Abstract
The cell cycle is driven by the kinase activity of cyclin·cyclin-dependent kinase (CDK) complexes, which is negatively regulated by CDK inhibitor proteins. Recently, we identified INCA1 as an interaction partner and a substrate of cyclin A1 in complex with CDK2. On a functional level, we identified a novel cyclin-binding site in the INCA1 protein. INCA1 inhibited CDK2 activity and cell proliferation. The inhibitory effects depended on the cyclin-interacting domain. Mitogenic and oncogenic signals suppressed INCA1 expression, whereas it was induced by cell cycle arrest. We established a deletional mouse model that showed increased CDK2 activity in spleen with altered spleen architecture in Inca1−/− mice. Inca1−/− embryonic fibroblasts showed an increase in the fraction of S-phase cells. Furthermore, blasts from acute lymphoid leukemia and acute myeloid leukemia patients expressed significantly reduced INCA1 levels highlighting its relevance for growth control in vivo. Taken together, this study identifies a novel CDK inhibitor with reduced expression in acute myeloid and lymphoid leukemia. The molecular events that control the cell cycle occur in a sequential process to ensure a tight regulation, which is important for the survival of a cell and includes the detection and repair of genetic damage and the prevention of uncontrolled cell division.
Keywords: CDK (Cyclin-dependent Kinase), Cell Cycle, Cyclins, Leukemia, Protein-Protein interactions, ALL, AML, Growth Suppression, Proliferation
Introduction
Activation of complexes of cyclins and cyclin-dependent kinases (CDK)5 and subsequent phosphorylation of their substrates constitute the major regulatory mechanism of cell cycle control (1, 2). CDKs as the catalytic subunit are inactive in the absence of its regulatory subunit, the cyclin. When activated by a bound cyclin, CDKs become phosphorylated and thus activate or inactivate target proteins to coordinate entry into the next phase of the cell cycle. Different cyclin-CDK combinations determine the downstream proteins targeted. CDKs are constitutively expressed in cells, whereas cyclins are synthesized at specific stages of the cell cycle, in response to various molecular signals.
Cell cycle progression promoted by activated cyclin·CDK complexes is controlled negatively by another class of proteins, the CDK inhibitors (1). Until now, two gene families of CDK inhibitors were known, the cip/kip and the INK4a/ARF family. The cip/kip family includes the genes p21, p27, and p57, which modulate the activities of cyclin D·CDK, E·CDK, A·CDK, and B·CDK complexes (3). The INK4a/ARF family includes p16INK4a, p15INK4b, p18INK4c, and p19INK4d that specifically interact with CDK4 and CDK6 (3). All of these CDK inhibitors were identified in the early 1990s (4–12). A huge range of important information about the mechanisms of action and physiological functions of cell cycle regulators has been gathered since then, especially on their function as tumor suppressors (1, 3, 13–16). However, analysis of knock-out mouse models deficient for cell cycle regulators revealed surprising redundant functions of almost all of these proteins, because the vast majority of these mouse models were viable and could therefore live without the respective regulator (17, 18). For example, mice could unexpectedly compensate their deficiency endogenously for RB (19), CDK2 (20, 21), CDK4 (22), cyclin E (23, 24), or for CDK inhibitors (25–30). To our knowledge, the only cell cycle factor that leads to embryonic lethality, when deleted as singular factor, was cyclin A2 (31, 32). These findings indicate that the many open questions have not been answered yet concerning cell cycle regulation.
Recently, we identified INCA1 as an interaction partner of the cyclin A1·CDK2 complex in a yeast triple-hybrid approach (33). We cloned human INCA1 by 5′-rapid amplification of cDNA ends and identified two splice variants encoded on chromosome 17p13 giving rise to the same protein of 221 amino acids. In mice, we found four splice variants transcribed from chromosome 11B leading to a protein of 231 amino acids. In addition, we cloned the promoter sequence of human INCA1, which is dependent on the transcription factor Sp1 similar to its interaction partner cyclin A1. INCA1 interacts in vitro and in vivo with cyclin A1, which is overexpressed in acute myeloid leukemias (33). It is highly expressed in testis where it is also regulated during development (34). Intermediate expression was found in ovary, pancreas, liver, lung, and spleen (33).
Because the function of INCA1 was unknown, we aimed to identify the molecular pathways in which INCA1 was involved and to elucidate its cellular functions under physiological conditions. Here, we characterize INCA1 as a novel growth-suppressive CDK inhibitor that is suppressed in proliferating leukemic blasts.
EXPERIMENTAL PROCEDURES
Binding Assays
For the GST binding assays, the human INCA1 or uncoupled GST was expressed using pGEX-5X2 vectors in Escherichia coli BL21-DE3 and purification according to the manufacturer's recommendations (GST Gene Fusion System, Amersham Biosciences) using glutathione-agarose beads (Sigma). BL21 cells were induced overnight at 30 °C to produce the respective GST protein. To produce and label CDK and cyclin proteins, 1 μg of human CDK1, CDK2, CDK4, cyclin A1, cyclin A2, cyclin B1, cyclin D1, cyclin E1, or cyclin H cDNA (all in pDEST12.2) was transcribed and translated by using [35S]sulfuric methionine (PerkinElmer Life Sciences) and the TnT® quick-coupled transcription/translation system (Promega) to a final volume of 40 μl. GST-protein lysates were incubated overnight with glutathione beads at 4 °C. After washing, 20 μl of the slurry was run on an SDS-polyacrylamide gel to control the preparations for equal concentrations and protein degradation and stained with Coomassie Brilliant Blue.
Subsequently, 1–2 μg of GST fusion proteins were incubated with 7.5 μl of translated CDKs or cyclins in a total volume of 1 ml of GST-binding buffer (20 mm Tris-HCl, pH 7.4, 0.01% Nonidet P-40, 150 mm NaCl, glycerol 10%) for 1 h at room temperature. After washing with binding buffer five times, the slurry was run on an SDS-polyacrylamide gel. The gel was dried with a vacuum dryer and exposed on a BIOMAX MR-1 film (Eastman Kodak Co.). Mutagenesis with INCA1 constructs in pEntry or pGEX vectors was performed with the QuikChange II site-directed mutagenesis kit according to the manufacturer's recommendations (Stratagene).
For GST binding assays using mutant INCA1 proteins, human CDK2, and human cyclin A1 proteins were expressed in BL21-DE3 E. coli cells. Subsequently, 2–5 μg of GST fusion proteins were incubated with 20 μg of CDK2 and/or 50 μg of cyclin A1 containing Sf9 lysates in a total volume of 1 ml of GST-binding buffer (50 mm Tris-HCl, pH 7.5, 1.0% Nonidet P-40, 400 mm NaCl, 1 mm dithiothreitol) overnight at 4 °C. After washing with binding buffer and SDS-PAGE, the binding was analyzed by Western blotting using an antibody against cyclin A1 (Pharmingen).
In Vitro Kinase Reactions
The fusion proteins GST-RB and GST-B-Myb as substrates for kinase assays were expressed with pGEX-5X-2 vectors in E. coli BL21-DE3 overnight at 30 °C and purified according to the manufacturer's recommendations (GST Gene Fusion System, Amersham Biosciences) using glutathione-agarose beads (Sigma). Briefly, lysates were incubated overnight with glutathione beads at 4 °C. After washing, 20 μl of the slurry were run on an SDS-polyacrylamide gel, to control the preparations for equal concentrations and protein degradation, and stained with Coomassie Brilliant Blue.
For kinase assays performed with lysates of Sf9 insect cells, cells transfected baculovirally with human INCA1 or cyclin A1/CDK2 were lysed and subjected to kinase reactions using the conditions as described previously (33, 35). Briefly, 5 μCi of [α-32P]ATP (ICN Biomedicals) were added to 15 μl of GST fusion beads and 6 μg of insect cell lysate expressing cyclin and CDK2 as well as insect cell lysates expressing control vector or INCA1. This was then incubated for 30 min in 1× kinase buffer (10 μm ATP, 50 mm Hepes, pH 7.5, 1 mm DTT, 10 mm MgCl2, 0.1 mm Na3VO4, 1 mm NaF). After washing and SDS-PAGE, phosphorylation was detected by autoradiography.
