. Thus, a subset of pancreatic cancers with the KRAS mutation might also exhibit elevated KLF4, which in turn acts as an oncogene similar to the case of breast cancers with high levels of KLF4. As both KRAS and RREB1 are also targets of miR-143/145, the negative regulatory relationship between KRAS/RREB1 and miR-145/143 is similar to the relationship between KLF4 and miR-143/145 (51). Altogether, these observations place KLF4 and miR-143/145 as critical regulators of diverse processes and emphasize the importance of understanding the mechanisms that regulate their expression. Modulation of miR-143/145 expression may represent a therapeutic option to suppress tumorigenecity in various tumors. Alternatively, inhibition of miR-143/145 and up-regulation of KLF4 may be beneficial for the protection of cardiomyocytes during cardiac hypertrophy.
In VSMCs, KLF4 is known as a potent negative regulator of contractile genes through multiple actions, including antagonizing transcriptional activation of the CArG box by SRF-Myocd or MRTFs as well as down-regulating Myocd expression (12). In this study, we demonstrate a functional significance of down-regulation of KLF4 by TGF-β and BMP through induction of miR-143/145 during the VSMC phenotype switch (see Fig. 7 for summary). We found that siRNA-mediated knockdown of KLF4 suppresses the basal expression of not only contractile genes but also SRF cofactors: Myocd and MRTF-A. We speculate that constitutive induction of miR-143/145 and down-regulation of KLF4 may result in a feed-forward circuit that enhances expression of contractile genes through induction of Myocd and MRTF-A. Additionally, as knockdown of miR-143/145 prevents BMP4 or TGF-β-mediated induction of contractile genes, reduction of KLF4 expression by miR-143/145 induction may be especially critical to allow the SRF-Myocd or SRF-MRTF-A complexes to associate with the CArG box in response to cellular stimuli. Interestingly, it was previously reported that unlike in human PASMC, in rat aortic SMC (rAoSMC) (52, 53) and mouse mesenchymal C3H10T1/2 cells (3) TGF-β and BMP4 induce KLF4 expression at some time points. Treatment of primary rAoSMC with TGF-β or BMP4 did not induce pri-miR-143/145 and no repression of KLF4 was observed (supplemental Fig. S10). Together these results suggest that induction of miR-145 by BMP and TGF-β, and subsequent repression of KLF4, may be specific for human VSMC.
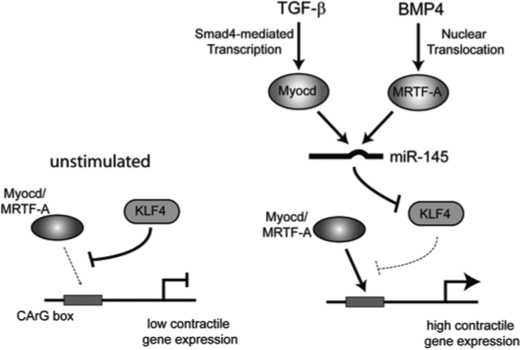
FIGURE 7.
Schematic of BMP and TGF-β induction of pri-miR-143/145 and inhibition of KLF4. Left, in unstimulated cells KLF4 represses contractile gene expression by preventing binding of the Myocd or MRTF-A-SRF complex to CArG boxes present in the promoter of contractile genes. Right, upon TGF-β stimulation, Myocd expression is induced through Smad4, whereas BMP4 promotes nuclear translocation of MRTF-A. Elevated expression of Myocd or nuclear MRTF-A induces transcription of pri-miR-143/145, which is processed into the mature miR-143 or miR-145. The mature miRNA then represses KLF4 expression allowing enhanced binding of Myocd or MRTF-A-SRF complexes to the CArG boxes of contractile genes and increased contractile gene expression. For simplicity, SRF is not depicted.
