Abstract
NMDA receptors are calcium-permeable ionotropic receptors that detect coincident glutamate binding and membrane depolarization and are essential for many forms of synaptic plasticity in the mammalian brain. The obligatory GluN1 subunit of NMDA receptors is alternatively spliced at multiple sites, generating forms that vary in N-terminal N1 and C-terminal C1, C2, and C2′ cassettes. Based on expression of GluN1 constructs in heterologous cells and in wild type neurons, the prevalent view is that the C-terminal cassettes regulate synaptic accumulation and its modulation by homeostatic activity blockade and by protein kinase C (PKC). Here, we tested the role of GluN1 splicing in regulated synaptic accumulation of NMDA receptors by lentiviral expression of individual GluN1 splice variants in hippocampal neurons cultured from GluN1 (−/−) mice. High efficiency transduction of GluN1 at levels similar to endogenous was achieved. Under control conditions, the C2′ cassette mediated enhanced synaptic accumulation relative to the alternate C2 cassette, whereas the presence or absence of N1 or C1 had no effect. Surprisingly all GluN1 splice variants showed >2-fold increased synaptic accumulation with chronic blockade of NMDA receptor activity. Furthermore, in this neuronal rescue system, all GluN1 splice variants were equally rapidly dispersed upon activation of PKC. These results indicate that the major mechanisms mediating homeostatic synaptic accumulation and PKC dispersal of NMDA receptors occur independently of GluN1 splice isoform.
Keywords: Membrane Trafficking, Neurotransmitter Receptors, Protein Kinase C (PKC), RNA Splicing, Synapses, GluN1 (−/−), NMDA Receptor, Homeostatic Plasticity, Postsynaptic Density, Synaptic Targeting
Introduction
N-Methyl-d-aspartate (NMDA)3 type ionotropic glutamate receptors are present at excitatory synapses in the nervous system. These receptors play roles in neuronal development, learning and memory and neurological diseases (1–4). They require the simultaneous binding of glutamate and membrane depolarization for the channel to open, which makes them coincidence detectors of pre- and postsynaptic activity. Calcium flux through NMDA receptors triggers signal transduction cascades that are necessary for inducing many forms of synaptic plasticity. NMDA receptors are heteromeric cation channels composed of GluN1 and GluN2 subunits, and more rarely GluN3 subunits (5). The obligatory GluN1 subunit, which binds the co-agonist glycine, is encoded by a single gene which gives rise to eight GluN1 variants through alternative splicing of exons 5, 21, and 22 (6). The expression of GluN1 splice variants changes during development and across brain regions (7, 8), and can be regulated by neuronal activity (9, 10). Therefore, the composition of NMDA receptor signaling complexes may be dynamically regulated through the splicing of GluN1.
At the region encoding the N-terminal domain of GluN1, alternative splicing of exon 5 gives rise to splice variants containing or lacking the N1 cassette which regulates receptor deactivation kinetics as well as sensitivity to pH, Zn2+, and spermine (11–13). Alternative splicing of exons 21 and 22 generates four different splice variants at the intracellular C terminus of GluN1, varying in C1, C2, or C2′ cassettes, and regulates protein-protein interactions, early receptor trafficking, and GluN1 phosphorylation. The C1 cassette is involved in GluN1 interaction with yotiao, a scaffold protein that physically attaches PP1 and PKA to NMDA receptors to regulate channel activity (14), with calmodulin (15), neurofilament-L (16), and importin α (17).
Expression of GluN1 constructs in heterologous cells and in wild type neurons has been used to identify alternatively spliced C-terminal motifs important for cellular trafficking (for review, see Ref. 3). The C1 cassette contains a RXR-type endoplasmic reticulum (ER) retention/retrieval motif (18), which is suppressed by phosphorylation of specific serine residues within the C1 domain (19), or by the presence of an adjacent alternatively spliced C2′ domain (18, 19), or by association with GluN2 (20, 21). The C-terminal TVV sequence of C2′ is a type I PDZ-binding sequence, which interacts with all PSD-95 family proteins (18, 19), and was also found to bind components of the COPII coats required for formation of vesicles destined for the Golgi, and therefore to accelerate the forward trafficking of GluN1 from the ER (10).
The synaptic accumulation of NMDA receptors has been shown to be regulated by activity in a homeostatic manner (22, 23). Long-term NMDA receptor blockade increases synaptic accumulation of NMDA receptors via a mechanism requiring PKA, whereas increased synaptic activity promotes receptor dispersal (22, 24). Chronic activity blockade increases the expression of C2′-containing GluN1 whereas increasing neuronal activity causes an increase in the expression of C2-containing GluN1 subunits (10). Taken together, these results lead to the hypothesis that the higher ratio of C2′ to C2 in GluN1 mediates the enhanced synaptic accumulation of NMDA receptors by homeostatic activity blockade. A second form of regulated trafficking is rapid dispersal of NMDA receptors from synaptic to extrasynaptic sites by activation of PKC (25). Studies in heterologous cells and wild type neurons again suggest a hypothesis involving C-terminal splice variants. PKC phosphorylates three residues in the C1 cassette, serines 890, 896, and 897 (26, 27). Phosphorylation of Ser-890 induces dispersal of GluN1 clusters in non-neuronal cells (28), and phosphorylation of Ser-896 and Ser-897 increases surface expression of GluN1 (19). Thus, dispersal of GluN1 by PKC activation in neurons may be mediated by phosphorylation of the C1 cassette. However, the hypotheses that the C2′/C2 ratio controls homeostatic synaptic accumulation and that C1 controls PKC dispersal have not been tested in neurons expressing single splice forms of GluN1. More generally, mechanisms responsible either for the long-term activity-regulated synaptic targeting of NMDA receptors or for the rapid dispersal of synaptic NMDA receptors triggered by PKC activation are not well understood, and it is not clear whether these regulatory mechanisms are specific for receptors containing particular GluN1 splice variants. Here, to assay the contribution of each of the GluN1 splice variants to synaptic NMDA receptors in a physiological context, we used hippocampal cultures from GluN1(−/−) mice and a lentiviral expression system to re-introduce the individual GluN1 splice forms.
EXPERIMENTAL PROCEDURES
Genotyping
GluN1(+/−) mice were kindly provided by Drs. Michisuke Yuzaki and Tom Curran (29). Genotyping of pups and embryos was performed according to a previously described protocol (29). Briefly, DNA was extracted with phenol/choloroform/isoamyl alcohol after tissue digestion with proteinase K (0.1 mg/ml). PCR amplification was performed with specific primers for GluN1 and for the neomycin cassette. Because GluN1(−/−) mice die shortly after birth, heterozygous mice were mated to obtain GluN1(−/−) and littermate control GluN1(+/+) embryos, which were used to culture hippocampal or cortical neurons.
Cell Culture
Hippocampal and cortical neuronal cultures were prepared from 17–18 day embryonic mice. Briefly, hippocampi and cortex were dissected and maintained in Hibernate E (Brain Bits) supplemented with B27 (Invitrogen or Stemcell Technologies) at 4 °C overnight while genotyping was performed. Tissues from the same genotype were pooled together and dissociated with papain (20 units/ml, 15 min, 37 °C) and deoxyribonuclease I (0.20 mg/ml).