For kinase assays using recombinant and purified INCA1, human GST-INCA and GST-INCA-del75–99 were cloned into the baculovirus-shuttle vector pDEST20, shuttled to the baculovirus via the Bac-to-Bac baculovirus expression system (Invitrogen), and transfected into Sf9 insect cells. Sf9 insect cells were cultured in Schneider's insect cell medium (Invitrogen) and High FiveTM cell line in Express Five SFM medium (Invitrogen) each supplemented with 10% FCS. Sf9 insect cells were infected by baculovirus constructs (baculovirus expression vector system, PharMingen), whereas High FiveTM cells were infected by supernatants from Sf9 insect cells that had been transfected with the constructs before. The cells were lysed on ice in 50 mm Tris-Cl, pH 7.5, 0.5% Nonidet P-40, 150 mm NaCl, 1 mm EDTA, and protease inhibitors and concentrations were determined by SDS-PAGE.
Purification was carried out according to the manufacturer's recommendations (GST Gene Fusion System, Amersham Biosciences) using glutathione-agarose beads (Sigma) as described below for the purification from bacteria. To control the preparations for equal concentrations and protein degradation, 2–5 μl of the slurry were run on an SDS-polyacrylamide gel and stained with Coomassie Brilliant Blue. Kinase assays were performed using 25 ng of recombinant CDK2·cyclin A (Biaffin GmbH and Co., Kassel, Germany) or 75 ng of PKCα (Cell Signaling) and the indicated amount of recombinant and purified GST-human INCA1 using the conditions as described above.
To evaluate kinase activity in spleens, 200 μg of whole cell lysate or murine spleen lysate were incubated with 20 μl of agarose bead-coupled CDK2 antibody or CDK1 antibody (both Santa Cruz Biotechnology) for 1 h at 4 °C and washed twice with PBS/1% Tween 20 and once with 1× kinase buffer, and kinase reaction was performed as described above. Densitometric analysis was performed using ImageJ software.
Real Time Quantitative RT-PCR, Cell Culture, and Transfection
Reverse transcription and real time quantitative RT-PCR were performed as described previously (33). The probes were labeled at the 5′ end with the fluorescent dye 6-carboxyfluorescein (“FAM” for hINCA1 and mInca1) or 6-carboxyrhodamine-6G (“VIC” for GAPDH) and at the 3′ end with the quencher 6-carboxytetramethylrhodamine (“TAMRA”).
Murine 32D myeloid progenitor cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine (all from Biochrom) and an additional 10% supernatant of IL-3-secreting WEHI cells. The establishment of stably transfected 32D cell lines expressing either FLT3 or FLT3-ITD has been described before (36).
32D cells were starved by depletion of either serum or IL-3. Added IL-3 concentrations refer to recombinant murine IL-3 (PeproTech) with the standard concentration of 2 ng/ml. 32D cells stably transfected with either FLT3-wt or FLT3-ITD (36) were stimulated with either 2 ng/ml recombinant murine IL-3 or 40 ng/ml FLT3 ligand (FL, PeproTech). The tyrosine kinase inhibitor D-65476 for FLT3-ITD was used at 1 μm (37).
Murine embryonic fibroblasts (MEF) were established by trypsinating E12.5 embryos derived from breeding Inca1+/− females and males. Immortalization of fibroblasts was achieved with the standard 3T3 protocol. MEF cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) supplemented with 10 or 20% fetal calf serum (FCS), 100 units/ml penicillin, 100 μg/ml streptomycin, and 2 mm l-glutamine (all from Biochrom). Sf9 insect cells were cultured in Schneider's insect cell medium (Invitrogen) and High FiveTM cell line in Express Five SFM medium (Invitrogen) each supplemented with 10% FCS. Mammalian cells were grown at 37 °C and 5% CO2 and insect cells at 27 °C.
Mammalian cells were transfected using SuperFect (Qiagen) or Nanofectin (PAA) according to the manufacturers' protocols. Sf9 insect cells were infected by baculovirus constructs (Baculovirus Expression Vector System, PharMingen), whereas High FiveTM cells were infected by supernatants from Sf9 insect cells that had been transfected with the constructs before. The cells were lysed on ice in 50 mm Tris-Cl, pH 7.5, 0.5% Nonidet P-40, 150 mm NaCl, 1 mm EDTA, and protease inhibitors and concentrations were determined by SDS-PAGE.
Retroviral Transduction, Colony Formation Assays, and Transplantation of Bone Marrow Cells
Murine Inca1 or mutant Inca1 was cloned into the pEntry vector for gateway system (Invitrogen) and then switched from the pEntry vector into PINCO-RFB destination vector that contains green fluorescence (GFP) expressed from an internal ribosomal entry site, by recombination reaction with LR-Clonase (Invitrogen).
For retroviral transduction, the packaging cell line Plat-E was transfected with the empty vector PINCO or PINCO-mInca1. Supernatants were collected every 12 h, starting 36 h after transfection. For transduction, viruses were bound to RetroNectin-coated plates by centrifugation as described previously (38). Lineage-depleted bone marrow cells were stimulated overnight, transduced by growth on the virus-coated plates for 24 h, and sorted by FACS for EGFP positivity. 1000 EGFP-positive cells per ml of methylcellulose M3434 (Stem Cell Technologies) were plated. The total number of EGFP-positive colonies was determined on day 14 after plating.
A total of 2 × 105 EGFP-positive cells without any preselection together with 105 wild type bone marrow cells were injected into the lateral tail vein of lethally irradiated (8 gray) C57Bl6/N mice. Cotrim was given in the drinking water for 3 weeks after bone marrow transplantation. A fraction of GFP-positive cells was determined by FACS in blood samples 8 weeks after transplantation.
Colony Formation Assays of 32D Cells
Murine 32D myeloid progenitor cells were cultured in RPMI 1640 medium (Invitrogen) supplemented as described above and an additional 10% supernatant of IL-3 secreting WEHI cells. The establishment of stably transfected 32D cell lines expressing FLT3 has been described before (36). Flt3-WT-expressing 32D cells were electroporated with 30 μg of PINCO-empty vector, PINCO-Inca1-wild type, or the mutant construct. The following day, cells were sorted for GFP positivity per FACS and seeded in triplicate 35 × 10-mm dishes containing Iscove's modified Dulbecco's medium (Invitrogen), 1% methylcellulose, 20% FCS, and 1 ng/ml IL-3. Colonies (>50 cells) were counted on days 7–10.
Cell Cycle Analysis and Growth Curves
Prior to cell cycle analysis, immortalized and primary MEFs were starved for 48 h in 0.1% FCS and released in the respective FSC concentrations for 24 h or the times indicated. Cell cycle analyses were performed as described using propidium iodide staining or BrdU incorporation combined with propidium iodide staining (39) and subsequent FACS analysis. If indicated, half of the cells were used for RT-PCR to determine Inca1 expression.
Generation of Inca1 Knock-out Mice
All animal experiments were performed with permission of the local veterinary administration (G15/2005 and 8.87-51.04.20.09-322).
To generate an Inca1-null mouse, we electroporated 129/SV ES cells with a targeting construct, resulting in the replacement of the Inca1 exons 3–7 with an IRES-lacZ cassette and the neomycin resistance gene flanked by loxP sites by homologous recombination. By genotyping of neomycin-resistant ES cell clones using flanking nested PCRs, we identified four different ES cell clones with the expected Inca1 mutated allele, which were all verified by Southern blot using a probe that hybridized 3′ of the recombination site, resulting in an 8-kb band for the wild type Inca1 allele and a 6.8-kb band for the Inca1 knock-out allele after PstI digest of the genomic ES cell DNA (supplemental Fig. S2A). Primers used for ES cell genotyping were as follows: PCR D-5′-CATCGCCTTCTATCGCCTTCTTGAC, PCR D-3′-TGCAGAGAGGCTAGGGTTGGGATAA, and PCR D-3′-nested CTAGGCAGTTTGCCAGGGTGCAGAG. Primers used for routinely genotyping were as follows: Inca1-geno-KO-f-GTCTTGTCGATCAGGATGATCTGG, Inca1-geno-WT-f-CTTATCCCCTGCTCTCAATTGCT, and Inca1-geno-WT-r-TTCATTCCCCTTCCATCACTACCT. Four clones were used to generate germ line chimeras following microinjection into C57Bl/6N blastocysts. Upon mating with C57Bl/6N mice, two chimeras of one clone gave offspring heterozygous for the Inca1-null allele. Intercrosses between heterozygous mice allowed the generation of Inca1 mutant MEFs (Fig. 2B) and living Inca1 homozygous knock-out (−/−) mice. The absence of Inca1 mRNA in Inca1 mutant mice was confirmed by real time RT-PCR using cDNA from wild type (+/+) and Inca1−/− of different organs and from Inca1+/+, Inca1+/−, and Inca1−/− MEFs (Fig. 2C and supplemental Fig. S2C). RNA isolation, reverse transcription, and real time quantitative RT-PCR for GAPDH expression were performed as described previously (33). Analysis of the expression of Inca1 expression was performed using primers mINCA1-F301 CAATTGGATGGTAGAACAGTACATTCC and mINCA1-R390 GGAGGGAGCCCCTCAAGAG and the probe mINCA1-T331 AATTCTGAGGGCCACTGATTGCTCCC. The probe was labeled at the 5′ end with the fluorescent dye 6-carboxyfluorescein and at the 3′ end with the quencher 6-carboxytetramethylrhodamine.
