Abstract
Monoamine oxidases (MAO-A, MAO-B) metabolize biogenic amines and have been implicated in neuronal apoptosis. Although apoptosis is an important process in embryo development, the role of MAO isoenzymes has not been investigated in detail. We found that expression of MAO-A and MAO-B can be detected early on during embryo development. Expression levels remained constant until around midgestation but then dropped to almost undetectable levels toward birth. Similar expression kinetics were observed in the brain. Isoform-specific expression silencing of MAO-A mediated by siRNA during in vitro embryogenesis induced developmental defects, as indicated by a reduction of the crown rump length and impaired cerebral development. These alterations were paralleled by elevated serotonin levels. Similar abnormalities were observed when embryos were cultured in the presence of the MAO-A inhibitor clorgyline or when the transcriptional inhibitor of MAO-A expression R1 was overexpressed. In contrast, no such alterations were detected when expression of MAO-B was knocked down. To explore the underlying mechanisms for the developmental abnormalities in MAO-A knockdown embryos, we quantified the degree of developmental apoptosis in the developing brain. MAO-A knockdown reduced the number of apoptotic cells in the neuroepithelium, which coincided with impaired activation of caspases 3 and 9. Moreover, we observed reduced cyclin D1 levels as an indicator of impaired cell proliferation in MAO-A knockdown embryos. This data highlights MAO-A as a vital regulator of embryonic brain development.
Keywords: Apoptosis, Development, Embryo, Neurobiology, Oxidase, siRNA, Monoamine Oxidase, Serotonin
Introduction
Monoamine oxidases (MAOs)2 are catalytically active flavoproteins that are widely expressed in the animal kingdom (1). They catalyze the oxidation of endogenous and xenobiotic amines to the corresponding aldehydes. This reaction requires molecular dioxygen and produces stoichiometric amounts of hydrogen peroxide and ammonia (2). In higher animals, there are two MAO isoforms, referred to as MAO-A and MAO-B (3), which share a high degree (70%) of amino acid conservation. The two isozymes can be distinguished with respect to their substrate specificity and their sensitivity toward inhibitors (4). A major site of MAO expression is the CNS. In adult rats, MAO-A is predominantly found in catecholaminergic neurons, and highest concentrations are detected in the locus coerulus (5). In contrast, MAO-B is most abundant in serotonergic and histaminergic neurons as well as in glial cells (5). In the periphery, distribution of MAO isoforms is variable, and there are species-specific differences (6).
The two MAO isoforms are encoded for by separate genes on the X chromosome. The promoter regions of the genes have been investigated (7), and the transcriptional repressor R1 was found to specifically bind and repress the MAO-A promoter (8). A functional promoter polymorphism of the MAO-A gene has been related to behavioral disorders (9). In humans, functional inactivation of MAO genes is associated with a severe form of Norrie disease (10). In addition, pharmacologic inactivation of MAO activity is a therapeutic approach for the treatment of neurological disorders that have been shown to involve MAO activity, such as depression or neurodegenerative disorders, including Parkinson's disease and Alzheimer's disease (11).
To study the in vivo role of MAO isoenzymes in cerebral function, MAO-deficient mice have been employed. Targeted knockout of the MAO-B gene leads to viable offspring that show an altered stress response (12). Unfortunately, to date targeted inactivation of the MAO-A gene has not been carried out. However, naturally occurring MAO-A-deficient mice strains have been identified. Incidental insertion of an interferon β minigene into exon 2 of the MAO-A gene abrogates the expression of a functional enzyme. These mice are viable and do not show obvious signs of functional defects in a resting state (13). However, they have significantly increased plasma levels of monoaminergic transmitters (serotonin, dopamine, norepinephrine), are more susceptible to different kinds of stress, and exhibit a tendency for aggressive behavior (13). Similar observations were made in mice bearing a spontaneous point mutation in exon 8, which introduced a premature stop codon analogous to a mutation found in humans suffering from Brunner syndrome (14). An identical mutation occurred in a colony of MAO-B-deficient mice, leading to MAO-A/B double knockout animals that are also viable but exhibit elevated monoamine levels and anxiety-like behavior (15).
Monoamines are essential for proper cerebral function. Monoaminergic transmitter systems appear early on in embryogenesis (16), and serotonin, the biologically most relevant MAO-A substrate, has been implicated in maturation of neuronal progenitors (17). MAO-A-deficient mice lack developmental clustering of layer IV granular neurons and suffer from aberrant maturation of the brainstem respiratory network (18, 19). Although the underlying mechanisms have not been studied in detail, these effects could be explained by the inability of the animals to metabolize serotonin (20). This data is consistent with the recent observation that transgenic overexpression of MAO-A in the forebrain of MAO-A knockout mice rescues their aggressive behavior (21).
Because the outcome of functional studies employing systemic stem cell knockouts is frequently impacted by genetic drifts and epigenetic phenomena (22, 23), we employed the siRNA technology to explore the role of MAO isoforms in embryogenesis. This experimental setup allows us to study the impact of MAO expression at greater spatial and temporal specificity. For this purpose, we explanted murine embryos at early developmental stages, injected siRNA probes or specific inhibitors into the amniotic cavity, and cultured the treated embryos for up to 72 h in vitro (24). Using these genetic and pharmacological intervention strategies, we found that knockdown of MAO-A expression disturbed embryonic brain development and dysregulated developmental apoptosis.
MATERIALS AND METHODS
Chemicals
The chemicals used were from the following sources: Superscript III reverse transcriptase and RNaseOUT from Invitrogen, BD Advantage 2 polymerase mix from BD Biosciences, dNTPs from Carl Roth GmbH (Karlsruhe, Germany), and PCR primers from BioTeZ Berlin-Buch GmbH (Berlin, Germany).
RNA Extraction and Reverse Transcription
Total RNA was extracted using the RNeasy mini kit (Qiagen, Germany) and was reversely transcribed according to standard protocols with oligo d(T)15 primers and SuperScript III reverse transcriptase (Invitrogen) according to the vendor's instructions.