Hippocampal neurons were plated on coverslips coated with poly-l-lysine in 60 mm Petri dishes in minimal essential medium (MEM) supplemented with 10% horse serum at an approximate density of 300,000 cells/dish. Once the neurons attached to the substrate, coverslips were inverted on top of a glial cell monolayer and maintained in serum-free MEM with N2 supplements, B27, and insulin (20 μg/ml), essentially as previously described (30, 31). Cortical neurons were plated on 6-well plates coated with poly-l-lysine in MEM supplemented with 10% horse serum at an approximate density of 870,000 cells/well. Cortical neurons were maintained in Neurobasal medium supplemented with B27, glutamine (0.5 mm), and insulin (20 μg/ml) at 37 °C. Neurons were maintained in a humidified incubator of 5% CO2. Cultures were used after 14–15 days in vitro (DIV). When indicated, 2-amino-5-phosphonovalerate (APV) (100 μm) and (+)-5-methyl-10,11-dihydro-5H-dibenzo[a,d] cyclohepten-5,10-imine maleate (MK-801) (5 μm) were added directly to the culture medium from 6 DIV, every 3 days. As indicated, neurons were treated with 200 nm 12-O-tetradecanoylphorbol-13-acetate (TPA) in culture medium containing 100 μm APV in the 37 °C incubator for 45 min and then immediately fixed.
Lentivirus Constructs and Transduction
The cDNAs encoding the GluN1 splice variants were PCR amplified from previously described constructs (GluN1-1a (U08261) and GluN1-2a (X63255) were provided by Dr. Morgan Sheng, GluN1-3a (U08265), GluN1-4a (U08267) and GluN1-1b (U08263) were provided by Dr. Jim Boulter). Rat GluN1 cDNAs were used since preliminary experiments were performed in rat neurons and there is 100% identity at the amino acid level with mouse. GluN1 cDNAs were subcloned into a derivative of pLL3.7 (32) under the control of the synapsin I promoter (a generous gift of Dr. A. El-Husseini). Plasmids for second generation lentiviral packaging, p8.9 and pVSVg, were a kind gift from Dr. D. Trono (33). For virus packaging, HEK293FT cells were transfected with the two packaging plasmids and the pLL-syn-GluN1 splice variants constructs using Lipofectamine 2000. The cell medium was harvested 48 h after transfection, centrifuged, and the filtered supernatant was used for viral transduction of neurons at 3 DIV.
RNA Extraction and Real-time PCR
Total RNA from 15 DIV cultured cortical neurons was extracted with Trizol reagent (Invitrogen), following the manufacturer's specifications. The total amount of RNA was quantified by optical density measurements at 260 nm, and the quality evaluated based on the ratio of OD at 260 and 280 nm and on the 18 S and 28 S ribosomal RNA bands in agarose gel.
For first strand cDNA synthesis, 1 μg of total RNA and SuperScript III reverse transcriptase (Invitrogen) were used. For quantitative gene expression analysis, 20 μl reactions were prepared with 2 μl of 1:100 diluted cDNA, 10 μl of 2× SYBR Green Master Mix (Bio-Rad), specific primers at 250 nm, and TaqDNA polymerase. The GluN1-specific primers, which recognize all the GluN1 splice variants, were 5′-CGGCTCTTGGAAGATACAG-3′ and 5′-GAGTGAAGTGGTCGTTGG-3′, and the primers for tubulin were 5′-CATCCTCACCACCCACAC-3′ and 5′-GGAAGCAGTGATGGAAGAC-3′. The fluorescent signal was measured after each elongation step of the PCR reaction, in the iQ5 Multicolor Real-Time PCR Detection System (Bio-Rad), and was used to determine the threshold cycle (Ct), as previously described (34). Melting curves were performed in order to detect nonspecific amplification products, a non-template control was included in all assays, and for each set of primers a standard curve was performed to assess primer efficiency. Reactions were run in duplicate. The level of expression of each splice variant was determined relative to the level of expression of all GluN1 splice variants combined (control) amplified from RNA isolated from wild type neurons from the same culture. The differences between the threshold cycle (Ct) of one condition and the control were measured after correction for primer efficiency and normalized to tubulin, the internal control (Pfaffl method).
Differential Detergent Solubilization, SDS-PAGE and Immunoblotting
Protein extracts were prepared according to a modified version of a previously described protocol (35). Briefly, cortical neurons were washed twice with ice-cold PBS and once more with PBS supplemented with a mixture of protease inhibitors (0.1 mm PMSF, 1 μg/ml leupeptin, 1 μg/ml aprotitin). The cells were then scraped into lysis buffer (50 mm Tris-HCl, pH 7.4, 5 mm EGTA, 1 mm DTT) supplemented with the mixture of protease inhibitors. After centrifugation at 16,100 × g for 30 min at 4 °C, supernatant was discarded and the pellet was denaturated with sample loading buffer (Bio-Rad) at 95 °C for 5 min.
For the differential detergent solubilization experiments, the cellular pellet was solubilized in 40 μl of lysis buffer (same as above) with 1% Triton X-100 and incubated for 5 min at 37 °C. After centrifugation for 30 min at 16,100 × g at 4 °C the resultant supernatant was collected (Triton X-100 soluble fraction). The remaining pellet was resuspended in 40 μl of lysis buffer with 1% sodium deoxycholate (DOC), pH 9.0, and incubated for 15 min at 37 °C. Samples were centrifuge again at 16,100 × g for 30 min, at 4 °C, and the supernatant was collected (DOC soluble fraction). The remaining pellet was resuspended in lysis buffer plus 2% SDS and incubated for 5 min at 37 °C (SDS soluble fraction). All the collected samples were denaturated with 5× denaturating buffer (125 mm Tris, pH 6.8, 100 mm glycine, 10% SDS, 200 mm DTT, 40% glycerol, 3 mm sodium orthovanadate, and 0.01% bromphenol blue), at 95 °C for 5 min, and the total volume was loaded in the gel for SDS-PAGE and Western blot analysis.
The extracts obtained were resolved by SDS-PAGE in 8% polyacrylamide gels and immunoblotted as previously described (36). Membranes were blocked for 1 h at room temperature in Tris-buffered saline (137 mm NaCl, 20 mm Tris-HCl, pH 7.6) containing 0.1% (v/v) Tween 20 (TBS-T), and 5% (w/v) low-fat milk and probed overnight with the primary antibodies (anti-GluN1 1:500, Millipore) diluted in TBS-T containing 0.5% low-fat milk at 4 °C. Following five washes in TBS-T, membranes were incubated for 1 h with the alkaline phosphatase-conjugated secondary antibody (anti-mouse 1:20000, Amersham Biosciences) at room temperature. The membranes were then washed again, incubated with chemifluorescence substrate for 5 min, and scanned with the Storm 860 scanner (Amersham Biosciences). Where indicated, the membranes were stripped and re-probed with mouse anti-transferrin receptor antibody (1:1000, Invitrogen) or mouse anti-tubulin antibody (1:300000, Sigma) 1 h at room temperature.