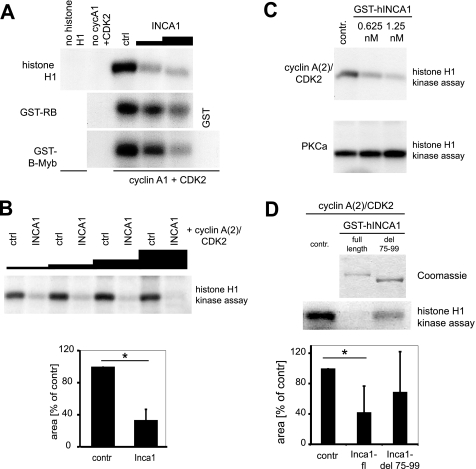
FIGURE 2.
INCA1 inhibits CDK activity in vitro. A, INCA1 dose-dependently inhibited CDK phosphorylation of histone H1, GST-Rb, GST-B-Myb. Baculovirally infected Sf9 cells expressing cyclin A1/CDK2 were incubated with [γ-32P]ATP and different CDK substrates and analyzed by autoradiography. No signals were detected without substrates (no histone H1 or GST alone) or in the absence of cyclin A1/CDK2. ctrl, control. B, upper panel, histone H1 kinase assays using different amounts of lysates from High5 insect cells infected with a baculovirus expressing INCA1 showed a dose-dependent inhibition of CDK2·cyclinA(2) complex activity compared with lysates from uninfected control cells (ctrl). Lower panel, densitometric analysis indicates the statistical significance of CDK2 inhibition by INCA1 (p < 0.001). C, purified full-length INCA1 inhibited CDK2-dependent kinase activity dose-dependently in the low nanomolar range (upper panel), but the activity of protein kinase Cα was not altered (lower panel). Recombinant and purified CDK2·cyclin A(2) complexes or PKCα were incubated with two concentrations of purified GST-INCA1 and analyzed in histone H1 kinase assays. contr, control. D, upper part, in histone H1 kinase assays, full-length but not the cyclin-binding mutant human INCA1-del 75–99 (see Fig. 1, B–F) inhibited cyclin A2/CDK2 activity. Recombinant and purified CDK2·cyclin A(2)-complexes alone (contr) or in combination with either GST-INCA1-full length (FL) or GST-INCA1-del75–99 were analyzed in histone H1 kinase assays (lower panel). A Coomassie staining confirmed equal expression (upper panel). Lower part, densitometric analysis underlines the significant decreased CDK2 activity upon inhibition by INCA1 (*, p = 0.006), whereas the mutated INCA1 could not significantly inhibit CDK2 (p = 0.23, n.s., not significant).
Inca1−/− mice were healthy, viable, and fertile. They were born according to the Mendelian ratio of 1:2:1 (seven different Inca1+/− intercrosses gave birth to 243 pups in total, out of these 60 were wild type, 120 Inca1+/−, and 63 Inca1−/−). Mice used for transplantations were crossed for at least four generations to the C57Bl/6N background.
Histology and Antibody Concentration Measurements
Histological sections and hematoxylin and eosin staining of paraffin-embedded tissues were performed according to standard procedures. NACE (naphthol and ASD chloroacetate esterase) staining was performed as described previously (40). Whole mount β-galactosidase staining of organs was performed as described previously (41).
The levels of immunoglobulin isotypes were determined from a 1:1000 plasma dilution using a competitive Luminex assay. Microbeads (Bio-Rad Bio-Plex®) were cross-linked to isotype detection antibodies (Sigma) with a bead coupling system (Bio-Rad Bio-Plex®). Plasma samples or standard dilutions were incubated to the mixture of microbeads, and then biotinylated standard antibodies (Sigma) were added. The solution was incubated with streptavidin-phycoerythrin, and a number of 100 beads was acquired with a Luminex System (Bio-Rad Bio-Plex®). The level of each isotype was calculated over a standard curve fitted with Four-Parameter-Logistic regression (Bio-Plex® manager software, Bio-Rad).
Flow Cytometry
For the analysis of Inca1 expression in different hematopoietic cells, bone marrow was flushed with PBS, 10% FCS from tibia and femur of C57Bl/6N wild type mice, and red blood cells were lysed with AKC buffer (0.15% NH4Cl, 0.1 m EDTA, 1 mm KHCO3, pH 7.4) for 5 min at room temperature and incubated with the respective antibody (c-Kit, B220, GR1, CD11b, Ter119, CD41, all form BD Biosciences) for 30 min on ice in the dark. Lineage depletion was obtained using the Lineage cell depletion kit, mouse (Miltenyi Biotec, Bergisch Gladbach, Germany), according to the manufacturer's protocol. FACS analyses of spleens were performed as described previously (38).
Patient Samples
Patient samples were collected from the bone marrow of acute myeloid leukemia (AML) and acute lymphoid leukemia (ALL) patients enrolled in different treatment optimization trials. Written informed consent was obtained from all individuals. The use of the human material for scientific purposes was approved by the human ethics committees of the participating institution.
RNA isolation and cDNA production were performed as described previously (42). Quantitative real time RT-PCR was performed using the ABI probe for human INCA1 (Hs01652223_m1) and normalized to GAPDH expression as described previously (34). Overall, 62 samples of patients with AML were analyzed (divided into FAB types as follows: M0, n = 4; M1, n = 6; M2, n = 8; M3, n = 8; M4, n = 6; M4eo, n = 4; M5, n = 12; M5a, n = 5; M5b, n = 1; M6, n = 5; unspecified, n = 3; FLT3-ITD positive, n = 11, 3 unspecified, 2 of the subtype M1, 1 of M2, 1 of M4, and 4 of M5). 10 samples of patients with normal bone marrow were analyzed in comparison with the AML samples.
46 samples of patients with ALL were isolated, 10 with B-ALL, 25 with c-ALL, 7 with pre-B-ALL, and 4 with pro-B-ALL and compared with normal B-cells FACS-sorted using an antibody against CD19 (Miltenyi Biotec) from blood samples. 16 samples from patients with T-ALL were compared with 10 samples from normal T-cells FACS-sorted using an antibody against CD3 (Miltenyi Biotec) from blood samples.
RESULTS
Amino Acids 75–99 of the Human INCA1 Are Required for Cyclin Binding
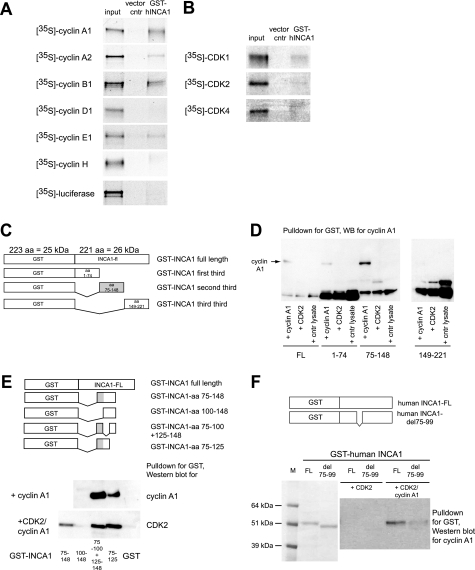
The interaction of INCA1 with the cyclin A1·CDK2 complex (33) prompted us to determine the specificity of INCA1 binding to the cyclin A1·CDK2 complex. GST pulldown assays using 35S-labeled cyclins or CDKs and GST-INCA1 revealed binding of human INCA1 predominantly to the CDK2-specific cyclins A1, A2, B1, and E1 (Fig. 1A). INCA1 did not bind to cyclin D1, cyclin H, or luciferase used as negative control. In contrast, INCA1 binding to purified CDKs in the absence of cyclins was very weak (Fig. 1B).
FIGURE 1.