miR-143 and miR-145 are processed from a single transcript that is regulated by an evolutionarily conserved CArG box located upstream of the miR-143/145 gene cluster. Myocd or MRTFs in complex with SRF associate with the CArG box to promote transcription of pri-miR-143/145 and subsequent elevation of the mature miR-143/145 (18, 20). Both miR-143 and miR-145 are highly enriched in VSMCs during development in mouse and deletion of miR-143 and miR-145 in mice results in a significant reduction in vascular tone, emphasizing the essential role of miR-143/145 in VSMC phenotype regulation and function (18, 20). Here, we show that both TGF-β and BMP4 induce expression of miR-143/145 through activation of Myocd and MRTF-A, respectively. Unlike Myocd, which is strictly expressed in cardiovascular tissues and is primarily localized in the nucleus, MRTF-A is ubiquitously expressed in different cell types and localized primarily in the cytoplasm. The activity of MRTF-A is regulated at the step of cytoplasmic to nuclear translocation. We previously demonstrated that BMP4 signaling induces nuclear translocation of MRTF-A in PASMCs (3). Our results indicate that induction of pri-miR-143/145 by TGF-β is rapid (within 2 h after treatment) in comparison with BMP4-mediated induction of pri-miR-143/145, which takes more than 6 h. As Myocd and MRTF-A are equally potent transcriptional activators of VSMC-specific genes, we speculate that nuclear accumulation of MRTF-A may be a slow process compared with induction of Myocd by TGF-β. It is interesting that TGF-β and BMP4 utilize distinct cofactors of SRF to induce miR-143/145 to promote contractile gene expression. Mutations in the TGF-β and BMP4 pathways both result in vascular disorders, however, the specific vascular compartments that are affected differ. For example, mutation in the BMP receptor leads to idiopathic pulmonary arterial hypertension, whereas mutations in TGF-β receptor leads to hereditary hemorrhagic telangiectasia (30). It is interesting to speculate that differences in the induction of miR-143/145 through TGF-β or BMP may contribute to the differential presentation of these vascular disorders.
In fibroblasts, it has been demonstrated that TGF-β mediates cell cycle control and differentiation to a myofibroblast-like phenotype by mediating the expression of Myocd, which exhibits tumor suppressor activity (54). In this study, we elucidate that in the TGF-β signal transducers, the Smads directly associate with the Myocd gene promoter and activate transcription, and indirectly promote differentiation into the VSMC phenotype similarly to the case of fibroblasts. As Myocd expression is restricted to cardiovascular tissues, unlike the TGF-β signaling molecules, which are ubiquitously expressed, we speculate that TGF-β activates Myocd gene transcription in concert with a tissue-specific cofactor. Recently, slow induction of Myocd mRNA by TGF-β (>18 h after TGF-β stimulation) is reported in mouse mesenchymal C3H10T1/2 cells (55). This induction is mediated by PI3K signaling, which activates the Myocd promoter through induction of Nkx2.5 (55). Interestingly, in this report Smad4 acts to inhibit Nkx2.5 activity. Several lines of evidence suggest that the mechanism of Myocd regulation differs between PASMC and mesenchymal cells: (i) unlike in 10T1/2 cells, in PASMC, induction of Myocd is rapid and occurs within 2 h following TGF-β stimulation (Fig. 5B); (ii) treatment with PI3K inhibitors, LY294002 or wortmanin, do not inhibit the induction of Myocd by TGF-β in PASMCs (supplemental Fig. S11), and (iii) Nkx2.5 expression is very low in PASMC and it is not altered by TGF-β (supplemental Fig. S12).
It is reported that activation of Notch signaling in ESC induces expression of Myocd and VSMC genes, and mediates differentiation into VSMCs (56). Furthermore, Notch-mediated induction of Myocd requires Smad3 (56). Thus, it is plausible that Smads and an intracellular domain of Notch might cooperatively activate the Myocd gene. It is important to note, however, that the TGF-β-Smad2 pathway could mediate the repression of Notch3 in fibroblasts, thus, cooperative action of Notch and TGF-β on Myocd expression might be cell type-specific (57). Forced expression of Myocd in ESC induces expression of multiple endogenous VSMC-specific genes and induces VSMC-specific cytoskeletal and contractile proteins. In VSMC, following resolution of vascular injury, extrinsic and intracellular cues lead to an activation of Myocd-SRF and modulate the VSMC phenotype to perform injury repair. More recently, transient expression of Myocd in skeletal muscle cells has also been observed during early development (58, 59). Overexpression of Myocd in skeletal muscle cells is found to promote a cell type switch toward VSMC by suppressing a subset of skeletal muscle genes and inducing VSMC-specific genes (58, 59). This observation raises the interesting possibility that Myocd is a critical determinant of VSMC lineage. It has been demonstrated that augmented expression of Myocd during heart failure plays a critical role as Myocd induces cardiomyocyte survival and maintenance of heart function (60).