Immunocytochemistry and Quantitation
Hippocampal neurons were fixed at 14–15 DIV in cold methanol for 10 min at −20 °C. The coverslips were incubated with 10% bovine serum albumin (BSA) in PBS for 1 h at 37 °C to block nonspecific staining and incubated overnight with the primary antibodies in 3% BSA at room temperature, with mild shaking. GluN1 splice variants were labeled with mouse anti-GluN1 antibody (1:1500, Invitrogen mAb 54.1), pre-synaptic sites were labeled with guinea pig anti-VGLUT1 antibody (1:4000, Millipore AB5905), post-synaptic sites were labeled with rabbit anti-SynGAP antibody (1:3000, Thermo PA1–046), and dendrites were stained with a chicken antibody against MAP2 (1:100, Abcam ab5392). Secondary antibodies (anti-mouse-Alexa 568, anti-rabbit-Alexa 488, anti-guinea pig-Alexa 647 (Molecular Probes) and anti-chicken-AMCA (Jackson ImmunoResearch)) were also prepared in 3% BSA and incubated at 37 °C for 1 h. The coverslips were mounted in elvanol (Tris-HCl, glycerol, and polyvinyl alcohol with 2% 1,4-diazabi-cyclo[2,2,2]octane).
Fluorescence images of neurons were obtained with a Zeiss Axioplan microscope with a 63×, 1.4 numerical aperture oil objective and a Photometrics Sensys cooled CCD camera, using MetaVue imaging software (Molecular Devices) with customized filter sets. Images in each channel were captured using the same exposure time across all fixed cells; images were acquired as gray scale from individual channels and pseudocolor overlays were prepared using Adobe Photoshop software. To quantitate the data from the immunocytochemistry, hippocampal neurons were chosen randomly for image acquisition and processed using Metamorph imaging software (10–15 cells each from three-four separate experiments for paired control and NMDAR blockade). For each neuron, two dendrites were chosen for analysis from the dendritic marker image, and their length was measured. To quantify GluN1 or SynGAP clusters per dendrite length, the digital images were subjected to a user-defined intensity threshold to select clusters and measured for cluster intensity, number, and area for the selected region. The number of synaptic clusters was determined as the number of clusters overlapping thresholded VGLUT dilated by 2 pixels. The ratio of GluN1-positive area to MAP2-positive area was measured with an absolute intensity threshold for GluN1. All imaging and analysis was done blind to rescue construct and treatment condition.
RESULTS
Rescue of GluN1 (−/−) Neurons by Lentiviral Expression of Individual Splice Variants
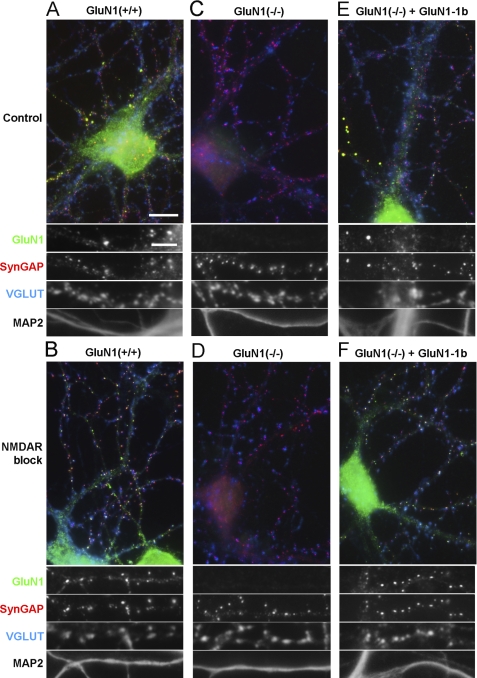
Hippocampal neurons were cultured at low density with glial feeder layers to optimize for GluN1 imaging (22, 31). Neurons from wild type mice grown for 14–15 DIV showed dendritic clusters of GluN1 (Fig. 1). The majority of clusters were synaptic, as they colocalized with the postsynaptic component SynGAP opposite terminals bearing the presynaptic vesicular glutamate transporter VGLUT. Some cells also exhibited a few bright clusters of GluN1 with SynGAP that were not apposed to VGLUT and thus non-synaptic (Fig. 1A), as described previously for GluN1, PSD-95, and GKAP (37). Such non-synaptic clusters were present in low numbers and appeared to be intracellular, thus we focused our study on the potentially functional synaptic clusters. As also previously described in rat neurons (22), synaptic clustering of GluN1 largely increased upon prolonged treatment with NMDA receptor antagonists APV and MK-801 (Fig. 1B). Hippocampal neurons cultured from GluN1(−/−) mice for 14–15 DIV presented no GluN1 clusters as expected (Fig. 1, C and D). Neurons from GluN1(−/−) mice showed robust staining for MAP2, a dendritic marker, with no apparent changes in dendritic morphology, and showed no apparent changes in clustering of the pre- and post-synaptic markers VGLUT and SynGAP compared with wild type neurons.
FIGURE 1.
Rescue of GluN1 synaptic clustering in GluN1(−/−) neurons transduced with lentivirus expressing GluN1-1b. Low density hippocampal cultures from wild type (A, B) or GluN1(−/−) (C–F) mice were fixed at 14–15 DIV and immunostained for GluN1, the excitatory postsynaptic component SynGAP, the excitatory presynaptic marker VGLUT, and the dendritic marker MAP2. As indicated, cultured neurons were treated with a mixture of the NMDAR blockers APV (100 μm) and MK801 (5 μm) from 6 DIV, to homeostatically increase synaptic clustering of NMDA receptors (B, D, F). GluN1(−/−) hippocampal neurons exhibited no immunoreactivity for GluN1 (C, D). GluN1(−/−) neurons transduced at 3 DIV with lentivirus expressing GluN1-1b under the control of the synapsin I promoter showed successful rescue of GluN1 immunofluorescence (E, F). No changes in SynGAP, VGLUT or MAP2 immunoreactivity were observed in the GluN1(−/−) transduced neurons compared with wild type neurons. Quadruple staining of hippocampal cultures from GluN1 wild type and GluN1(−/−) mice transduced with GluN1-1b showed in both cases a similar distribution of GluN1. In control conditions, GluN1-1b like wild type GluN1 formed a few bright non-synaptic clusters co-localizing with SynGAP but not VGLUT, shown enlarged in panels A and E, and a greater number of synaptic clusters co-localizing with SynGAP and VGLUT1 (shown enlarged in Fig. 3A for GluN1-1b). Long-term NMDAR blockade increased synaptic accumulation of rescued GluN1-1b (F) like that of wild type GluN1 (B). Scale bars: 10 μm for color overlays, 5 μm for single channel enlarged regions.