Quantitative RT-PCR
RT-qPCR was carried out with a RotorGene 3000 system (Corbett Research, Australia) using ImmoMix/SYBR Green (Bioline, Germany). Isoform-specific amplification primers were designed, and their sequences are given in supplemental Table S1. Absence of cross-amplification between the two isoforms was ensured, and the standard protocol has been described before (24). RNA preparations were analyzed at least in triplicate, and mean values are given. The experimental raw data were evaluated with the RotorGene Monitor software (version 4.6). To generate standard curves for quantification of expression levels, specific amplicons were used as external standards for each target gene (5 × 103 to 3 × 106 copy numbers). GAPDH mRNA was used as the internal standard to normalize expression of the target transcripts.
Preparation of Mouse Embryos and in Vitro Culture
All animal experiments were performed in strict adherence to the guidelines for experimentation with laboratory animals set in institutions. Inbred Institute for Cancer Research pregnant mice were obtained from the animal house, and embryos at different developmental stages (gestational day 6.5 (E6.5) to E17.5) were prepared under a stereomicroscope (Olympus). For RT-qPCR, preparations were kept in PBS (0.1% diethyl pyrocarbonate), and extraembryonic tissue was removed. For in vitro culture, the embryos were dissected in modified phosphate-buffered saline medium (5% FBS) as described before (24).
Transfection Experiments
For our knockdown strategy, we designed isoform-specific siRNA using the StealthTM RNAi program (BLOCK-iTTM RNAi Designer, Invitrogen). Among the designed probes we selected those constructs supplemental Table S1) exhibiting highest estimated knockdown probability and a GC content below 40%. No sequence cross-homology was allowed. Control siRNA duplexes with no homology to any vertebrate transcript were used as random control probes (Stealthtm RNAi negative controls, Invitrogen). For R1 overexpression, the R1 coding sequence was cloned into the mammalian pcDNA3.1 expression vector supplemental Table S1). Empty vector was used as a control. For knockdown experiments, murine embryos were explanted at E7.5 and transfected with siRNA constructs. 10 nl of annealed double-stranded siRNA (25–100 nm) were mixed with 0.01% LipofectamineTM 2000 (Invitrogen) and then microinjected with an Application Solution Transgenic platform micromanipulator (Leica, Germany) into the amniotic cavity. After microinjection the embryos were placed in a whole embryo culture roller incubator (BTC Engineering, UK). The embryos at early gastrulation stage (E7.5) were allowed to develop for up to 72 h until early organogenesis stages E9.5-E10.5 in 100% heat-inactivated rat serum with a continuous flow of gas mixtures (24).
Evaluation of Developmental Milestones
The impact of the treatments on embryonic growth (embryo size measured by the crown rump and head length) and on development of the brain, heart, and limbs was quantified by a microscopic scoring procedure. To judge cerebral embryogenesis, we assessed the degree of brain maturation by a number of morphological parameters (25). A score of 0 represented strong developmental retardations, whereas a score of 5 indicated normal development. The following morphological criteria were applied for the different parts of embryonic brain: Forebrain: prosencephalon invisible (score 0), V-shape (score 1), U-shape (score 2), partially fused (score 3), completely fused (score 4), telencephalic evaginations (score 5). Midbrain: mesencephalic invisible (score 0), V shape (score 1), U shape (score 2), partially fused (score 3), completely fused (score 4), discernible division between mesencephalon and diencephalons (score 5). Hindbrain: rhombencephalon invisible (score 0), V shape (score 1), U shape (score 2), folds fused with anterior neuropore (score 3), anterior neuropore closure (score 4), dorsal flexion develops with 4th ventricle roof (score 5). Significances were calculated with Students' t test.
MAO Activity Assays
Enzymatic activity of MAO isoforms was assayed in vitro with 14C-labeled tyramine hydrochloride as a substrate, as described previously (26). The relative share contributed by the two MAO isoforms was quantified using isoform-specific irreversible inhibitors. Clorgyline was used for specific inhibition of MAO-A and deprenyl for inhibition of MAO-B.
In Situ Hybridization
In situ hybridization was performed according to Wilkinson's methods and as outlined previously (24). Suitable riboprobes (sense and antisense probes) were prepared by PCR (supplemental Table S1), cloned, and transcribed using T7 RNA polymerase. To label the RNA probes, digoxigenin-11-UTP (Roche) was incorporated using the AmpliScribe T7 kit (Epicenter Technologies). Treated embryos and corresponding controls were fixed in 4% p-formaldehyde, dehydrated in graded methanol solutions, and stored at −20 °C prior to whole mount in situ hybridization. For section in situ hybridization, the fixed embryos were dehydrated in graded ethanol/xylene mixtures and then embedded in paraffin (24). Sagittal sections (5 μm thick) of the embryos were prepared and stored at room temperature prior to in situ hybridization.
Immunohistochemistry
Immunohistochemical staining was performed on paraffin-embedded embryo sections employing the immunoperoxidase method. The antigenic epitopes were first exposed by heating the sections in 10 mm citrate buffer (pH 6.0) containing 0.05% Tween 20. After quenching the endogenous peroxidase with 3% hydrogen peroxide, sections were incubated in TBS (5% normal sheep serum) containing either anti-human MAO-A (clone C-19) or anti-human MAO-B (clone C-17) monoclonal antibodies (Santa Cruz Biotechnology, Inc.). The sections were then incubated with a peroxidase-labeled secondary antibody (Biocare Medical Kit), and slides were developed with diaminobenzidin (brown color). Negative controls lacked the primary antibodies.
Apoptotic cells were stained with both the supravital dye staining method and standard TUNEL technique. The whole mount supravital dye Nile blue sulfate visualizes non-viable cells. After cultivation, embryos at E10.5 were washed in Ringer's solution, bathed in a Nile blue sulfate solution for 15–20 min at 37 °C, and then washed in Ringer's solution before microscopic examination. TUNEL staining was performed on embryo sagittal sections using the Chemicon kit according to the vendor's instructions. Nuclear DNA was counterstained with 0.5% (w/v) methyl green for microscopic examination.