GST-pulldown assays revealed a cyclin A1-binding site at amino acids 75–99 in the human INCA1 protein. A, GST pulldown assays using 35S-labeled cyclins and GST-INCA1 were performed to identify INCA1-binding cyclins. INCA1 interacted directly with the CDK2-specific cyclins A1, A2, B1, and E1 but not with cyclin D1 or H. B, GST pulldown assays as in A revealed only weak binding of GST-INCA1 to CDKs. C, constructs used to determine cyclin-CDK-binding sites in initial experiments. Because the domain structure of INCA1 is unknown, three different deletion mutants were generated and tested. The gray box indicates the binding site. aa, amino acids. D, cyclin A1 bound to amino acids 75–148 of human INCA1. The weak band in this blot at amino acids 1–74 was not consistently reproducible. No binding to amino acids 149–221 was observed. The lower bands are nonspecific. WB, Western blot. E, upper panel, further GST-INCA1 deletions were constructed to localize the region responsible for CDK and cyclin binding. The gray box indicates the binding site. Lower panel, pulldown assays using further deletion mutants indicate that amino acids at positions 75–99 of INCA1 are required and sufficient for binding of cyclin A1 and CDK2. F, amino acids 75–99 that are required for cyclin-CDK binding were deleted in the full-length construct (upper panel). Coomassie staining of the proteins expressed in E. coli from the constructs to determine the equal amounts of both proteins for the pulldown assays depicted (lower panel, left side). Upon incubation with CDK2 and cyclin A1, the INCA1 deletion mutant del75–99 showed strongly decreased binding affinity for cyclin A1 (lower panel, right side). M, marker; FL, full-length INCA1; del 75–99, deletion mutant of full-length INCA1 lacking amino acids 75–99.
To further analyze the importance of the INCA1 binding to cyclin A1 for its function as a CDK inhibitor, we identified the cyclin-binding sites of human (Fig. 1, C–F) and murine INCA1 (supplemental Fig. S1B). GST pulldown assays were performed with various deletion mutants (as depicted in Fig. 1C), which identified a cyclin A1-binding site in the middle part of the human protein (Fig. 1, D and E; amino acids 75–148). In small fragments of this part of INCA1 as well as in mutated full-length INCA1, we identified amino acids 75–99 (Fig. 1E) and the homologous amino acids 88–116 of murine Inca1 (supplemental Fig. S1, A and B) to be required for cyclin binding. These findings underline the specificity of Inca1 binding to cyclin A1 in complex with CDK2.
Binding to Cyclins Is Critical for INCA1 Function as CDK Inhibitor
The interaction of INCA1 with CDK2-associated cyclins led us to analyze the effects of INCA1 on CDK2 kinase activity. We therefore performed in vitro experiments to analyze the inhibitory effects of INCA1 that were produced within Sf9 insect cells. Indeed, lysates containing human INCA1 inhibited CDK2·cyclin A1 complex kinase activity in vitro toward the different CDK2 substrates histone H1, RB, and B-Myb in a dose-dependent manner (Fig. 2A), whereas Sf9 lysates expressing GST alone did not inhibit CDK2 activity (Fig. 2A, ctrl). In addition, we used lysates containing INCA1 produced by another insect cell line (High5) in combination with a purified human CDK2·cyclin A (which corresponds to murine cyclin A2), because INCA1 also bound to cyclin A2 (Fig. 1A). In this setting, INCA1-containing lysates significantly inhibited purified CDK2·cyclin A activity dose-dependently compared with the same amount of lysates of empty cells (Fig. 2B). These results suggest that INCA1 functionally behaves as a CDK inhibitor without sequence homology to any known family of CDK inhibitors.
To further underline the specificity of INCA1 as a CDK inhibitor, we produced recombinant and purified GST-INCA1 (Fig. 2D, upper panel) from insect cells. Kinase assays using recombinant CDK2·cyclin A complexes revealed that recombinant GST-INCA1 inhibited CDK2·cyclin A function in a dose-dependent manner, whereas GST alone had no effect (Fig. 2C, upper panel, and data not shown). INCA1 did not inhibit PKCα activity, which belongs to a different class of kinases (Fig. 2C, lower panel). INCA1 was active at low nanomolar concentrations, which were in a comparable range as determined for other CDK inhibitors (43) and indicated a strong inhibitory effect.
Remarkably, when we compared the kinase inhibitory function of purified full-length INCA1 and the cyclin-binding mutant GST-INCA1-del75–99 identified in previous experiments described in Fig. 1, we determined a strong and significant decrease in CDK2 inhibitory activity of the mutant compared with full-length INCA1 protein (Fig. 2D, lower panel). The capacity of INCA1 to bind cyclins was therefore required for its full kinase inhibitor activity. These results further substantiated the assumption that INCA1 is a CDK specific kinase inhibitor.
Endogenous Inca1 Expression Is Regulated by Mitogenic Signals and Binding to Cyclin A1 Is Critical for INCA1 Function as a Suppressor of Growth
To uncover mechanisms regulating Inca1 expression, we assessed the effects of mitogenic signals on endogenous Inca1 expression using serum starvation and cytokine depletion experiments in different cell types.
We first examined Inca1 mRNA expression by real time quantitative RT-PCR in interleukin-3 (IL-3)-dependent 32D myeloid progenitor cells. In these cells, presence of IL-3 prevented Inca1 induction by serum depletion, whereas IL-3 starvation induced Inca1 expression (Fig. 3A).
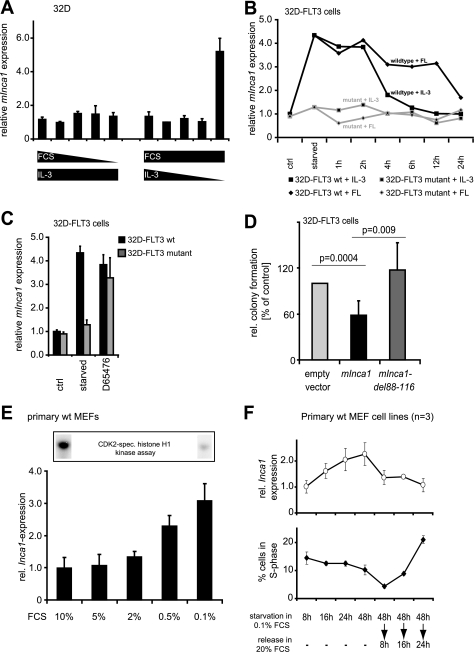
FIGURE 3.
Inca1 expression is induced by serum starvation and repressed by pro-proliferative signaling. A, regulation of Inca1 mRNA expression was analyzed in IL-3-dependent 32D cells by quantitative RT-PCR. Serum starvation alone in the presence of IL-3 did not induce Inca1 mRNA expression, whereas depletion of IL-3 in the presence of serum resulted in increased Inca1 mRNA expression, indicating the dependence of Inca1 induction on the inhibition of mitogenic signaling rather than serum concentration. B, mitogenic signaling from a constitutively active mutant FLT3 receptor FLT3-ITD (36) blocked Inca1 induction as determined by quantitative RT-PCR. Stable expression of the FLT3 wild type tyrosine kinase receptor in 32D cells did not alter Inca1 induction after IL-3 depletion. Inca1 mRNA induction after IL-3 depletion was reversible either by addition of IL-3 or by addition of FLT3 ligand (FL). Expression of leukemogenic, constitutively active FLT3-internal tandem duplications (FLT3-mutant) inhibited Inca1 induction. Addition of IL-3 or FL after IL-3 starvation did not change expression of Inca1 in 32D-FLT3 mutant. C, tyrosine kinase inhibitor D65476 increased Inca1 expression in 32D-FLT3-ITD to a level similar to that detected in 32D-FLT3 after starvation. Thus, Inca1 suppression through FLT3 mutant was reversible upon tyrosine kinase inhibition, and Inca1 was induced upon inhibition of mitogenic signaling. D, inhibition of colony formation in hematopoietic progenitor cells 32D-FLT3 requires the cyclin-binding site. Colony formation assays were performed to analyze the growth inhibitory functions of murine Inca1-full length and the cyclin binding-deficient mutant del88–116. The homologous sites required for cyclin binding were mapped and deleted in murine Inca1 (see supplemental Fig. S1). Expression of murine Inca1-full length (mInca1) led to significantly decreased colony formation (p = 0.0004). No growth inhibitory effect was observed for the cyclin binding-deficient mutant mInca1-del88–116. Indicated are means ± S.D. of three independent experiments. E, serum starvation of primary MEFs dose-dependently induced expression of Inca1 mRNA. MEFs were starved for 48 h in 0.1% FCS and released for 24 h in the indicated FCS concentrations. Lower panel, expression levels were determined by quantitative RT-PCR and normalized to GAPDH expression. Upper panel, lysates from cells grown at 0.1 or 10% FCS were subjected to CDK2-specific histone H1 kinase assays and revealed high CDK2 kinase activity in cells grown with 10% FCS. F, time course of the regulation of Inca1 expression and cell cycle distribution during starvation and release of primary wild type MEFs (n = 3). The regulation of Inca1 expression preceded the re-entry of cells into S-phase after refeeding of the starved cells. The Inca1 expression was determined as in E, and cell cycle distribution was assessed by FACS analysis after BrdU incorporation and propidium iodide staining. Please note that the mean ± S.D. was derived from three independent batches of MEF cells.