Finally, we demonstrate that the basal expression of both Myocd and MRTF-A is negatively regulated by KLF4 (12). This is interesting as it indicates an autoregulatory loop among positive regulators (Myocd, MRTF-A, or miR-143/145) and a negative regulator (KLF4) of contractile genes. It is plausible that the autoregulation mechanism is necessary to maintain homeostasis. The mechanism of inhibition of Myocd or MRTF-A by KLF4 is currently unclear and warrants future study. The expression of Myocd has also been recently reported to be negatively regulated by miR-9 (61). Reduction of miR-9 in myocardium in response to hypertrophic stimulation results in elevated expression of Myocd (61). It is plausible that KLF4 and miR-9 additively or synergistically modulate Myocd expression in VSMCs. Further understanding of the intricate regulatory network of Myocd, KLF4, and miRNAs may allow the development of effective therapies for the prevention of various human disorders, including vascular injury, cancer, or heart failure.
Supplementary Material
Acknowledgments
We thank Dr. M. Mendelsohn for sharing Ao184 cells and Dr. J. J. Castellot for sharing rAoSMC. We thank all members of the Hata lab for helpful suggestions and critical discussion.
Note Added in Proof
After this paper was accepted we became aware of a related study showing a similar mechanism of regulation of the miR-143/145 gene by TGF-β1 (Long, X., and Miano, J. M. (June 28, 2011) J. Biol. Chem. 10.1074/jbc.M111.258814).
This work was supported, in whole or in part, by National Institutes of Health Grants HD042149 and HL082854 (to A. H.), HL078869 (to M. D. L.), HL086572 (to G. L.), Training Grant T32 HL069770 (to B. N. D.-D.), and American Heart Association Grant 0940095N (to A. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S12.
- VSMC
- vascular smooth muscle cell
- TGF-{β}
- transforming growth factor-{β}
- PDGF
- platelet-derived growth factor
- PAI-1
- plasminogen activator inhibitor-1
- PDCD4
- programed cell death 4
- BMP
- bone morphogenetic protein
- MRTF
- Myocd-related transcription factor
- PASMC
- pulmonary artery smooth muscle cell
- ESC
- embryonic stem cell
- SRF
- serum response factor
- Myocd
- myocardin
- miRNA
- microRNA
- pri-miRNA
- primary miRNA
- SBE
- Smad-binding element
- rAoSMC
- rat aortic SMC
- qRT
- quantitative reverse transcriptase.
REFERENCES
- 1. Owens G. K., Kumar M. S., Wamhoff B. R. (2004) Physiol. Rev. 84, 767–801 [DOI] [PubMed] [Google Scholar]
- 2. Barst R. J. (2005) J. Clin. Invest. 115, 2691–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lagna G., Ku M. M., Nguyen P. H., Neuman N. A., Davis B. N., Hata A. (2007) J. Biol. Chem. 282, 37244–37255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yoshida T., Sinha S., Dandré F., Wamhoff B. R., Hoofnagle M. H., Kremer B. E., Wang D. Z., Olson E. N., Owens G. K. (2003) Circ. Res. 92, 856–864 [DOI] [PubMed] [Google Scholar]
- 5. Wang Z., Wang D. Z., Hockemeyer D., McAnally J., Nordheim A., Olson E. N. (2004) Nature 428, 185–189 [DOI] [PubMed] [Google Scholar]
- 6. Miano J. M. (2004) Circ. Res. 95, 340–342 [DOI] [PubMed] [Google Scholar]
- 7. Han Z., Li X., Wu J., Olson E. N. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 12567–12572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jeon E. S., Park W. S., Lee M. J., Kim Y. M., Han J., Kim J. H. (2008) Circ. Res. 103, 635–642 [DOI] [PubMed] [Google Scholar]
- 9. Medjkane S., Perez-Sanchez C., Gaggioli C., Sahai E., Treisman R. (2009) Nat. Cell Biol. 11, 257–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Miralles F., Posern G., Zaromytidou A. I., Treisman R. (2003) Cell 113, 329–342 [DOI] [PubMed] [Google Scholar]