To study each NMDA receptor splice variant individually in a neuronal background, we optimized a lentiviral expression system to rescue GluN1 expression in hippocampal neurons cultured from GluN1(−/−) mice. At 3 DIV, GluN1(−/−) hippocampal neurons were transduced with lentivirus expressing individual GluN1 splice variants in the pLentiLox3.7 vector (32) under the control of the synapsin I promoter. We first tested the GluN1-1b splice variant, which contains both the C1 and the C2 cassettes at the C-terminal domain, and the N1 cassette at the N-terminal domain (Fig. 2A). At 2 weeks in culture, GluN1(−/−) hippocampal neurons transduced with GluN1-1b showed immunoreactivity for GluN1 similar to sister wild type neurons using an antibody against a constitutively expressed domain (Fig. 1). In addition to low diffuse levels in dendrites, there were many synaptic GluN1-1b clusters colocalizing with SynGAP apposed to VGLUT and a few bright non-synaptic clusters of GluN1-1b colocalizing with SynGAP but not apposed to VGLUT (Fig. 1E). The number of synaptic GluN1-1b clusters in GluN1(−/−) transduced neurons (13.6 ± 2.6 clusters/100 μm dendrite length) was close to the number of GluN1 clusters in wild type mouse hippocampal neurons (16.7 ± 2.9 clusters/100 μm dendrite length), and these GluN1 cluster densities are similar to what has been reported in low density rat hippocampal cultures (22). In cells chronically incubated with the NMDA receptor blockers APV and MK801, the expressed GluN1-1b subunit showed a dramatic increase in the number of bright clusters, similarly to wild type GluN1 (Fig. 1), with no visible changes in SynGAP, VGLUT or MAP2 staining. Moreover the GluN1 clusters in rescued GluN1(−/−) neurons chronically treated with APV and MK801, similarly to GluN1 wild type clusters, colocalized with pre- and post-synaptic markers along dendrites (Fig. 1F). Thus the developed lentiviral system successfully rescues GluN1 staining in GluN1(−/−) hippocampal neurons and the expressed GluN1 protein is integrated in receptors which are properly addressed to the synapse. Importantly, the expressed GluN1 subunit showed regulation of its synaptic localization by activity.
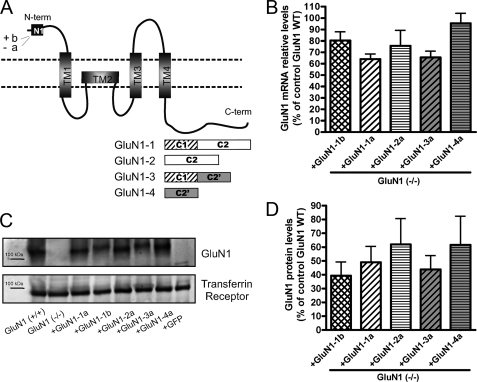
FIGURE 2.
The lentiviral expression system drives expression of all GluN1 splice variants at similar levels in GluN1(−/−) neurons. A, GluN1 subunit schematic representation indicating the alternatively spliced cassettes, both in the N and C terminus and the transmembrane domains. B–D, high density cortical cultures from GluN1(−/−) mice were transduced at 3 DIV with lentivirus expressing each of 5 different GluN1 splice variants (GluN1-1b, GluN1-1a, GluN1-2a, GluN1-3a, and GluN1-4a) under the control of the synapsin I promoter. B, relative mRNA levels for the GluN1 splice variants were assessed by quantitative real-time PCR. No significant differences (p > 0.05 ANOVA, n = 3 independent experiments) in the mRNA levels were found among GluN1 splice variants. C and D, total GluN1 protein levels were assessed by Western blot using an antibody against the GluN1 N-terminal region and normalized using the transferrin receptor as a loading control (D). No significant differences among the protein levels of all splice variants were found (D, p > 0.05 ANOVA, n = 6 independent experiments). In this system, GluN1 splice variants were expressed at about 50% of the expression level of total GluN1 in wild type cultures. Data are presented as mean ± S.E. Graph bar fills here and in Figs. 3 and S1 indicate cassette usage: cross-hatch contains N1 and C1, diagonal lines contain C1, horizontal lines lack C1, white background contain C2, and gray background contain C2′.
All GluN1 Splice Variants Are Expressed at Near Native GluN1 Levels in Transduced GluN1 (−/−) Neurons
The GluN1 gene undergoes alternative splicing to generate eight different splice variants, which differ in their N- and C-terminal domains (Fig. 2A; (38)). At the N-terminal domain of GluN1, two configurations (a or b) are generated by alternative splicing of exon 5, which encodes the N1 segment. The variability between splice variants is higher at the C-terminal domain, because of the alternative splicing of exons 21 and 22, which encode the C1 and C2 cassettes, respectively. GluN1-1 and GluN1-3 contain the C1 domain (GluN1-C1) while GluN1-2 and GluN1-4 lack this cassette (GluN1-ΔC1). The C2 cassette is present in GluN1-1 and GluN1-2 (GluN1-C2), while GluN1-3 and GluN1-4 lack this domain but express instead another cassette named C2′ (GluN1-C2′).
In the lentiviral expression system optimized to rescue expression of GluN1 individual splice variants in hippocampal cultures from GluN1(−/−) mice, we routinely achieved expression in >50% of transduced neurons. To compare the expression pattern of different GluN1 splice variants, we started by comparing their total expression levels. Due to limiting numbers of hippocampal neurons, for these assays we used high density cortical cultures. Evidence suggests that the high density cortical cultures regulate NMDA receptor trafficking in a similar manner as the low density hippocampal cultures (39). The mRNA levels of every splice variant individually expressed in GluN1(−/−) cortical neurons were quantified relative to the mRNA level of GluN1 in wild type neurons by quantitative real time PCR (Fig. 2B). All splice variants exhibited mRNA levels, which are 64 to 85.5% of the mRNA levels for total GluN1 splice variants combined expressed in wild type neurons. Furthermore, no significant differences in the mRNA levels between any of the different GluN1 splice variants were observed (Fig. 2B, ANOVA p > 0.05).
The total protein levels for the GluN1 splice variants expressed from the lentiviral system were analyzed in cortical extracts of GluN1(−/−) neurons transduced with the different splice variants. Western blot using an antibody against the GluN1 N-terminal region confirmed similar expression levels for every splice variant (Fig. 2C). Quantification of GluN1 levels normalized to the levels of the transferrin receptor confirmed no significant differences among the analyzed splice variants, which are expressed to ∼40–60% of the expression level of all GluN1 splice variants combined in wild type cortical neurons (Fig. 2D, ANOVA p > 0.05). Nor was there any significant difference in mRNA or protein level comparing GluN1 splice variants grouped according to cassette usage, GluN1-N1 with GluN1-ΔN1, GluN1-C1 with GluN1-ΔC1, or GluN1-C2′ with GluN1-C2 (t-tests p > 0.05). The developed lentiviral system, which avoids overexpression of the GluN1 splice variants, provides a suitable model for assessing the individual contribution of each splice variant to the synaptic localization of NMDA receptors in a neuronal background.