We next analyzed the regulation of Inca1 expression in 32D cell lines, which stably express either a constitutively active and leukemogenic mutant FLT3 receptor (FLT3-ITD) or its wild type form (36, 44). Mitogenic signaling from the mutant FLT3 receptor prevented Inca1 induction upon growth factor depletion (Fig. 3B, starved). In contrast, in the 32D-FLT3 wild type cells, Inca1 expression increased after growth factor starvation and decreased time-dependently following stimulation with either IL-3 or FLT3 ligand (FL).
The suppression of Inca1 induction by mutant FLT3 directly depended on FLT3 kinase activity, because its inhibition by the specific FLT3 inhibitor D-65476 (37) led to re-induction of Inca1 mRNA to expression levels comparable with 32D-FLT3 wild type cells (Fig. 3C) corroborating a negative correlation between active mitogenic signaling and proliferation versus Inca1 expression.
To further determine the putative function of Inca1 as a bona fide CDK inhibitor, we analyzed its involvement in the control of proliferation. We therefore compared the proliferative capacity of the murine Inca1 full-length with the CDK2·cyclin A1-binding Inca1-mutant protein (mInca1-del88–116) in colony-forming unit assays of the hematopoietic progenitor cell line 32D-FLT3. The cyclin-binding mutant form of the murine Inca1 protein was identified in analogy to the human mutant (supplemental Fig. 1). Indeed, transient transfection of full-length Inca1 decreased colony formation by almost 50% (p = 0.0004), whereas no inhibitory effect of the binding-deficient mutant was observed (Fig. 3D). Both proteins were expressed efficiently in 32D-FLT3 cells as determined by Western blot detecting murine Inca1 (data not shown). The inhibition of colony formation by Inca1 was associated with decreased proliferation of 32D-FLT3 cells (data not shown). These experiments revealed the antiproliferative functions of Inca1 that clearly depended on cyclin binding.
For further functional analyses, we established primary MEF cell lines. In primary MEFs, low serum concentrations consistently induced Inca1 mRNA expression (Fig. 3E), which correlated with a high CDK2-specific kinase activity in the presence of high FCS concentrations and low activity under FCS-limiting conditions, as we determined in kinase assays using lysates from primary MEFs grown at the respective FCS concentration (Fig. 3F, upper panel). Similar time- and dose-dependent results were obtained for cell lines such as NIH3T3 and HeLa verifying the general induction of Inca1 expression upon serum starvation (data not shown).
Time course experiments were performed to correlate Inca1 mRNA expression and cell cycle distribution. Upon serum starvation, Inca1 expression in primary MEFs steadily increased (Fig. 3F, upper panel), and Inca1 expression rapidly decreased upon serum addition. This occurred long before the cells re-entered S-phase (Fig. 3F, lower panel). These experiments implicate regulation of Inca1 expression during cell cycle and a function of Inca1 as regulator of proliferation.
Inca1 Knock-out Mice Are Viable and Fertile
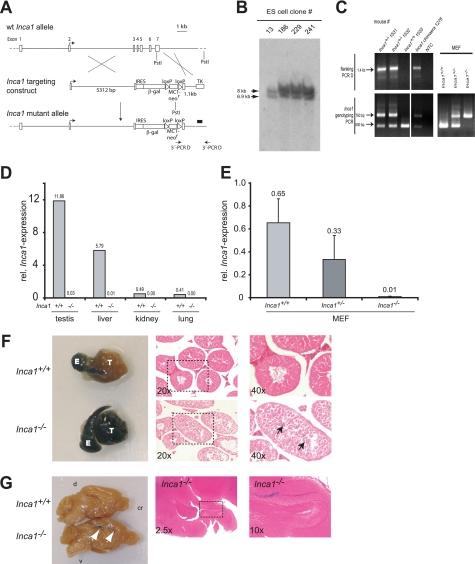
The analyses of relevant molecular and cellular functions are very limited in wild type cells. A knock-out mouse model provides a singular opportunity to reveal physiological mechanisms in which the respective protein is indispensable. We therefore generated a knock-out mouse model replacing five of the six coding exons of the murine Inca1 gene by the β-galactosidase gene (Fig. 4, A–C). Inca1-null mice were viable and did not express Inca1 mRNA in all organs tested (Figs. 4D and 5A) nor in MEFs (Fig. 4E) as determined by quantitative RT-PCR.
FIGURE 4.
Establishment of the Inca1 knock-out mouse, which is viable and fertile. A, generation of Inca1 knock-out (−/−) mice. The Inca1-targeting construct was used to obtain homologous recombination in ES cells between exon 3 and exon 7 of the Inca1 gene. The black bar indicates the Southern blot probe. B, Southern blot revealed that the ES cell clones 13, 186, 229, and 241 were heterozygous for the Inca1 allele, as indicated by the correct bands at 7 kb for the mutant (ko) and 8 kb for the wild type (wt) allele. C, offspring of Inca1 chimeric mice as well as MEFs that were generated by breeding heterozygous Inca1 mice were genotyped by PCR with flanking (PCR D) and internal (genotyping) primers, respectively. C and D, real time RT-PCR using a primer/probe combination specific for the 5′-UTR of the Inca1 mRNA, which was not replaced by the knock-out construct, revealed complete loss of Inca1 mRNA in MEFs (D) and all organs tested (C). The expression of Inca1 was normalized to GAPDH expression. F and G, β-galactosidase expression pattern in Inca1+/+ and Inca1−/− testis (F) and brain (G). Organs were whole mount stained using X-gal staining (left side) and embedded in paraffin, and sections were subjected to eosin counter staining (right side). Specific β-galactosidase expression driven by the Inca1 knock-out construct can be observed in inner layer cells of the seminiferous tubules of Inca1−/− testis (black arrows in F) which cannot be detected in the wild type testis. Unspecific β-galactosidase activity occurs in the epididymis of wild type and knock-out mice. Marked areas in the middle panel (magnification ×20) are shown as higher magnifications in the righthand panels (×40). E, epididymis; T, testis. G, defined region of the dentate gyrus is β-galactosidase-positive in Inca1−/− brain (white arrows in G) that is negative in wild type brains. Marked areas in the middle panel (magnification ×2.5) are shown as higher magnifications in the right panels (×10). d, dorsal side; cr, cranial side; v, ventral side of the brain.
FIGURE 5.
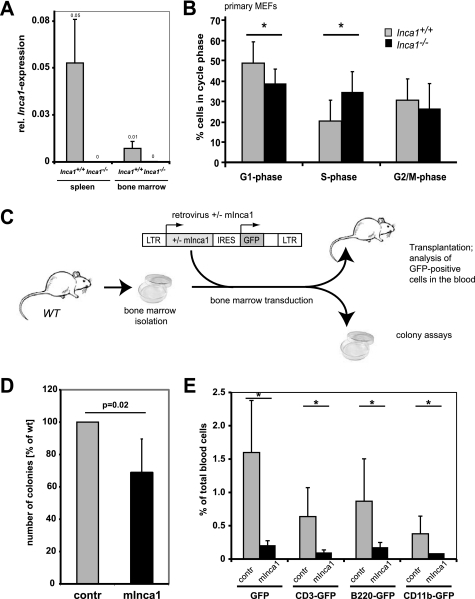
Inca1 represses growth in primary MEFs and bone marrow cells. A, real time RT-PCR using a primer/probe combination specific for the region of the Inca1 mRNA, which was replaced by the knock-out construct, revealed complete loss of Inca1 mRNA in bone marrow and spleen of Inca1−/−mice. The expression of Inca1 was normalized to GAPDH expression. B, cell cycle analysis of Inca1+/+ and Inca1−/− MEFs. Cells were starved for 72 h in 0.1% FCS and analyzed for cell cycle distribution by propidium iodide staining. The number of Inca1−/− cells in G1-phase was significantly decreased (p = 0.03, t test), whereas the number of Inca1−/− cells in S-phase increased compared with Inca1+/+ cells in low serum condition (p = 0.01, t test). The data show mean ± S.D. of four wild type and two Inca1−/− MEF preparations, each analyzed independently at least two times. C, schematic overview about the performed transduction and transplantation experiments. Bone marrow isolated from wild type (WT) mice was retrovirally transduced with either empty vector or with mInca1-IRES-GFP. Equal numbers of positive cells were transplanted into lethally irradiated recipients, which were then subjected to different analyses. D, colony formation assay of wild type bone marrow cells, which were retrovirally transduced with empty vector (contr) or murine Inca1-cDNA (mInca1) (n = 3). Bone marrow transduced with mInca1 formed a significantly decreased number of colonies compared with the empty vector transduced cells (p = 0.02). Shown here are the results of three independent experiments in triplicate as mean ± S.D. E, fraction of wild type bone marrow cells that were transduced with empty vector (contr) or mInca1 was determined by FACS analysis of GFP-positive cells in the blood of transplanted mice 8 weeks after transplantation. The fraction of GFP-positive cells was significantly lower in the blood of recipients transplanted with mInca1-transduced bone marrow cells in all lineages (*, p < 0.01).