Western Blotting
For caspase 3, caspase 8, caspase 9, cyclin D1, E2F1, and β-actin immunoblotting, whole embryo lysates were obtained by homogenizing embryos in lysis buffer (50 mm Tris HCl, 0.3 m NaCl, 1 mm EDTA, 1 mm DTT, and 1 mm PMSF). Aliquots with equal amounts of total protein (100 μg) were separated on 12% SDS-PAGE gels, and separated proteins were transferred to nitrocellulose membranes (Amersham Biosciences). The membranes were blocked overnight with an ovalbumin solution (Sigma) and then incubated at 37 °C for 1 h with primary antibodies. The membranes were then washed three times with TBS and incubated with HRP-labeled secondary antibody (Dako) at 1:1000 dilution (30 min at room temperature). Blots were finally developed using the ECL kit (GE Healthcare). Immunoreactive protein bands were imaged using the Quantity One® quantification software (Bio-Rad) and normalized to β-actin expression. For MAO and R1 immunoblotting, primary antibodies were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), but for quantification of caspase 3, caspase 8, caspase 9, and β-actin, antibodies were obtained from Abcam, Inc. (Cambridge, UK).
Serotonin Assay
Serotonin concentration in the embryos were measured using the Serotonin enzyme immunoassay (Ultra Sensitive) kit (Immuno Biological Laboratory, Inc.) according to the manufacturer's instructions. In brief, after treatments in vitro cultured embryos were washed and homogenized, and aliquots of the lysates were applied for quantification. The lower detection limit of the kit was 1.0 nmol/liter, and a linear calibration plot was established between 1.45 and 100 nmol/liter (r2 = 0.9807 ± 0.0012). The intra- and interassay variation coefficients were < 3.02%, and the recovery range varied between 100.4 and 102.3%.
RESULTS
Expression of MAO mRNA During the Time Course of Murine Embryogenesis
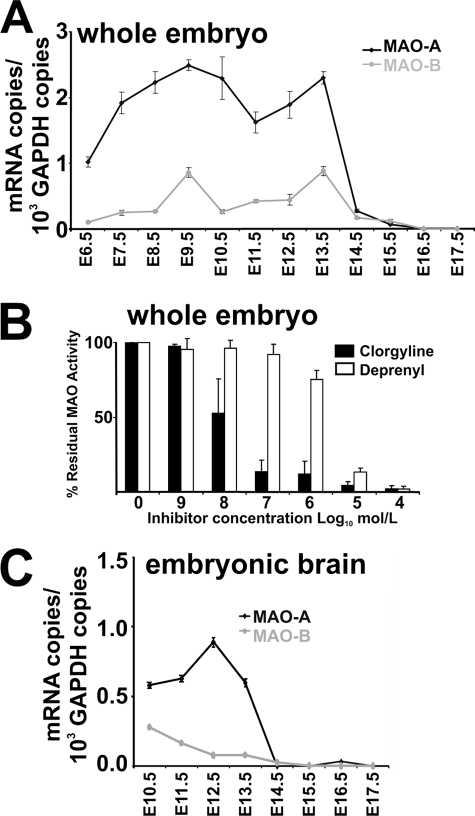
Previous in situ hybridization indicated that MAO isoforms are expressed in embryonic mouse brain as early as at E12 (17). Employing a more sensitive quantitative RT-PCR (RT-qPCR) strategy, we detected MAO mRNA species much earlier on during mouse embryogenesis. At E6.5, MAO-A mRNA was clearly detectable, and its concentration increased until E9.5. Then it remained at constant levels until E13.5 (Fig. 1A). At E14.5, MAO-A mRNA levels dropped and did not recover until birth. MAO-B mRNA follows similar expression kinetics, but its average steady state concentration was 4-fold lower than MAO-A mRNA. The prevalence of MAO-A over MAO-B in murine embryos was confirmed by activity assays (Fig. 1B). More than 90% inhibition of total MAO activity was observed when total embryonic lysates were assayed in the presence of 10−7 m clorgyline, a specific MAO-A inhibitor. In contrast, when the MAO-B-specific inhibitor deprenyl was used, a similar degree of inhibition was only observed at 10−5 m.
FIGURE 1.
Expression kinetics of MAO isoforms during mouse embryo development. A, MAO expression in whole embryos. Mouse embryos (n = 5) were dissected at different developmental stages (E6.5 to E17.5) and pooled together. Total RNA was extracted, and expression levels of MAO isoforms were quantified by RT-qPCR in triplicates (see “Materials and Methods”). B, MAO activity assay of whole embryos. Mouse embryos of gestation day E13.5 were homogenized, and total enzymatic activity was assayed as described under “Material and Methods” using tyramine as a substrate. The relative shares of the two different MAO isoforms contributing to the total activity were quantified by adding isoform-specific inhibitors (clorgyline is MAO-A specific, deprenyl is MAO-B specific) at different concentrations. C, MAO expression in embryonic brain. Mouse embryos were removed at different developmental stages (E10.5-E17.5), brains were isolated, and preparations from five different individuals were pooled. Total RNA was extracted from these pools, and expression levels of MAO isoforms were quantified by RT-qPCR in triplicates (see “Materials and Methods”). The error bars mirror the accuracy of the assay system. The biological variability was averaged by pooling the tissue of five different individuals.
To follow cerebral expression of MAO isoforms during embryo development, we prepared embryonic brains at different developmental stages and recorded the concentration profiles for MAO mRNAs (Fig. 1C). Cerebral MAO-A mRNA follows similar expression kinetics as in the whole embryo, with constant levels from E10.5 to E13.5, and dropped down to lower levels when the embryos approached birth. MAO-B mRNA concentrations are lower than those of MAO-A and decline further toward delivery.