The C2 and C2′ Cassettes Regulate Synaptic Accumulation and Detergent Extraction of NMDA Receptors under Basal Conditions
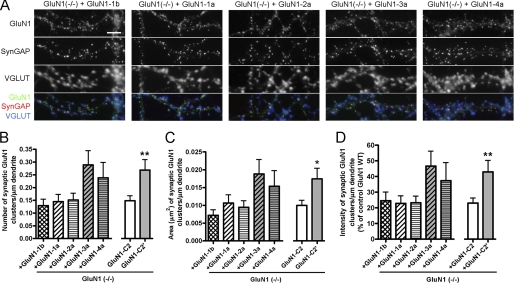
We next compared synaptic accumulation of GluN1 splice variants in GluN1(−/−) neurons following lentiviral-mediated expression of GluN1-1b, GluN1-1a, GluN1-2a, GluN1-3a, or GluN1-4a. All splice variants were able to rescue GluN1 immunoreactivity in transduced GluN1(−/−) hippocampal neurons, forming clusters colocalizing with SynGAP and apposed to VGLUT (Fig. 3A). The synaptically localized GluN1 clusters were selected based on their colocalization with VGLUT and quantified. There was a significant difference among GluN1 splice variants in number and area of synaptic GluN1 clusters (Fig. 3, B and C, ANOVA p < 0.05). To further assess the role of specific cassettes in synaptic accumulation, GluN1 splice variants were grouped according to cassette usage. The presence or absence of the N1 cassette (comparing GluN1-1b with GluN1-1a) or C1 cassette (comparing GluN1-1a + GluN1-3a with GluN1-2a + GluN1-4a) did not affect the number, area or integrated intensity of synaptic GluN1 clusters (t test p > 0.1). However, GluN1-3a and GluN1-4a containing the C2′ cassette had significantly higher number, area, and integrated intensity of synaptic GluN1 clusters, by 1.5-fold to 1.9-fold, compared with GluN1-1a and GluN1-2a containing the C2 cassette (Fig. 3, B–D, t test p < 0.05 and p < 0.01). Thus, consistent with data from heterologous cells and expression in wild type neurons (10, 18, 19), GluN1 C2′ mediates higher basal synaptic accumulation than GluN1 C2 upon rescue in GluN1(−/−) neurons.
FIGURE 3.
The GluN1 C2 and C2′ cassettes regulate synaptic accumulation of NMDA receptors under basal conditions. A, low density hippocampal cultures generated from GluN1(−/−) mice were transduced at 3 DIV with lentiviral vectors encoding the GluN1--1b, -1a, -2a, -3a, and -4a splice variants. Neurons were fixed at 14–15 DIV and immunostained for the GluN1 subunit, SynGAP and VGLUT (A) and for MAP2 (not shown). All splice variants show some co-clustering at synapses with SynGAP and VGLUT, although synaptic clustering of GluN1-3a and GluN1-4a appears highest. Scale bar, 5 μm. B–D, quantification of the GluN1 signal in randomly selected cells transduced with GluN1 splice variants. The number (B), total area (C), and integrated intensity (D) of synaptic (opposed to VGLUT) GluN1 clusters per dendritic length were analyzed. Significant differences among GluN1 splice variants were found, p < 0.05 ANOVA for number and area, p = 0.06 ANOVA for intensity, n≥28 neurons from 3–4 independent experiments. To determine the role of specific splice cassettes, data were then analyzed by grouping GluN1 variants according to cassette usage. Only groups showing a significant difference are graphed, specifically GluN1-C2′ (GluN1-3a + GluN1-4a) compared with GluN1-C2 (GluN1-1a + GluN1-2a) (*, p < 0.05; **, p < 0.01 t test).
To further assess differences in behavior of GluN1 splice variants by an independent method, we used differential detergent extraction. Differential detergent extraction is part of the larger procedure to prepare postsynaptic density fractions, and alone is feasible with the limiting amount of material generated by the molecular rescue approach. A difference among splice variants in the fraction of GluN1 extracted by a specific detergent would support a difference in molecular associations. Cell lysates of GluN1(−/−) cortical neuron cultures transduced with GluN1-1b, GluN1-1a, GluN1-2a, GluN1-3a, or GluN1-4a were sequentially extracted with buffer containing non-ionic detergent 1% Triton X-100, then ionic detergent 1% DOC, and finally with 2% SDS to solubilize remaining GluN1 (supplemental Fig. S1). Western blot analysis using an antibody against a common N-terminal region of GluN1 showed no significant difference among the five GluN1 splice variants in solubility in Triton X-100 (supplemental Fig. S1, A and B, p > 0.05, ANOVA), a fraction thought to represent mainly GluN1 in the endopasmic reticulum not associated with GluN2 (40). However, there was a significant difference among splice variants in solubility in 1% DOC (supplemental Fig. S1, A and C, ANOVA p < 0.005). Specifically, the C2′-containing splice variants showed higher solubility in DOC than splice variants containing the C2 cassette (supplemental Fig. S1C; t test p < 0.001). The C2′ variants also showed lower subsequent solubility in 2% SDS than C2 variants (supplemental Fig. S1D; t test p < 0.005), presumably because of the higher solubility in DOC in the sequential assay. The presence or absence of the N1 or C1 cassettes did not affect detergent solubility (t-tests p > 0.1). These experiments support a difference in molecular associations of GluN1 C2′ variants compared with C2 variants.
All GluN1 Splice Variants Show Homeostatic Regulation of Synaptic Targeting upon Rescue in GluN1(−/−) Neurons
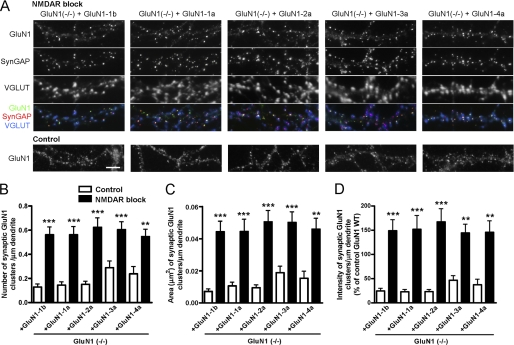
A hypothesis based on previous studies (10) is that alternative splicing increasing incorporation of C2′ and reducing incorporation of C2 in GluN1 mediates enhanced synaptic accumulation upon prolonged NMDA receptor blockade. Indeed, our results above showing increased synaptic incorporation of GluN1-C2′ relative to GluN1-C2 in part supports this idea. Another prediction of this hypothesis is that NMDA receptor blockade would have no effect on synaptic accumulation of individual GluN1 splice variants expressed in a GluN1(−/−) background. We tested this prediction by incubating GluN1(−/−) hippocampal cultures transduced with GluN1-1b, GluN1-1a, GluN1-2a, GluN1-3a, or GluN1-4a splice variants with NMDA receptor antagonists APV and MK-801 from 6 DIV until analysis at 14–15 DIV. However, hippocampal neurons expressing each individual splice variant showed a strong homeostatic increase in GluN1 clustering (Fig. 4A). Quantification of the number, area, and integrated intensity of synaptic GluN1 clusters showed a significant increase upon NMDA receptor blockade for all splice variants (Fig. 4, B–D, ANOVA p < 0.0001 and pairwise post-hoc Bonferroni p < 0.01). Synaptic clustering of all splice variants increased by more than 2-fold upon prolonged NMDA receptor blockade. Comparison among GluN1-1b, GluN1-1a, GluN1-2a, GluN1-3a, or GluN1-4a just under conditions of NMDA receptor blockade revealed no significant difference in synaptic accumulation of splice variants (ANOVA p > 0.1 for number, area, and integrated intensity). This evidence taken together indicates that the different GluN1 splice domains do not contribute major differences to the homeostatic regulation of NMDA receptor synaptic targeting upon chronic blockade of NMDA receptors.
FIGURE 4.