Although Inca1 expression is highest in testis and liver (Fig. 4D), loss of Inca1 did not lead to infertility or to histological liver defects (Fig. 4F and data not shown). Inca1-deficient mice were fertile in both sexes. Specific β-galactosidase expression driven by the Inca1 knock-out construct could be observed in inner layer cells of the seminiferous tubules of Inca1−/− testis (Fig. 4F, arrows in the lower panel), which were not stained in the wild type testis (Fig. 4F, upper panel). A defined region of the dentate gyrus was β-galactosidase-positive in Inca1−/− brain that is negative in wild type brains (Fig. 4G). Because no other Inca1−/− organs were positively stained for β-galactosidase, protein levels were putatively below the β-galactosidase threshold.
Inca1 Suppresses Cell Growth in Vivo
For further functional analyses, we established primary MEFs from Inca1+/+ and Inca1−/− littermates. We first compared the cell cycle distribution of different primary wild type and Inca1 knock-out MEFs by propidium iodide staining (Fig. 5B). After 24 h of starvation in 0.1% FCS, the fraction of cells in S-phase was significantly higher (p = 0.03), and the fraction of cells in G1-phase was significantly lower (p = 0.01) in Inca1−/− MEFs than in wild type cells. G2/M-phase was not altered. These effects were also detected at 0.5% FCS but disappeared at higher serum concentrations (supplemental Fig. S2).
To further unravel the proliferation restrictive function of Inca1 in vivo, we retrovirally transduced wild type bone marrow cells with murine Inca1 and compared it with cells transduced with the empty control vector in colony formation assays and upon transplantation in mice (Fig. 5C). Remarkably, expression of murine Inca1 in wild type bone marrow decreased the number of colonies in retrovirally transduced wild type bone marrow (Fig. 5D; p = 0.02).
Moreover, less GFP-positive cells transduced with mInca1 were detected in the blood 8 weeks after transplantation compared with empty vector transduced cells (Fig. 5E; p < 0.01 for all lineages). Interestingly, this effect was seen in all lineages, indicating an equal impact of Inca1 on the proliferation of all lineages (Fig. 5E).
Inca1 Knock-out Spleen Show Increased CDK Activity and Histological Changes
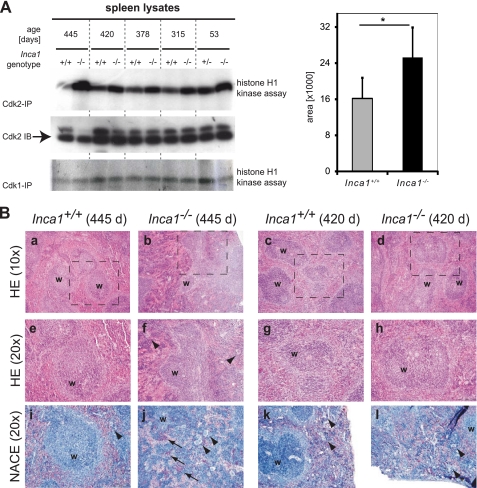
To further determine the relevance of the kinase inhibitory function of Inca1 in vivo, we performed kinase assays. For this purpose, we used lysates from different organs from wild type and Inca1 knock-out mice. Protein levels were normalized to total CDK2 protein. We consistently observed higher CDK2 activity in spleen lysates lacking Inca1 than in the wild type lysates at all ages (Fig. 6A, upper panel), whereas CDK1 activity was not changed (Fig. 6A, lower panel). Densitometric analysis determined this effect as significant (Fig. 6A, right side; p = 0.035).
FIGURE 6.
Inca1 is required for Cdk2 inhibition in vivo in the spleen. A, CDK associated kinase activity in Inca1+/+ and Inca1−/− spleens. Cdk2- or Cdk1-containing complexes were immunoprecipitated from lysates of control and Inca1−/− spleens and subjected to histone H1 kinase assays. In the absence of Inca1, Cdk2 activity was consistently increased compared with lysates from wild type animals (upper panel). Cdk1 activity was unchanged (lower panel). Cdk2 Western blot (middle panel) confirmed equal amounts of Cdk2 in the lysates. Densitometric analysis determined that the Cdk2 activity was significantly increased in Inca1−/− compared with the Inca1+/+ spleens (*, right panel; p = 0.035). B, H&E (HE) staining (panels a–h) and naphthol and ASD chloroacetate esterase (NACE) staining of granulopoietic cells (panels i–l). The Inca1-deficient spleen from a 445-day-old mouse showed a disorganized structure of the white pulp (“w” in panels b and f), an increased number of megakaryocytes (arrowheads in panels f and j), and prominent extramedullary hematopoiesis (arrows in panel j hint at red-stained granulocytes). Other spleens also tested in the kinase assays depicted in Fig. 5C were represented by the spleens of the 420-day-old pair of mice, which did not differ significantly concerning their white pulp structure (w in panels c and d, g and h) and the occurrence of megakaryocytes (arrowheads in panels k and l) or extramedullary hematopoiesis, respectively. Marked areas in panels a–d are shown with higher magnifications in panels e–h.
Investigation of potential histological alterations of 13 wild type and 17 matched Inca1−/− spleens revealed disruption of the splenic architecture, with disturbance of the lymphoid follicles and increased extramedullary hematopoiesis in five Inca1-deficient mice, which were all older than 14 months, but not in any wild type control (Fig. 6, B, panels b, f, j, and d, h, l compared with B, panels a, e, i and c, g, k). Interestingly, the highest CDK2 kinase activity was found in the lysate from a 445-day-old Inca1−/− mouse with altered spleen morphology (see sections of this spleen in Fig. 6B, panels b, f, and j). However, increased kinase activity was present even in Inca1-deficient spleen with normal physiological composition. Cellular proliferation was not generally increased as determined by proliferating cell nuclear antigen staining, and spleen sizes were not altered (data not shown), indicating intrinsic cellular effects of Inca1. Furthermore, FACS analysis revealed an unchanged composition of Inca1 knock-out spleens compared with their wild type controls (supplemental Fig. S3A). Also, the concentration of antibodies was mostly unaffected in Inca1−/− plasma samples (supplemental Fig. S3B).
Moreover, CDK2 activity was not significantly altered in unfractionated bone marrow, liver, or testis suggesting cell type-specific regulation of CDK2 activity by Inca1 (data not shown). These results indicate that the loss of the CDK-inhibitory Inca1 protein is associated with increased CDK2 activity in specific cell types in vivo.
INCA1 Expression Is Significantly Decreased in Blasts of AML and ALL Patients
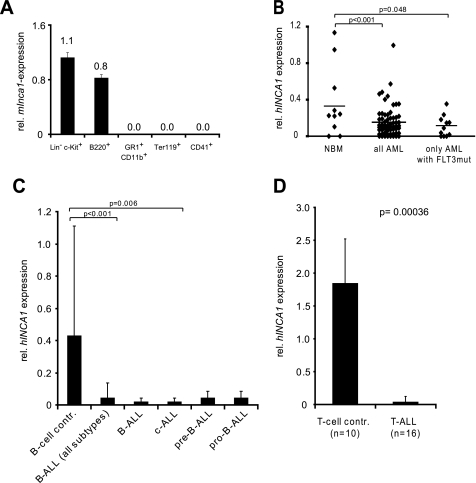
We identified INCA1 as an interacting protein for cyclin A1. Cyclin A1 is a tissue-specific cyclin that is highly expressed in human myeloid leukemias. Overexpression of cyclin A1 has been shown to induce leukemia in mice (45). The fact that Inca1 deficiency obviously affected spleen function (Fig. 6) and that Inca1 was expressed in murine hematopoietic progenitors (Fig. 7A, lin−c-kit+) as well as lymphocytic progenitor cells (Fig. 7A, B220+) challenged us to further analyze the expression of Inca1 in different human leukemic entities. We therefore analyzed expression of INCA1 in human leukemia cells.