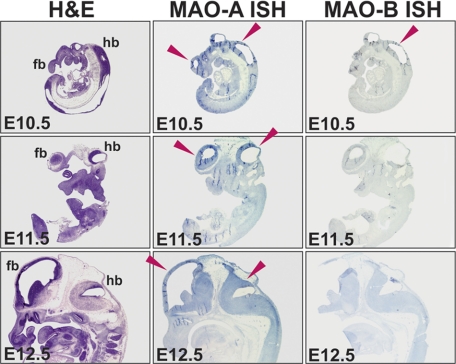
To explore the spatial distribution of MAO mRNA expression during embryogenesis, we performed in situ hybridization (Fig. 2). Between E10.5 and E12.5, specific signals for MAO-A mRNA expression (blue staining indicated by the red arrows) were mainly observed in developing brains. Low levels of in situ hybridization signals for MAO-B mRNA were detected in the fore- and hindbrain at E10.5. At later stages (E11.5 and E12.5), the signals were even less intense.
FIGURE 2.
In situ hybridization of embryonic brain sections at different developmental stages. In situ hybridization for MAO-A and MAO-B was performed as outlined under “Material and Methods.” Positive hybridization signals are indicated by the blue staining, and red arrows indicate brain regions with high MAO expression. fb, forebrain; hb, hindbrain.
Silencing of MAO-A Expression Induced Developmental Defects
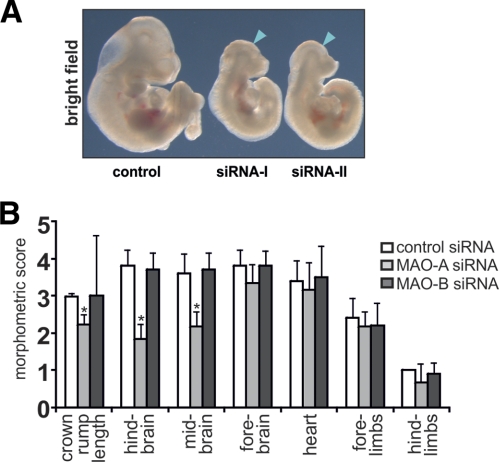
To silence expression of MAO-A and MAO-B during in vitro embryogenesis, we designed different specific siRNA probes for each MAO-isoform (supplemental Table S1). In an initial experiment, different siRNA species were injected into the amniotic cavity, and embryos were recovered after a culturing period of 72 h. Following knockdown of MAO-A using two independent siRNA constructs, we observed severe developmental retardations for both siRNA species, as indicated by the reduced embryo size compared with embryos treated with scrambled siRNAs (Fig. 3A). In contrast, embryos treated with siRNAs targeted at MAO-B expression did not have an apparent impact on embryo development (data not shown).
FIGURE 3.
Knockdown of MAO-A expression using two independent siRNA constructs. For knockdown studies, MAO-A-specific siRNA constructs (siRNA-I and siRNA-II) or corresponding controls were mixed with an appropriate volume of transfection reagent (see “Materials and Methods”). Murine embryos were explanted at E7.5, and the transfection mixture (10 nl with 50 nm probe) was injected into the amniotic cavity. Embryos were then maintained in in vitro cultures for 72 h (E10.5), and the silencing of MAO-A expression was assessed. A, macroscopy. The blue arrows indicate developmental defects of the brain. The figures are representative of five independent experiments. B, developmental scoring. After the culturing period, the crown-rump length and developmental defects in different organ system were evaluated by a numeric scoring procedure or in millimeters (mm) for crown-rump length (see “Materials and Methods”). Significances were calculated using Student's t test. n = 10. *, p < 0.01.
To characterize the siRNA-induced developmental defects in different organ systems in more detail, we employed a morphometric scoring system (25). This scoring system enabled us to translate the developmental progress at organ and tissue levels into a quantifiable matrix of scoring points. Using this system, we evaluated the siRNA-induced developmental defects of the developing brain, heart, and limbs in addition to general growth retardations (crown rump length). We found that siRNA-mediated knockdown of MAO-A expression reduced the crown-rump length and significantly attenuated the developmental scores for the fore-, mid-, and hindbrain (Fig. 3B). On the other hand, development of the heart and the limbs was not significantly altered. Although all siRNA-treated embryos at E10.5 exhibited normal neural tube closure, MAO-A knockdown embryos showed retarded development (lower morphometrics scores), mainly because of delayed segmentation of the rhombomeres in the hindbrain and of the diencephalon in the midbrain. In contrast, knockdown of MAO-B expression did not induce developmental defects in any of the analyzed embryo parts within our experimental time window (Fig. 3B).
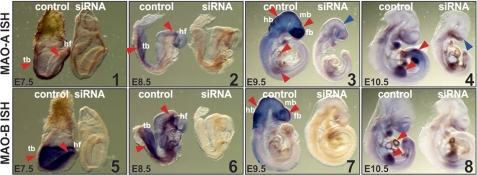
The success of the knockdown strategy was verified by whole mount in situ hybridization. Strong expression of both MAO isoforms was observed in the head region, as indicated by the dark blue in vitro hybridization signals (Fig. 4). Embryos treated with siRNA species targeted at MAO-A expression exhibited strongly reduced in situ hybridization signals after 72 h incubation (Fig. 4, panels 1–4). Similar results were observed when embryos were treated with MAO-B siRNA (Fig. 4, panels 5–8). These data suggest successful knockdown of the expression of either MAO isoform.
FIGURE 4.
Impact of MAO expression silencing on embryo development. For knockdown studies, siRNA constructs or corresponding control probes were mixed with an appropriate volume of transfection reagent (see “Materials and Methods”). Murine embryos were explanted at E7.5, and the transfection mixture (10 nl with 50 nm probe) was injected into the amniotic cavity. The embryos were then maintained in in vitro cultures for 2 h (E7.5), 24 h (E8.5), 48 h (E9.5) and 72 h (E10.5), and silencing of MAO expression was tested. Whole mount in situ hybridization: Mouse embryos were removed from in vitro culture and in situ hybridization with MAO mRNA antisense probes was carried out. The red arrows indicate MAO mRNA expression. The blue arrows indicate developmental defects of the brain. fb, forebrain; mb, midbrain; hb, hindbrain; hf, headfold; tb, tail bud.