- 11. Qiu P., Ritchie R. P., Fu Z., Cao D., Cumming J., Miano J. M., Wang D. Z., Li H. J., Li L. (2005) Circ. Res. 97, 983–991 [DOI] [PubMed] [Google Scholar]
- 12. Liu Y., Sinha S., McDonald O. G., Shang Y., Hoofnagle M. H., Owens G. K. (2005) J. Biol. Chem. 280, 9719–9727 [DOI] [PubMed] [Google Scholar]
- 13. Chan M. C., Hilyard A. C., Wu C., Davis B. N., Hill N. S., Lal A., Lieberman J., Lagna G., Hata A. (2010) EMBO J. 29, 559–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis B. N., Hilyard A. C., Lagna G., Hata A. (2008) Nature 454, 56–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Davis B. N., Hilyard A. C., Nguyen P. H., Lagna G., Hata A. (2009) J. Biol. Chem. 284, 3728–3738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Davis B. N., Hilyard A. C., Nguyen P. H., Lagna G., Hata A. (2010) Mol. Cell 39, 373–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kim V. N., Han J., Siomi M. C. (2009) Nat. Rev. Mol. Cell. Biol. 10, 126–139 [DOI] [PubMed] [Google Scholar]
- 18. Xin M., Small E. M., Sutherland L. B., Qi X., McAnally J., Plato C. F., Richardson J. A., Bassel-Duby R., Olson E. N. (2009) Genes Dev. 23, 2166–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheng Y., Liu X., Yang J., Lin Y., Xu D. Z., Lu Q., Deitch E. A., Huo Y., Delphin E. S., Zhang C. (2009) Circ. Res. 105, 158–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cordes K. R., Sheehy N. T., White M. P., Berry E. C., Morton S. U., Muth A. N., Lee T. H., Miano J. M., Ivey K. N., Srivastava D. (2009) Nature 460, 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cen B., Selvaraj A., Burgess R. C., Hitzler J. K., Ma Z., Morris S. W., Prywes R. (2003) Mol. Cell. Biol. 23, 6597–6608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen C. H., Wu M. L., Lee Y. C., Layne M. D., Yet S. F. (2010) Arterioscler. Thromb. Vasc. Biol. 30, 835–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Neuman N. A., Ma S., Schnitzler G. R., Zhu Y., Lagna G., Hata A. (2009) J. Biol. Chem. 284, 13202–13212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hata A., Seoane J., Lagna G., Montalvo E., Hemmati-Brivanlou A., Massagué J. (2000) Cell 100, 229–240 [DOI] [PubMed] [Google Scholar]
- 25. Laiho M., Saksela O., Keski-Oja J. (1987) J. Biol. Chem. 262, 17467–17474 [PubMed] [Google Scholar]
- 26. Hollnagel A., Oehlmann V., Heymer J., Rüther U., Nordheim A. (1999) J. Biol. Chem. 274, 19838–19845 [DOI] [PubMed] [Google Scholar]
- 27. Xie C., Huang H., Sun X., Guo Y., Hamblin M., Ritchie R. P., Garcia-Barrio M. T., Zhang J., Chen Y. E. (2011) Stem Cells Dev. 20, 205–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tian Y., Luo A., Cai Y., Su Q., Ding F., Chen H., Liu Z. (2010) J. Biol. Chem. 285, 7986–7994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuhn A. R., Schlauch K., Lao R., Halayko A. J., Gerthoffer W. T., Singer C. A. (2010) Am. J. Respir. Cell Mol. Biol. 42, 506–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. ten Dijke P., Arthur H. M. (2007) Nat. Rev. Mol. Cell Biol. 8, 857–869 [DOI] [PubMed] [Google Scholar]
- 31. Garrett-Sinha L. A., Eberspaecher H., Seldin M. F., de Crombrugghe B. (1996) J. Biol. Chem. 271, 31384–31390 [DOI] [PubMed] [Google Scholar]
- 32. Yoshida T., Kaestner K. H., Owens G. K. (2008) Circ. Res. 102, 1548–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 34. Xu N., Papagiannakopoulos T., Pan G., Thomson J. A., Kosik K. S. (2009) Cell 137, 647–658 [DOI] [PubMed] [Google Scholar]
- 35. Chivukula R. R., Mendell J. T. (2009) Cell 137, 606–608 [DOI] [PubMed] [Google Scholar]
- 36. Watabe T., Miyazono K. (2009) Cell Res. 19, 103–115 [DOI] [PubMed] [Google Scholar]
- 37. Choi B. J., Cho Y. G., Song J. W., Kim C. J., Kim S. Y., Nam S. W., Yoo N. J., Lee J. Y., Park W. S. (2006) Pathol. Res. Pract. 202, 585–589 [DOI] [PubMed] [Google Scholar]