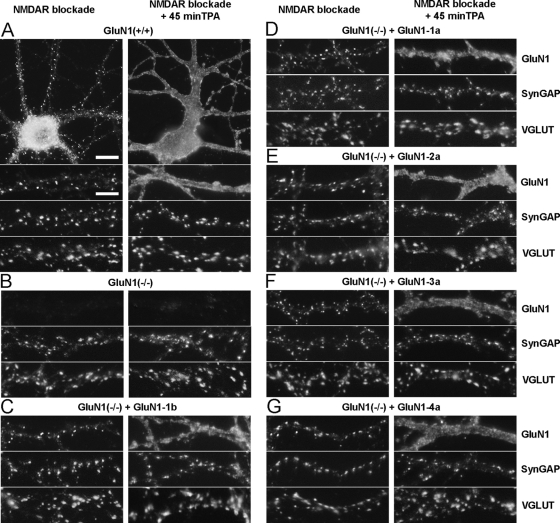
NMDA receptor blockade increases synaptic clusters of all GluN1 splice variants. A, hippocampal cultures from GluN1(−/−) mice were transduced at 3 DIV with lentiviral vectors encoding each GluN1 splice variant. Where indicated, cultures were treated with NMDAR antagonists APV (100 μm) and MK801 (5 μm) from 6 DIV. Neurons were fixed at 14–15 DIV and immunostained for GluN1, SynGAP, VGLUT (co-localization appears white in the triple overlay) and MAP2 (not shown). After the prolonged treatment with NMDAR antagonists, all splice variants show increased synaptic clustering of GluN1. The distribution of GluN1 splice variants under control culture conditions is reproduced from Fig. 3A for comparison. Scale bar, 5 μm. B–D, GluN1(−/−) neurons transduced with GluN1 splice variants were selected randomly and analyzed for number (B), area (C), and integrated intensity (D) of GluN1 synaptic clusters per dendritic length, identified by their apposition to VGLUT staining. All measures showed an increase in synaptic GluN1 for all splice variants under conditions of NMDA receptor blockade (black bars) compared with control (white bars), p < 0.0001 ANOVA, ***, p < 0.001, **, p < 0.01 by pairwise post-hoc Bonferroni's test.
All GluN1 Splice Variants Show Rapid PKC-induced Dispersal upon Rescue in GluN1(−/−) Neurons
Activation of PKC induces rapid dispersal of NMDA receptors from synaptic to extrasynaptic sites (25). Phosphorylation of a residue in the C1 cassette mediates similar NMDA receptor cluster dispersal in non-neuronal cells (28). Using the same strategy as described above, we studied the individual role of the C1 and other splice cassettes in the dispersal of synaptic NMDA receptors by PKC in neurons (Fig. 5). Low density hippocampal cultures from GluN1(−/−) mice were transduced at 3 DIV with lentivirus encoding GluN1-1b, GluN1-1a, GluN1-2a, GluN1-3a, or GluN1-4a and cultured in the presence of NMDAR antagonists to maximize synaptic clustering of the expressed NMDA receptor subunits. PKC was activated with 200 nm TPA; as shown previously, the effects of TPA on NMDA receptor distribution were blocked by bisindolylmaleimide an inhibitor of PKC and did not occur with 4α-TPA which does not activate PKC (25). As before with rat hippocampal neurons, we saw a dispersal of GluN1 from synaptic clusters to a nearly uniform plasma membrane distribution within 45 min of TPA treatment in wild type mouse hippocampal neurons (Fig. 5A). The distribution of SynGAP, VGLUT, and MAP2 was not affected by the TPA treatment. Surprisingly, TPA induced a similar rapid dispersal of all rescued GluN1 splice variants from synaptic clusters to extrasynaptic plasma membrane (Fig. 5, C–G). GluN1-2a and GluN1-4a, which lack the C1 cassette bearing the known PKC phosphorylation sites showed a similar redistribution as GluN1-1a, GluN1-3a, and GluN1-1b, which contain the C1 cassette (Fig. 5, E and G versus Fig. 5, C, D, F).
FIGURE 5.
All GluN1 splice variants show similar rapid redistribution to extrasynaptic plasma membrane upon activation of protein kinase C. Cultures from the hippocampus of wild type (A) or GluN1(−/−) (B–G) mice were fixed at 14–15 DIV and immunostained for GluN1, SynGAP, VGLUT (A–G), and MAP2 (not shown). GluN1(−/−) neurons were transduced at 3 DIV with lentiviral vectors encoding GluN1-1b (C), GluN1-1a (D), GluN1-2a (E), GluN1-3a (F), and GluN1-4a (G). Cultures were treated with NMDAR antagonists from 6 DIV to maximize synaptic clustering of NMDA receptors, and then treated as indicated for 45 min with TPA to activate PKC. All splice variants show a similar basal synaptic clustering of NMDA receptors, as assessed by the GluN1 clusters co-localized with SynGAP and apposed to VGLUT. Activation of PKC induced a rapid dispersal of all GluN1 splice variants to fill the extrasynaptic dendritic membrane, with no change in distribution of SynGAP, VGLUT (A–G), or MAP2 (not shown). Scale bars: 10 μm for whole cell images in A, 5 μm for dendrite regions.
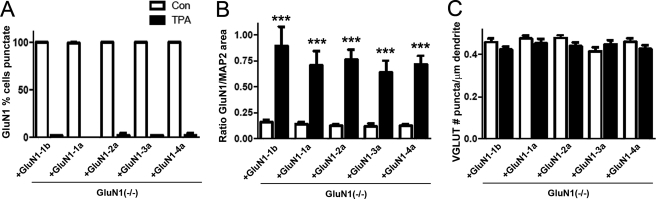
To quantify the effect of PKC activation on GluN1 splice variants, we first classified transduced GluN1-expressing cells as bearing or lacking GluN1 clusters. While essentially all transduced neurons had GluN1 clusters under the baseline NMDA receptor blockade conditions, TPA reduced the percent of neurons with any clusters of GluN1 to <5% for all GluN1 splice variants (Fig. 6A). Thus, synaptic clusters as measured in the previous analyses were essentially eliminated by PKC activation. As an objective measure to compare the extent of GluN1 redistribution among splice variants, we chose an absolute intensity and measured the fraction of dendrite area with GluN1 at or above this intensity. The baseline values for this measure, mainly representing synaptic NMDA receptor clusters, did not differ significantly among splice variants (white bars in Fig. 6B), consistent with the previous figures under activity blockade conditions. Importantly, the fraction of dendrite area with GluN1 above this set intensity increased >5-fold for all splice variants (ANOVA p < 0.0001 and post-hoc Bonferroni p < 0.001, Fig. 6B). Nor was there any significant difference in fraction of dendrite area above threshold for GluN1 comparing GluN1 splice variants grouped according to cassette usage, GluN1-N1 with GluN1-ΔN1, GluN1-C1 with GluN1-ΔC1, or GluN1-C2′ with GluN1-C2 (t-tests p > 0.05). Moreover, the fraction of dendrite area with GluN1 above threshold following PKC activation did not differ among GluN1-1b, GluN1-1a, GluN1-2a, GluN1-3a, or GluN1-4a (black bars in Fig. 6B, ANOVA p > 0.1). As a control, VGLUT clusters measured in the same cells showed no change with TPA treatment (Fig. 6C, ANOVA p > 0.1). Thus, PKC induces a robust and equal dispersal of NMDA receptors irrespective of GluN1 splice isoform composition and independently from phosphorylation sites in the C1 cassette.