Next, we aimed to analyze the effects of MAO knockdown at different time points during our experimental time frame. Both MAO isoforms are expressed at E7.5 and E8.5 in the headfold region and in the caudal neural tube, as indicated by the dark in situ hybridization signals (Fig. 4). At these developmental stages injection of siRNA constructs strongly reduced expression of the corresponding MAO isoform without causing apparent developmental alterations. At E9.5 mRNA of both MAO isoforms was mainly detected in the developing fore-, mid-, and hindbrain and the forelimbs. Faint in situ hybridization signals found in the developing heart indicate expression of MAO-A in this organ (Fig. 4, red arrows). Treatment with specific siRNA selectively reduced MAO isoform expression. However, at this stage (Fig. 4, upper panel), we observed developmental defects as indicated by the reduced size of the whole embryo and of the developing brain (blue arrow). In conclusion, developmental retardations induced by MAO-A siRNA are apparent by day E9.5 but become most pronounced at day E10.5.
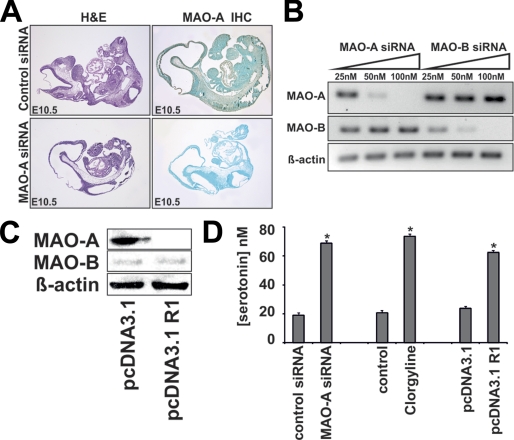
To confirm siRNA-mediated knockdown of MAO isoforms on the protein level, we performed immunohistochemistry and Western blotting (Fig. 5, A and B). Here we detected isoform-specific and dose-dependent siRNA-induced reduction of MAO protein. This indicates successful down-regulation of target gene expression as well as a high isoform specificity of our siRNA constructs.
FIGURE 5.
Knockdown of MAO expression is isoform-specific and induces elevated embryonic 5-HT levels. For knockdown studies, siRNA constructs or corresponding control probes were mixed with an appropriate volume of transfection reagent (see “Materials and Methods”). A, immunohistochemistry. Cross sections were prepared from cultured mouse embryos (E10.5) and stained with a commercial antibody raised against MAO-A. This antibody was tested in our lab to recognize recombinant MAO-A and cross-reacts with MAO-B at a lower intensity. Immunoreactive material is indicated by the brown color. B and C, Western blotting. Mouse embryos were explanted at E7.5 and transfected with increasing amounts of siRNA constructs. After 72 h in culture, the embryos were homogenized, and aliquots were subjected to Western blotting. D, serotonin concentrations in mouse embryos. Embryos were explanted at E7.5, and the siRNA or plasmid constructs indicated were injected into the amniotic cavity. Clorgyline was supplemented in the culture medium. The embryos were then cultured in vitro for 72 h, homogenized, and the endogenous serotonin concentration was assayed as described under “Materials and Methods.” *, p < 0.01.
To make sure that our knockdown data is not the results of methodological artifacts (off-target effects of the siRNA constructs) we employed two additional experimental approaches. Firstly, we used the specific and irreversible inhibitor of MAO-A activity clorgyline. Secondly, we transfected the developing embryos with a mammalian expression plasmid that drives the overexpression of the transcriptional inhibitor of MAO-A expression R1. Indeed, our Western blotting data indicates specific suppression of MAO-A but not of MAO-B expression following R1 overexpression (Fig. 5C).
Reduced expression and/or activity of MAO-A has been reported to be associated with increased systemic levels of its substrate serotonin (13, 15). This prompted us to measure embryonic serotonin levels following MAO-A knockdown. Indeed, we observed that embryonic serotonin levels were elevated almost 4-fold 72 h after MAO-A siRNA treatment (Fig. 5D). Similarly, serotonin levels were also elevated when MAO-A activity was inhibited by clorgyline or when MAO-A gene transcription was suppressed by R1 overexpression.
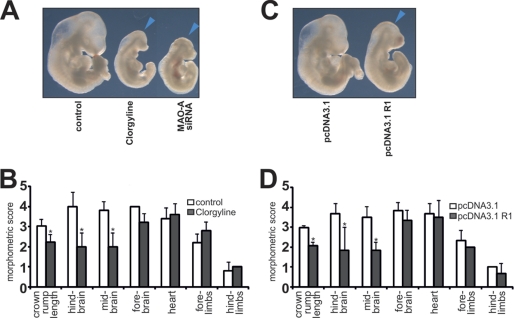
Inhibition of MAO-A Activity Induces Developmental Defects Similar to MAO-A Knockdown
If the developmental abnormalities observed following siRNA-mediated knockdown of MAO-A expression are specific, pharmacological inhibition of MAO-A activity or suppression of MAO-A gene expression by R1 overexpression should induce similar developmental alterations as siRNA treatment. Indeed, when embryos were cultured in the presence of clorgyline or when R1 was overexpressed, embryo growth was impaired to a similar extent as observed for MAO-A knockdown embryos (Fig. 6, A and C). Clorgyline treatment resulted in retarded development of the hind- and midbrain, whereas development of the forebrain was less affected (Fig. 6B). Development of the limbs and the heart proceeded normally. Similar observations were made when R1 was overexpressed (Fig. 6D). Thus, embryos treated with clorgyline or in whom R1 was overexpressed phenotypically resemble MAO-A knockdown embryos.
FIGURE 6.
Developmental defects of mouse embryos cultured in vitro in the presence of the inhibitor of MAO-A activity clorgyline or when the inhibitor of MAO-A transcription R1 was artificially overexpressed. Expression plasmids (pcDNA3.1) or siRNA constructs were mixed with an appropriate volume of transfection reagent (see “Materials and Methods”), or embryos were incubated in the presence of 1 μm clorgyline in the culture medium. A and C, macroscopy. Representative images of differently treated mouse embryos are shown. The blue arrows indicate brain regions with defective development. B and D, developmental scoring. Mouse embryos were explanted at E7.5 and cultured for 72 h either in the presence of 1 μm clorgyline (B) or transfected with an R1 overexpression plasmid (pcDNA3.1 R1) or the backbone vector (pcDNA3.1) (D). After the culturing period, embryos, the crown-rump length, and developmental defects in different organ system were evaluated by a numeric scoring procedure or in millimeters (mm) for crown-rump length (see “Materials and Methods”). Significances were calculated using Student's t test. n = 10. *, p < 0.01.