FIGURE 7.
Inca1 expression is suppressed in human AML and ALL blasts. A, Inca1 mRNA expression was determined by real time quantitative RT-PCR using cDNA from murine bone marrow cells, which were sorted by FACS. GAPDH served as housekeeping gene for normalization. Inca1 was expressed in hematopoietic stem/progenitor cells (Lin− c-Kit+) and lymphoid progenitor cells (B220+). Granulocytic (CD11b+/GR1+), erythrocytic (Ter119+), or megakaryocytic progenitors (CD41+) did not express Inca1. B, INCA1 expression is significantly down-regulated in bone marrow cells from human AML patients compared with normal bone marrow cells (NBM). In bone marrow cells from AML patients, which were positively tested for the FLT3-ITD mutation, INCA1 expression was specifically and significantly less expressed compared with normal bone marrow samples. INCA1 expression was determined by quantitative RT-PCR and normalized to GAPDH expression level. C, expression of INCA1 was also significantly repressed in B-ALLs, especially in the subtype c-ALL, compared with normal B-cells. B-cell control, n = 12; ALL (all subtypes), n = 46; B-ALL, n = 10; c-ALL, n = 25; pre-B-ALL, n = 7; pro-B-ALL, n = 4. D, T-ALL bone marrow cells expressed significantly less INCA1 than normal T-cells (p = 0.00036).
In human leukemia samples obtained at the time of diagnosis, INCA1 was expressed at significantly lower levels compared with control cells. In AML, INCA1 was expressed at reduced levels compared with normal bone marrow (Fig. 7B). In addition, reduced expression levels were observed in AML blasts that harbored an FLT3-ITD mutation (Fig. 7B). INCA1 levels were not related to specific cytogenetic subtypes (data not shown). Suppression of INCA1 levels were also observed in ALL blasts: INCA1 levels were significantly decreased compared with normal B or T cells in progenitor and mature B-ALLs (Fig. 7C) as well as in T-ALLs (Fig. 7D). These findings indicated that INCA1 is consistently down-regulated in human acute leukemias.
DISCUSSION
In this study, we describe INCA1 as a novel CDK inhibitor. INCA1 binds via a novel cyclin interaction domain to CDK1- and CDK2-bound cyclins. Mutations of this site inhibit the interaction with the cyclin·CDK complex and significantly reduce the CDK inhibitory activity. This domain is also important for the proliferation inhibitory effects.
INCA1 is induced by serum starvation and is suppressed by growth factor signaling. In line, it is suppressed by oncogenic mutants of the FLT3 receptor tyrosine kinase, and expression is significantly reduced in acute leukemia samples compared with normal bone marrow.
In general, despite many discoveries in the field of cell cycle regulation, it appears recently that many riddles remain to be solved and that the complex interplay of pro- and anti-proliferative mechanisms is far from being entirely understood as illustrated by the unexpected viability of mice lacking Cdk2 (20, 21). Thus, the identification of a novel bona fide CDK inhibitor of a yet unknown family is another piece of the jigsaw puzzle for understanding cell cycle regulation.
Comparing INCA1 to the known CDK inhibitor families on the molecular level reveals an intriguing analogy: Only cip/kip CDK inhibitors are found in complex with cyclin and CDK (1, 3) and therefore behave similarly to INCA1, whereas the INK4 family members exclusively bind to the CDK hindering its heterodimerization with the cyclin (1, 3). In line with this observation, INCA1 binds to CDK1/2-specific cyclins but not to the CDK4/6-specific cyclin D (Fig. 1A). Remarkably, a single domain in the INCA1 protein confers binding to cyclins and eventually to CDKs. Because this domain is very small, this could hint at a structurally relevant amino acid sequence. It will be interesting to analyze the nature of this domain to further dissect the regulation of INCA1 function
Because Inca1 knock-out mice are viable and fertile, Inca1 deficiency can be endogenously compensated as also observed for many other cell cycle regulators (17, 18, 25–30). Also, INCA1 overexpression effects on cell cycle regulation were rather weak most likely due to the direct down-regulation of INCA1 upon growth factor signals. However, primary MEF cell lines revealed an increased number of cells in S-phase in the absence of Inca1 (Fig. 5B), which corroborates a direct function of Inca1 in cell cycle control. Taken together, our cell cycle analyses suggest a direct effect of INCA1 onto cell cycle progression.
INCA1 mRNA expression levels in AML blasts are lower than in normal bone marrow cells. Because low levels of Inca1 predispose cells to unrestricted cycling, it is intriguing to speculate that AML cells harbor decreased levels of INCA1. Indeed, our data show that Inca1 is significantly down-regulated in bone marrow from AML and ALL patients as compared with normal bone marrow or control blood cells, which could hint at a function of Inca1 as a tumor suppressor. Notably, many other CDKIs possess tumor-suppressive functions (25, 46–50). Further analysis will provide an insight into the putative role of Inca1 in tumorigenesis and especially in leukemogenesis.
In summary, we have identified a novel growth-suppressive CDK inhibitor that is induced by cell cycle arrest and lost in human tumors. INCA1 thereby constitutes another player in the complex network of cell cycle regulation that controls proliferation and development.
Supplementary Material
Acknowledgments
We are grateful to Frank Berkenfeld, Beate Lindtner, Annika Krause, Linda Kerstiens, Sandra Gelsing, and Britta Körner for excellent technical assistance. We highly appreciate the help of Dr. Thure Adler from the German Mouse Clinic, Neuherberg, Germany, for help with the antibody concentration determination. We thank Helen Piwnica-Worms (St. Louis) and Boris Sarcevic (Sydney, Australia) for providing constructs.
This work was supported by the Interdisciplinary Center for Clinical Research (IZKF) at University of Münster and Deutsche Forschungsgemeinschaft Grants Mu1328/6-1.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- CDK
- cyclin-dependent kinase
- AML
- acute myeloid leukemia
- ALL
- acute lymphoid leukemia
- MEF
- murine embryonic fibroblast
- EGFP
- enhanced GFP.