FIGURE 6.
Activation of protein kinase C induces rapid dispersal of clusters of all splice variants of GluN1. GluN1(−/−) neurons transduced with GluN1 splice variants were selected randomly and analyzed in conditions of NMDAR blockade with (black bars) or without (white bars) 45 min treatment with TPA to activate PKC, as in Fig. 5. In blind classification, TPA treatment reduced the percentage of neurons bearing clusters of GluN1 for all splice variants (A; 200–400 cells each, statistics not applicable). Because there were few or no GluN1 clusters to measure following TPA treatment, the area of GluN1 immunoreactivity above a set intensity threshold was measured per MAP2-positive dendrite area (B), ANOVA p < 0.0001, ***, p < 0.001 by pairwise post-hoc Bonferroni's test. The same cells were analyzed for number of VGLUT clusters per dendritic length (C), ANOVA p > 0.1.
DISCUSSION
In this work we developed a lentiviral system for rescuing GluN1 clustering at synapses in GluN1(−/−) hippocampal neurons in culture. Using this system, we studied individually the role of five GluN1 splice variants (GluN1-1b, GluN1-1a, GluN1-2a, GluN1-3a, or GluN1-4a) in the synaptic expression of NMDA receptors, in the homeostatic regulation of their synaptic clustering, and in their rapid dispersal by activation of PKC. The lentiviral delivery system used transduced >50% of the neurons in the culture, and the protein levels obtained for each GluN1 splice variant were 40–60% of the protein levels for GluN1 in wild-type neurons (Fig. 2). Therefore, little if any overexpression of the different GluN1 splice variants occurs in this system, which also avoids artifacts that can arise from using GluN1 modified constructs in a wild-type background, or working with heterologous systems.
We demonstrated that all splice variants are able to rescue GluN1 clustering at synapses in GluN1(−/−) hippocampal neurons (Figs. 1 and 3). Synaptic clustering under basal conditions was significantly higher for GluN1-C2′ variants compared with GluN1-C2, consistent with differential detergent extraction specifically of these variants (supplemental Fig. S1). However, contrary to expectations based on previous studies in non-neuronal cells and in wild type neurons, all GluN1 splice variants showed significantly enhanced synaptic accumulation upon prolonged activity blockade (Fig. 4) and rapid dispersal upon activation of PKC (Figs. 5 and 6). Thus, the major mechanisms mediating homeostatic activity and PKC regulation of synaptic NMDA receptor accumulation occur independently of GluN1 splice variant.
The prevailing view is that the N1 domain at the N-terminal region of the GluN1 subunit influences receptor sensitivity to modulators as well as its desensitizing properties. Accordingly, we observed no major differences in the synaptic targeting or detergent extraction of NMDA receptors containing either the GluN1-1b or the GluN1-1a splice variants (Figs. 1–3, supplemental Fig. S1). We also demonstrate that the N1 cassette does not contribute to the homeostatic or PKC regulation of NMDA receptor distribution (Figs. 4–6).
GluN1-1 subunits are retained in the ER when expressed individually in heterologous systems (21), because of the presence of the ER retention signal in the C1 cassette (18, 19). Upon receptor assembly in the ER, the GluN1 retention signal is masked by GluN1 oligomerization with the GluN2 subunits, which promotes forward traffic of the heteromeric receptors to the cell surface (20). Accordingly, once assembled with GluN2, GluN1 subunits are delivered to the cell surface both in heterologous cells and in neurons (21, 41) and are targeted to synapses (42). This mechanism ensures that only correctly assembled NMDA receptors pass the ER quality control and reach the cell surface. Shorter GluN1 splice variants, lacking the C1 domain or containing the C2′ cassette, are less retained in the ER when expressed alone in heterologous systems (21). However, in our GluN1(−/−) cultured hippocampal neurons, GluN1 clustering at synaptic sites was similar for neurons expressing the C1-containing or C1-lacking splice variants of GluN1 (Fig. 3). These results indicate that in this neuronal system where GluN2 subunits are available, the GluN1 splice variants, independently of the presence of the C1 cassette, were assembled into heteromeric complexes that could efficiently traffic to the cell surface and to the synapse. In these experiments, the level of expressed GluN1 individual splice variant per GluN1(−/−) neuron is estimated to be a little lower than the combined level of all GluN1 splice variants in wild type neurons, based on comparing immunofluorescence intensity (e.g. Fig. 1B versus Fig. 1F) and based on total Western blot signal (Fig. 2, C and D) relative to transduction efficiency. Thus, in this rescue system, GluN2 levels may be sufficient to form heteromers with much of the expressed GluN1, and we propose that under such conditions the C1 cassette may not limit forward trafficking to control steady state levels of synaptic NMDA receptors. On the other hand, in neurons where GluN1 is in greater excess relative to GluN2 (43) or in response to stimuli where rapid forward trafficking is required, a role of the C1 cassette in limiting forward trafficking of GluN1 may be more apparent.
The major identified PKC phosphorylation sites in NMDA receptor subunits are in the C1 cassette, and phosphorylation at Ser-890 in the C1 cassette disperses recombinant GluN1 clusters in fibroblasts (27, 28). However, we show here that the C1 cassette plays no role in the dispersal of NMDA receptors in neurons upon activation of PKC; this form of regulated trafficking is completely independent of GluN1 splice variant (Figs. 5 and 6). Thus PKC must be acting on GluN2 subunits or accessory trafficking proteins. GluN2A and GluN2B can be phosphorylated by PKC (44) but the sites and functional consequences are not yet known; phosphorylation in the PDZ-binding domain of GluN2B that regulates its trafficking is controlled by casein kinase 2 but not by PKC (45, 46). PKC induces plasma membrane insertion of NMDA receptors in heterologous cells and extrasynaptically in neurons (47), a process that likely contributes to the redistribution we observed. Importantly, it was recently found that PKC promotion of NMDA receptor cell surface delivery occurs via PKC phosphorylation of the SNARE protein SNAP-25 (48). This enhanced insertion of NMDA receptors into extrasynaptic plasma membrane via PKC phosphorylation of SNAP-25 at Ser-187 is likely to apply to all GluN1 splice variants. It is possible that this recently discovered mechanism (48), in addition to directly increasing extrasynaptic levels of all GluN1 variants, may contribute to dispersal of synaptic NMDA receptors, for example by recruiting receptor-associated proteins away from the synapse, proteins that might normally function to stabilize synaptic receptors.
Despite the fact that the presence of the C1 cassette did not significantly influence regulated synaptic accumulation of NMDA receptors, the macromolecular complexes and thus signaling by NMDA receptors may be influenced by the presence of the C1 cassette. Indeed, a rescue approach in young cultured GluN1(−/−) neurons comparing NR1-1a and NR1-2a showed that the presence of the C1 cassette increased CREB phosphorylation and CRE-dependent reporter gene expression (49).