Knockdown of MAO-A Expression Inhibited Developmental Apoptosis
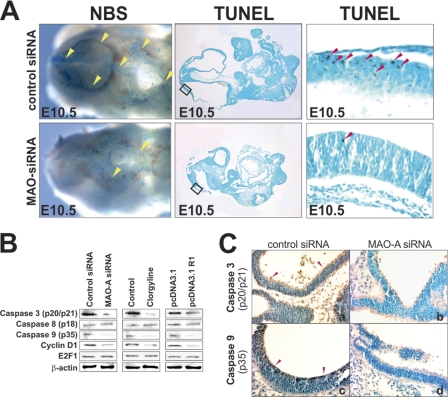
Apoptosis is a vital process in embryo development (27) and MAO-A has been implicated in the regulation of cell proliferation and programmed cell death (26, 28). To test whether knockdown of MAO-A expression impacts developmental apoptosis during murine embryogenesis, we first monitored embryonic cell death by whole mount Nile blue staining of embryos at E10.5 (30-somite stage) and by the TUNEL technique in microscopic cross-sections (Fig. 7A). When we knocked down embryonic expression of MAO-A, the number of dead (Nile blue staining) and apoptotic cells (TUNEL staining) in the neuroepithelium of the developing brain was decreased significantly (yellow and red arrows, respectively) (Fig. 7A).
FIGURE 7.
Silencing of MAO-A expression impairs developmental apoptosis in the murine brain. Mouse embryos were explanted at E7.5. Expression plasmids (pcDNA3.1) or siRNA constructs were injected into the amniotic cavity, and the embryos were further cultured in vitro for 72 h, or embryos were incubated in the presence of 1 μm clorgyline in the culture medium. Representative experiments are shown (n = 5). A, left panels, embryos were treated with Nile blue sulfur (NBS) to stain (dark blue) death cells in whole mount. Dorsal brain areas with dominant staining are indicated by yellow arrows. Center panels, TUNEL assays were performed on microscopic cross-sections to stain (dark brown) apoptotic cells. Right panels, magnified section of TUNEL assays shown in the center panels as indicated. Apoptotic cells with dominant staining are indicated by red arrows in the neuroepithelial lining of the developing midbrain. B, Western blotting. Embryos were homogenized, and aliquots were applied to Western blotting using antibodies recognizing the active forms of caspase 3 (p20/17), caspase 8 (p18), caspase 9 (p35), cyclin D1, and E2F1. β-actin was used as a control protein. C, immunostaining against caspase-3 (p20/17) and caspase-9 (p35). Immunoreactive material is indicated by dark brown staining in the entire neuroepithelial linings of the developing brain in control embryos only (a and c) but not in siRNA-treated embryos (b and d).
For more detailed information on the mechanisms of the developmental defects, we repeated the knockdown experiments and evaluated the impact of the siRNA constructs on the activation state of various caspases (Fig. 7, B and C). MAO-A is thought to modulate the intrinsic branch of the apoptotic pathway involving caspase 3 and caspase 9 (26). When we knocked down embryonic MAO-A expression, the activation states of caspase 3 and caspase 9 were reduced significantly, whereas activation of caspase 8 was not altered (Fig. 7, B and C). These data suggest that knockdown of MAO-A expression reduces apoptotic cell death by interfering with the intrinsic pathway of apoptosis.
Neural development relies on the fine balance between apoptotic cell death on one hand and cell proliferation and differentiation on the other (29). MAO-A has been shown to modulate cell proliferation by regulating intracellular levels of cyclin D1 and E2F1 (28). Cyclin D1 specifically associates with selected cyclin-dependent kinases, phosphorylating the retinoblastoma protein 1 (Rb1). This phosphorylation suppresses the growth inhibitory activity of hypophosphorylated Rb1 and thus induces cell proliferation by releasing E2F1 (30). We found that MAO-A knockdown was paralleled by a reduction in cellular cyclin D1 levels (Fig. 7B). In contrast, cellular levels of the transcriptional regulator E2F1 whose activity is associated with proliferation (31) remained unaffected by any treatment (Fig. 7B). Similar observations were made when MAO-A activity was inhibited using clorgyline or when MAO-A expression was suppressed by R1 overexpression.
DISCUSSION
In vertebrates MAO isoforms metabolize neurotransmitters and thus are important for the functionality of the adult brain. However, the role of MAO in murine embryonic development is less well understood. To fill this gap of knowledge, we employed an in vitro embryogenesis model (24). Mouse strains that exhibit incidental inactivation of the MAO-A gene are fertile and only show minor developmental alterations (13, 14), which appears to conflict with the robust changes we observed. However, our experimental setup avoids limitations of stem cell-based approaches, such as adaptive or compensational effects (23). Such effects may be responsible for the lack of developmental defects in MAO-A-deficient mouse strains (13, 14). Moreover, our experimental approach causes a sudden reduction of MAO-A expression at a certain developmental stage in embryos that have grown in the presence of MAO-A until expression knockdown was initiated. Knockout embryos, in contrast, might have activated compensatory mechanisms during early embryo development that are not available anymore at later developmental stages. In this respect, the two experimental models (knockout versus knockdown) are different and may lead to different experimental data. To make sure that our knockdown data are not the result of methodological artifacts, we combined genetic (siRNA-mediated MAO-A knockdown and R1 overexpression) and pharmacological (MAO-A inhibitor clorgyline) approaches and obtained identical results. In fact, recent observations of impaired cell proliferation of the telencephalon during late embryo development in an MAO-A/B-deficient mouse line supports our line of argumentation that MAO-A affects prenatal development (32).