REFERENCES
- 1. Besson A., Dowdy S. F., Roberts J. M. (2008) Dev. Cell 14, 159–169 [DOI] [PubMed] [Google Scholar]
- 2. Gao C. Y., Zelenka P. S. (1997) BioEssays 19, 307–315 [DOI] [PubMed] [Google Scholar]
- 3. Sherr C. J., Roberts J. M. (1999) Genes Dev. 13, 1501–1512 [DOI] [PubMed] [Google Scholar]
- 4. Harper J. W., Adami G. R., Wei N., Keyomarsi K., Elledge S. J. (1993) Cell 75, 805–816 [DOI] [PubMed] [Google Scholar]
- 5. Xiong Y., Hannon G. J., Zhang H., Casso D., Kobayashi R., Beach D. (1993) Nature 366, 701–704 [DOI] [PubMed] [Google Scholar]
- 6. Polyak K., Lee M. H., Erdjument-Bromage H., Koff A., Roberts J. M., Tempst P., Massagué J. (1994) Cell 78, 59–66 [DOI] [PubMed] [Google Scholar]
- 7. Toyoshima H., Hunter T. (1994) Cell 78, 67–74 [DOI] [PubMed] [Google Scholar]
- 8. Lee M. H., Reynisdóttir I., Massagué J. (1995) Genes Dev. 9, 639–649 [DOI] [PubMed] [Google Scholar]
- 9. Hannon G. J., Beach D. (1994) Nature 371, 257–261 [DOI] [PubMed] [Google Scholar]
- 10. Guan K. L., Jenkins C. W., Li Y., Nichols M. A., Wu X., O'Keefe C. L., Matera A. G., Xiong Y. (1994) Genes Dev. 8, 2939–2952 [DOI] [PubMed] [Google Scholar]
- 11. Hirai H., Roussel M. F., Kato J. Y., Ashmun R. A., Sherr C. J. (1995) Mol. Cell. Biol. 15, 2672–2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. el-Deiry W. S., Tokino T., Velculescu V. E., Levy D. B., Parsons R., Trent J. M., Lin D., Mercer W. E., Kinzler K. W., Vogelstein B. (1993) Cell 75, 817–825 [DOI] [PubMed] [Google Scholar]
- 13. Evan G. I., Vousden K. H. (2001) Nature 411, 342–348 [DOI] [PubMed] [Google Scholar]
- 14. Hanahan D., Weinberg R. A. (2000) Cell 100, 57–70 [DOI] [PubMed] [Google Scholar]
- 15. Morgan D. O. (1995) Nature 374, 131–134 [DOI] [PubMed] [Google Scholar]
- 16. Sherr C. J. (1996) Science 274, 1672–1677 [DOI] [PubMed] [Google Scholar]
- 17. Gladden A. B., Diehl J. A. (2003) Cancer Cell 4, 160–162 [DOI] [PubMed] [Google Scholar]
- 18. Sherr C. J., Roberts J. M. (2004) Genes Dev. 18, 2699–2711 [DOI] [PubMed] [Google Scholar]
- 19. Walkley C. R., Orkin S. H. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9057–9062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Berthet C., Aleem E., Coppola V., Tessarollo L., Kaldis P. (2003) Curr. Biol. 13, 1775–1785 [DOI] [PubMed] [Google Scholar]
- 21. Ortega S., Prieto I., Odajima J., Martín A., Dubus P., Sotillo R., Barbero J. L., Malumbres M., Barbacid M. (2003) Nat. Genet. 35, 25–31 [DOI] [PubMed] [Google Scholar]
- 22. Tsutsui T., Hesabi B., Moons D. S., Pandolfi P. P., Hansel K. S., Koff A., Kiyokawa H. (1999) Mol. Cell. Biol. 19, 7011–7019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Geng Y., Yu Q., Sicinska E., Das M., Schneider J. E., Bhattacharya S., Rideout W. M., Bronson R. T., Gardner H., Sicinski P. (2003) Cell 114, 431–443 [DOI] [PubMed] [Google Scholar]
- 24. Parisi T., Beck A. R., Rougier N., McNeil T., Lucian L., Werb Z., Amati B. (2003) EMBO J. 22, 4794–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bai F., Pei X. H., Godfrey V. L., Xiong Y. (2003) Mol. Cell. Biol. 23, 1269–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheng T., Rodrigues N., Dombkowski D., Stier S., Scadden D. T. (2000) Nat. Med. 6, 1235–1240 [DOI] [PubMed] [Google Scholar]
- 27. Cheng T., Rodrigues N., Shen H., Yang Y., Dombkowski D., Sykes M., Scadden D. T. (2000) Science 287, 1804–1808 [DOI] [PubMed] [Google Scholar]
- 28. Janzen V., Forkert R., Fleming H. E., Saito Y., Waring M. T., Dombkowski D. M., Cheng T., DePinho R. A., Sharpless N. E., Scadden D. T. (2006) Nature 443, 421–426 [DOI] [PubMed] [Google Scholar]
- 29. van Os R., Kamminga L. M., Ausema A., Bystrykh L. V., Draijer D. P., van Pelt K., Dontje B., de Haan G. (2007) Stem Cells 25, 836–843 [DOI] [PubMed] [Google Scholar]
- 30. Zindy F., van Deursen J., Grosveld G., Sherr C. J., Roussel M. F. (2000) Mol. Cell. Biol. 20, 372–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kalaszczynska I., Geng Y., Iino T., Mizuno S., Choi Y., Kondratiuk I., Silver D. P., Wolgemuth D. J., Akashi K., Sicinski P. (2009) Cell 138, 352–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Murphy M., Stinnakre M. G., Senamaud-Beaufort C., Winston N. J., Sweeney C., Kubelka M., Carrington M., Bréchot C., Sobczak-Thépot J. (1997) Nat. Genet. 15, 83–86 [DOI] [PubMed] [Google Scholar]
- 33. Diederichs S., Bäumer N., Ji P., Metzelder S. K., Idos G. E., Cauvet T., Wang W., Möller M., Pierschalski S., Gromoll J., Schrader M. G., Koeffler H. P., Berdel W. E., Serve H., Müller-Tidow C. (2004) J. Biol. Chem. 279, 33727–33741 [DOI] [PubMed] [Google Scholar]
- 34. Diederichs S., Bäumer N., Schultz N., Hamra F. K., Schrader M. G., Sandstede M. L., Berdel W. E., Serve H., Müller-Tidow C. (2005) Int. J. Cancer 116, 207–217 [DOI] [PubMed] [Google Scholar]
- 35. Müller-Tidow C., Ji P., Diederichs S., Potratz J., Bäumer N., Köhler G., Cauvet T., Choudary C., van der Meer T., Chan W. Y., Nieduszynski C., Colledge W. H., Carrington M., Koeffler H. P., Restle A., Wiesmüller L., Sobczak-Thépot J., Berdel W. E., Serve H. (2004) Mol. Cell. Biol. 24, 8917–8928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mizuki M., Fenski R., Halfter H., Matsumura I., Schmidt R., Müller C., Grüning W., Kratz-Albers K., Serve S., Steur C., Büchner T., Kienast J., Kanakura Y., Berdel W. E., Serve H. (2000) Blood 96, 3907–3914 [PubMed] [Google Scholar]
- 37. Teller S., Krämer D., Böhmer S. A., Tse K. F., Small D., Mahboobi S., Wallrapp C., Beckers T., Kratz-Albers K., Schwäble J., Serve H., Böhmer F. D. (2002) Leukemia 16, 1528–1534 [DOI] [PubMed] [Google Scholar]
- 38. Agrawal S., Koschmieder S., Bäumer N., Reddy N. G., Berdel W. E., Müller-Tidow C., Serve H. (2008) Leukemia 22, 78–86 [DOI] [PubMed] [Google Scholar]
- 39. Ji P., Agrawal S., Diederichs S., Bäumer N., Becker A., Cauvet T., Kowski S., Beger C., Welte K., Berdel W. E., Serve H., Müller-Tidow C. (2005) Oncogene 24, 2739–2744 [DOI] [PubMed] [Google Scholar]
- 40. Mufti G. J., Flandrin G., Schaefer H., Sanberg A. A., Kanfer E. J. (1996) in An Atlas of Malignant Haematology (M. Dunitz M. ed) pp. 389–390, Martin Dunitz, London [Google Scholar]
- 41. Bäumer N., Marquardt T., Stoykova A., Ashery-Padan R., Chowdhury K., Gruss P. (2002) Development 129, 4535–4545 [DOI] [PubMed] [Google Scholar]
- 42. Tickenbrock L., Schwäble J., Wiedehage M., Steffen B., Sargin B., Choudhary C., Brandts C., Berdel W. E., Müller-Tidow C., Serve H. (2005) Blood 105, 3699–3706 [DOI] [PubMed] [Google Scholar]
- 43. Castaño E., Kleyner Y., Dynlacht B. D. (1998) Mol. Cell. Biol. 18, 5380–5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grundler R., Miething C., Thiede C., Peschel C., Duyster J. (2005) Blood 105, 4792–4799 [DOI] [PubMed] [Google Scholar]
- 45. Liao C., Wang X. Y., Wei H. Q., Li S. Q., Merghoub T., Pandolfi P. P., Wolgemuth D. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 6853–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Martín-Caballero J., Flores J. M., García-Palencia P., Serrano M. (2001) Cancer Res. 61, 6234–6238 [PubMed] [Google Scholar]
- 47. Fero M. L., Randel E., Gurley K. E., Roberts J. M., Kemp C. J. (1998) Nature 396, 177–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fero M. L., Rivkin M., Tasch M., Porter P., Carow C. E., Firpo E., Polyak K., Tsai L. H., Broudy V., Perlmutter R. M., Kaushansky K., Roberts J. M. (1996) Cell 85, 733–744 [DOI] [PubMed] [Google Scholar]
- 49. Sharpless N. E., Bardeesy N., Lee K. H., Carrasco D., Castrillon D. H., Aguirre A. J., Wu E. A., Horner J. W., DePinho R. A. (2001) Nature 413, 86–91 [DOI] [PubMed] [Google Scholar]
- 50. Zindy F., Nilsson L. M., Nguyen L., Meunier C., Smeyne R. J., Rehg J. E., Eberhart C., Sherr C. J., Roussel M. F. (2003) Cancer Res. 63, 5420–5427 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.