- 38. Hu D., Wan Y. (2011) J. Biol. Chem. 286, 6890–6901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ghaleb A. M., McConnell B. B., Nandan M. O., Katz J. P., Kaestner K. H., Yang V. W. (2007) Cancer Res. 67, 7147–7154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ohnishi S., Ohnami S., Laub F., Aoki K., Suzuki K., Kanai Y., Haga K., Asaka M., Ramirez F., Yoshida T. (2003) Biochem. Biophys. Res. Commun. 308, 251–256 [DOI] [PubMed] [Google Scholar]
- 41. Rowland B. D., Bernards R., Peeper D. S. (2005) Nat. Cell Biol. 7, 1074–1082 [DOI] [PubMed] [Google Scholar]
- 42. Rowland B. D., Peeper D. S. (2006) Nat. Rev. Cancer 6, 11–23 [DOI] [PubMed] [Google Scholar]
- 43. Tai S. K., Yang M. H., Chang S. Y., Chang Y. C., Li W. Y., Tsai T. L., Wang Y. F., Chu P. Y., Hsieh S. L. (2011) Cancer Sci. 102, 895–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang J., Place R. F., Huang V., Wang X., Noonan E. J., Magyar C. E., Huang J., Li L. C. (2010) Cancer Res. 70, 10182–10191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yet S. F., McA'Nulty M. M., Folta S. C., Yen H. W., Yoshizumi M., Hsieh C. M., Layne M. D., Chin M. T., Wang H., Perrella M. A., Jain M. K., Lee M. E. (1998) J. Biol. Chem. 273, 1026–1031 [DOI] [PubMed] [Google Scholar]
- 46. Yu F., Li J., Chen H., Fu J., Ray S., Huang S., Zheng H., Ai W. (2011) Oncogene [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liao X., Haldar S. M., Lu Y., Jeyaraj D., Paruchuri K., Nahori M., Cui Y., Kaestner K. H., Jain M. K. (2010) J. Mol. Cell Cardiol. 49, 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Akao Y., Nakagawa Y., Kitade Y., Kinoshita T., Naoe T. (2007) Cancer Sci. 98, 1914–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Michael M. Z., O'Connor S. M., van Holst Pellekaan N. G., Young G. P., James R. J. (2003) Mol. Cancer Res. 1, 882–891 [PubMed] [Google Scholar]
- 50. Takagi T., Iio A., Nakagawa Y., Naoe T., Tanigawa N., Akao Y. (2009) Oncology 77, 12–21 [DOI] [PubMed] [Google Scholar]
- 51. Kent O. A., Chivukula R. R., Mullendore M., Wentzel E. A., Feldmann G., Lee K. H., Liu S., Leach S. D., Maitra A., Mendell J. T. (2010) Genes Dev. 24, 2754–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li H. X., Han M., Bernier M., Zheng B., Sun S. G., Su M., Zhang R., Fu J. R., Wen J. K. (2010) J. Biol. Chem. 285, 17846–17856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. King K. E., Iyemere V. P., Weissberg P. L., Shanahan C. M. (2003) J. Biol. Chem. 278, 11661–11669 [DOI] [PubMed] [Google Scholar]
- 54. Shats I., Milyavsky M., Cholostoy A., Brosh R., Rotter V. (2007) Cell Cycle 6, 1141–1146 [DOI] [PubMed] [Google Scholar]
- 55. Xie W. B., Li Z., Miano J. M., Long X., Chen S. Y. (2011) J. Biol. Chem. 286, 15050–15057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kurpinski K., Lam H., Chu J., Wang A., Kim A., Tsay E., Agrawal S., Schaffer D. V., Li S. (2010) Stem Cells 28, 734–742 [DOI] [PubMed] [Google Scholar]
- 57. Kennard S., Liu H., Lilly B. (2008) J. Biol. Chem. 283, 1324–1333 [DOI] [PubMed] [Google Scholar]
- 58. Long X., Bell R. D., Gerthoffer W. T., Zlokovic B. V., Miano J. M. (2008) Arterioscler. Thromb. Vasc. Biol. 28, 1505–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Long X., Creemers E. E., Wang D. Z., Olson E. N., Miano J. M. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 16570–16575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Huang J., Min Lu M., Cheng L., Yuan L. J., Zhu X., Stout A. L., Chen M., Li J., Parmacek M. S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 18734–18739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Wang K., Long B., Zhou J., Li P. F. (2010) J. Biol. Chem. 285, 11903–11912 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.