Our studies indicate that both MAO-A and MAO-B are expressed at early stages when neuronal differentiation (gastrulation and neurulation) is induced and major subdivision of the brain (migration, specification, segmentation) takes place (33). MAO-A is a proapoptotic enzyme, and caspase 3 and caspase 9 of the intrinsic pathway of apoptosis have been related to MAO-A activity (26, 34). Moreover, MAO-A appears to modulate cell proliferation via controlling cytoplasmic cyclin D1 and E2F1 levels (28). The precise mechanisms of MAO-mediated regulation of cellular signaling cascades have not been explored in detail. A potential effector molecule is hydrogen peroxide, which is generated by MAO activity and affects both mitochondrial function and the cellular redox state (35, 36). On one hand, mitochondrial dysfunction is a feature of the intrinsic pathway of cell death signaling and is paralleled by increased lipid peroxidation of the mitochondrial membranes (37). On the other hand, changes of the cellular redox state affect a multitude of signaling cascades involved in the regulation of cellular proliferation and differentiation (38). However, more research is needed to understand the role of MAO-A derived hydrogen peroxide in the regulation of these signaling events.
Moreover, MAO-A has been implicated in cell cycle progression mediated by the oncogene c-myc (28). The myc protein acts as a transcription factor that modulates the expression of a variety of genes associated with cell cycle regulation, including E2F1 and cyclin D1 (39–41). In addition, c-myc regulates the expression as well as the activity of the transcription factor R1 (28). We observed similar developmental abnormalities in embryos overexpressing R1 compared with MAO-A knockdown embryos. Moreover, R1 overexpression similar to MAO-A knockdown or pharmacological inhibition of MAO-A activity coincides with elevated levels of the cyclin D1 protein. In conclusion, this data implicates MAO-A as an integral mediator of the c-myc proliferation signaling cascade via modulating cyclin D1 levels.
Developmental apoptosis is a well balanced process that is crucial for the formation of embryonic structures. Disturbance of this equilibrium induces morphological abnormalities, such as neural tube defects (29, 42). We found that MAO-A knockdown embryos and embryos exposed to clorgyline showed normal neural tube closure, which was followed by decreased apoptosis as well as impaired cell proliferation. Embryos of caspase 3-deficient mice die before birth and exhibit profoundly impaired brain development and cellular hyperplasia (43). In our MAO-A knockdown studies, caspase 3 activation was significantly impaired, which is in line with caspase 3 knockout data.
A hallmark of MAO-A deficiency in previously reported animal models are elevated levels of serotonin (13). We also observed elevated embryonic serotonin levels following MAO-A knockdown or following pharmacologic inhibition of MAO-A activity. These data underline the validity of the different experimental setups. Elevated 5-HT levels were also found in neural stem cells treated with Moclobemide, a reversible inhibitor of MAO-A activity. MAO-A inhibition resulted in an induction of the expression of the anti-apoptotic factor Bcl-2 in neural stem cells via ERK activation, and thereby inhibited apoptotic signaling and subsequently induced stem cell differentiation (34). Newborn mice lacking the cerebral vesicular monoamine transporter (VMAT2−/−) suffer from increased developmental apoptosis in the cerebral cortex (44). In these animals, caspase 3 and caspase 9 are hyperactive, and expression of antiapoptotic Bcl-XL was reduced. When these mice were crossed with MAO-A deficient animals (VMAT2−/−/MAO-A−/− double KO mice), 5-HT levels were elevated, and the increased cell death induced by VMAT2 deficiency was normalized (44). This finding is consistent with our data and also implicates MAO-A in proper brain development.
Although our data suggest MAO-A as an important player in the regulation of cerebral embryogenesis, more work is needed to identify the components of the associated signaling cascades. For instance, dysregulation of the finely tuned spatial and temporal balance between 5-HT production and breakdown may have fatal consequences for the developing embryo. Indeed, for several years it has been a matter of discussion of whether or not pharmacological intervention aimed at 5-HT homeostasis, such as selective serotonin reuptake inhibitors, during pregnancy has teratogenic effects (45, 46).
Supplementary Material
This work was supported in part by European Commission Grants FP6 and LSHM-CT-2004-0050333 (to H. K.), by Direct Grant 2008.1.052 and Special Equipment Grant for High-performance Microscopic Platform for Live Genetics in 2008 and for Micromanipulation System for Transgenics in 2004 and Li Ka Shing Institute of Health Sciences 6901988 (to C. C. W.), and by a Leverhulme Trust grant (to E. E. B. and C. U.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- MAO
- monoamine oxidase
- qPCR
- quantitative PCR
- E6.5
- gestational day 6.5.
REFERENCES
- 1. Shih J. C. (2007) Neurochem. Res. 32, 1757–1761 [DOI] [PubMed] [Google Scholar]
- 2. Scrutton N. S. (2004) Nat. Prod. Rep. 21, 722–730 [DOI] [PubMed] [Google Scholar]
- 3. Bach A. W., Lan N. C., Johnson D. L., Abell C. W., Bembenek M. E., Kwan S. W., Seeburg P. H., Shih J. C. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 4934–4938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Geha R. M., Rebrin I., Chen K., Shih J. C. (2001) J. Biol. Chem. 276, 9877–9882 [DOI] [PubMed] [Google Scholar]
- 5. Jahng J. W., Houpt T. A., Wessel T. C., Chen K., Shih J. C., Joh T. H. (1997) Synapse 25, 30–36 [DOI] [PubMed] [Google Scholar]
- 6. Billett E. E. (2004) Neurotoxicology 25, 139–148 [DOI] [PubMed] [Google Scholar]
- 7. Chen K. (2004) Neurotoxicology 25, 31–36 [DOI] [PubMed] [Google Scholar]
- 8. Chen K., Ou X. M., Chen G., Choi S. H., Shih J. C. (2005) J. Biol. Chem. 280, 11552–11559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Syagailo Y. V., Stöber G., Grässle M., Reimer E., Knapp M., Jungkunz G., Okladnova O., Meyer J., Lesch K. P. (2001) Am. J. Med. Genet 105, 168–171 [DOI] [PubMed] [Google Scholar]
- 10. Suarez-Merino B., Bye J., McDowall J., Ross M., Craig I. W. (2001) Hum. Mutat. 17, 523. [DOI] [PubMed] [Google Scholar]
- 11. Youdim M. B., Edmondson D., Tipton K. F. (2006) Nat. Rev. Neurosci. 7, 295–309 [DOI] [PubMed] [Google Scholar]
- 12. Grimsby J., Toth M., Chen K., Kumazawa T., Klaidman L., Adams J. D., Karoum F., Gal J., Shih J. C. (1997) Nat. Genet. 17, 206–210 [DOI] [PubMed] [Google Scholar]
- 13. Cases O., Seif I., Grimsby J., Gaspar P., Chen K., Pournin S., Müller U., Aguet M., Babinet C., Shih J. C. (1995) Science 268, 833–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scott A. L., Bortolato M., Chen K., Shih J. C. (2008) Neuroreport 19, 739–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen K., Holschneider D. P., Wu W., Rebrin I., Shih J. C. (2004) J. Biol. Chem. 279, 39645–39652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Luo X., Persico A. M., Lauder J. M. (2003) Dev. Neurosci. 25, 173–183 [DOI] [PubMed] [Google Scholar]
- 17. Vitalis T., Fouquet C., Alvarez C., Seif I., Price D., Gaspar P., Cases O. (2002) J. Comp. Neurol. 442, 331–347 [DOI] [PubMed] [Google Scholar]
- 18. Burnet H., Bevengut M., Chakri F., Bou-Flores C., Coulon P., Gaytan S., Pasaro R., Hilaire G. (2001) J. Neurosci. 21, 5212–5221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Upton A. L., Salichon N., Lebrand C., Ravary A., Blakely R., Seif I., Gaspar P. (1999) J. Neurosci. 19, 7007–7024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Popova N. K., Maslova L. N., Morosova E. A., Bulygina V. V., Seif I. (2006) Psychoneuroendocrinology 31, 179–186 [DOI] [PubMed] [Google Scholar]
- 21. Chen K., Cases O., Rebrin I., Wu W., Gallaher T. K., Seif I., Shih J. C. (2007) J. Biol. Chem. 282, 115–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Beglopoulos V., Shen J. (2004) Neuromolecular Med. 6, 13–30 [DOI] [PubMed] [Google Scholar]
- 23. Senechal Y., Larmet Y., Dev K. K. (2006) Neurodegener Dis. 3, 134–147 [DOI] [PubMed] [Google Scholar]
- 24. Ufer C., Wang C. C., Fähling M., Schiebel H., Thiele B. J., Billett E. E., Kuhn H., Borchert A. (2008) Genes Dev. 22, 1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Van Maele-Fabry G., Delhaise F., Picard J. J. (1990) Toxicol. In Vitro 4, 149–156 [DOI] [PubMed] [Google Scholar]
- 26. Fitzgerald J. C., Ufer C., De Girolamo L. A., Kuhn H., Billett E. E. (2007) J. Neurochem. 103, 2189–2199 [DOI] [PubMed] [Google Scholar]
- 27. Bortner C. D., Cidlowski J. A. (2002) Annu. Rev. Pharmacol. Toxicol. 42, 259–281 [DOI] [PubMed] [Google Scholar]
- 28. Ou X. M., Chen K., Shih J. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 10923–10928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Copp A. J. (2005) J. Anat. 207, 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Guo Y., Yang K., Harwalkar J., Nye J. M., Mason D. R., Garrett M. D., Hitomi M., Stacey D. W. (2005) Oncogene 24, 2599–2612 [DOI] [PubMed] [Google Scholar]
- 31. McClellan K. A., Slack R. S. (2007) Cell Cycle 6, 2917–2927 [DOI] [PubMed] [Google Scholar]
- 32. Cheng A., Scott A. L., Ladenheim B., Chen K., Ouyang X., Lathia J. D., Mughal M., Cadet J. L., Mattson M. P., Shih J. C. (2010) J. Neurosci. 30, 10752–10762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang C. C. (2005) Neuroembryol. Aging 3, 78–91 [Google Scholar]
- 34. Chiou S. H., Ku H. H., Tsai T. H., Lin H. L., Chen L. H., Chien C. S., Ho L. L., Lee C. H., Chang Y. L. (2006) Br. J. Pharmacol. 148, 587–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Naoi M., Maruyama W., Yi H., Inaba K., Akao Y., Shamoto-Nagai M. (2009) J. Neural Transm. 116, 1371–1381 [DOI] [PubMed] [Google Scholar]
- 36. Cohen G., Farooqui R., Kesler N. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 4890–4894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Imai H., Nakagawa Y. (2003) Free Radic. Biol. Med. 34, 145–169 [DOI] [PubMed] [Google Scholar]
- 38. Ufer C., Wang C. C., Borchert A., Heydeck D., Kuhn H. (2010) Antioxid. Redox Signal. [DOI] [PubMed] [Google Scholar]
- 39. Cole M. D., McMahon S. B. (1999) Oncogene 18, 2916–2924 [DOI] [PubMed] [Google Scholar]
- 40. Mateyak M. K., Obaya A. J., Sedivy J. M. (1999) Mol. Cell Biol. 19, 4672–4683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Santoni-Rugiu E., Jensen M. R., Thorgeirsson S. S. (1998) Cancer Res. 58, 123–134 [PubMed] [Google Scholar]
- 42. Cecconi F., Piacentini M., Fimia G. M. (2008) Cell Death Differ. 15, 1170–1177 [DOI] [PubMed] [Google Scholar]
- 43. Kuida K., Zheng T. S., Na S., Kuan C., Yang D., Karasuyama H., Rakic P., Flavell R. A. (1996) Nature 384, 368–372 [DOI] [PubMed] [Google Scholar]
- 44. Stankovski L., Alvarez C., Ouimet T., Vitalis T., El-Hachimi K. H., Price D., Deneris E., Gaspar P., Cases O. (2007) J. Neurosci. 27, 1315–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Alwan S., Friedman J. M. (2009) CNS Drugs 23, 493–509 [DOI] [PubMed] [Google Scholar]
- 46. Borue X., Chen J., Condron B. G. (2007) Int. J. Dev. Neurosci. 25, 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.