The C2′ cassette contains the C-terminal -STVV signal, which is a binding site for PDZ-domain scaffold proteins such as PSD-95, SAP102, PSD-93, SAP97 (18) and for the coatomer COPII (10), which coats export vesicles from the ER destined for the Golgi. Interactions of C2′ both with COPII and with PDZ domain proteins might be expected to increase synaptic accumulation of NMDA receptors, and indeed we observed significantly higher number, area, and integrated intensity of synaptic GluN1-C2′ compared with GluN1-C2 (Fig. 3). Furthermore, detergent extraction showed a significant difference for GluN1-C2′ compared with GluN1-C2 (supplemental Fig. S1). Naively, one might expect the greater synaptic accumulation of C2′ compared with C2 containing GluN1 splice variants observed by imaging to result in lower solubility in DOC, rather than greater solubility in DOC as found here. However, non-synaptic GluN1 is partially sequestered in extremely dense extrasynaptic clusters in poorly characterized organelles in hippocampal dendrites, as shown in Fig. 1, A and E. Immunoreactivity for such extrasynaptic clusters was abolished in GluN1(−/−) neurons, indicating these are true GluN1 pools, and GluN1 associated with dendritic vesicular organelles is also observed in vivo (50). According to our previous measures, nearly half of the extrasynaptic GluN1 is sequestered in these dense non-synaptic clusters in control low density hippocampal cultures (22). We suggest that GluN1 in these extrasynaptic dendritic clusters may be resistant to extraction with 1% DOC, in contrast to synaptic pools which are partially solubilized by 1% DOC (40).
The prevalent idea, proposed by Mu et al. (10), is that homeostatic accumulation of NMDA receptors occurs via regulation of GluN1 splicing at exon 22. Chronic activity blockade promotes the generation of C2′ domain variants proposed to accumulate strongly at synapses, and chronic activity promotes generation of C2 variants proposed to exhibit lower synaptic accumulation. Indeed, the proposed difference in synaptic accumulation of these variants was confirmed here under basal conditions (Fig. 3). However, both GluN1-C2 and GluN1-C2′ showed significantly enhanced synaptic targeting upon prolonged NMDA receptor blockade, with >2-fold increase in number, area, and integrated intensity of synaptic clusters (Fig. 4). Thus, clearly the C2′/C2 ratio is not the primary mechanism mediating the homeostatic blockade-induced increase in synaptic accumulation of NMDA receptors. Indeed, a mechanism independent of new protein synthesis (and thus independent of altering C2′/C2 ratio) was suggested previously (24). This previous study (24) also found that the effects of activity blockade on NMDA receptor synaptic accumulation required cAMP-dependent protein kinase (PKA), and that activation of PKA alone increased synaptic accumulation of NMDA receptors, mimicking the effects of activity blockade. The key target of PKA that mediates this effect is still not identified, it could be GluN1 (in a region common to all splice variants, not at the PKA site in C1 based on the results shown here), GluN2A, or GluN2B, all of which can be phosphorylated by PKA and show some basal phosphorylation in hippocampal tissue (27, 44), or an NMDA receptor interacting protein. The blockade-induced increase in synaptic NMDA receptor is accompanied by increased surface association of GluN1 (24) and loss of the dense non-synaptic aggregates in dendrites (22), suggesting a change in exocytic trafficking or recycling.
All the splice cassettes in the GluN1 subunit are regionally and developmentally regulated (7) and all are found in cortical and cerebellar PSD fractions (51). As discussed above, specific binding partners for the splice cassettes in GluN1 have been described which impart specific characteristics to the NMDA receptor signaling complex and may regulate functional properties. A preference for pairing GluN2 subunits with different GluN1 splice variants has been suggested (52). Although biochemical studies from whole forebrain do not support preferential GluN1/GluN2 co-assembly (40), immunogold localization in retina suggests that preferential association and localization can occur in some neurons. At bipolar to ganglion cell synapses in the rat retina, C2-containing GluN1 together with GluN2B localize perisynaptically, whereas C2′-containing GluN1 together with GluN2A are more abundant at the PSD (53). The lentiviral delivery system that we developed for expressing GluN1 splice variants in a GluN1-null neuronal background may be useful for further testing selective association with GluN2 and GluN3 subunits. More generally, this approach may be useful for analyzing the composition and physiological roles of macromolecular signaling complexes brought together by receptors formed by individual splice variants. Our rescue approach showing that all GluN1 splice variants undergo similar homeostatic and PKC regulation of trafficking indicates the importance of assessing GluN1 trafficking and function in a neuronal context with physiological stoichiometry of interacting partners.
Supplementary Material
Acknowledgment
We thank Xiling Zhou and Nazarine Fernandes for excellent technical assistance.
This work was supported by the Canadian Institutes of Health Research (CIHR) MOP-69096 (to A. M. C.), by the Portuguese Foundation for Science and Technology (FCT) PTDC/BIA-BCM/71789/2006 and PTDC/SAU-NEU/099440/2008 (to A. L. C.), and by salary awards CIHR Frederick Banting and Charles Best Canada Graduate Scholarship (to K. S.), Canada Research Chair and Michael Smith Foundation for Health Research Senior Scholarship (to A. M. C.), and FCT Graduate Scholarships (SFRH/BD/37522/2007, to J. S. F. and SFRH/BD/47879/2008, to L. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- NMDA
- N-methyl-d-aspartate
- DOC
- deoxycholate
- TPA
- 12-O-tetradecanoylphorbol-13-acetate
- APV
- 2-amino-5-phosphonovalerate
- DIV
- days in vitro.
REFERENCES
- 1. Cull-Candy S., Brickley S., Farrant M. (2001) Curr. Opin. Neurobiol. 11, 327–335 [DOI] [PubMed] [Google Scholar]
- 2. Wenthold R. J., Prybylowski K., Standley S., Sans N., Petralia R. S. (2003) Annu. Rev. Pharmacol. Toxicol. 43, 335–358 [DOI] [PubMed] [Google Scholar]
- 3. Lau C. G., Zukin R. S. (2007) Nat. Rev. Neurosci. 8, 413–426 [DOI] [PubMed] [Google Scholar]
- 4. Stephenson F. A., Cousins S. L., Kenny A. V. (2008) Mol. Membr. Biol. 25, 311–320 [DOI] [PubMed] [Google Scholar]
- 5. McBain C. J., Mayer M. L. (1994) Physiol. Rev. 74, 723–760 [DOI] [PubMed] [Google Scholar]
- 6. Moriyoshi K., Masu M., Ishii T., Shigemoto R., Mizuno N., Nakanishi S. (1991) Nature 354, 31–37 [DOI] [PubMed] [Google Scholar]
- 7. Laurie D. J., Seeburg P. H. (1994) J. Neurosci. 14, 3180–3194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Monyer H., Burnashev N., Laurie D. J., Sakmann B., Seeburg P. H. (1994) Neuron 12, 529–540 [DOI] [PubMed] [Google Scholar]
- 9. Xie J., Black D. L. (2001) Nature 410, 936–939 [DOI] [PubMed] [Google Scholar]
- 10. Mu Y., Otsuka T., Horton A. C., Scott D. B., Ehlers M. D. (2003) Neuron 40, 581–594 [DOI] [PubMed] [Google Scholar]
- 11. Durand G. M., Gregor P., Zheng X., Bennett M. V., Uhl G. R., Zukin R. S. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 9359–9363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Traynelis S. F., Hartley M., Heinemann S. F. (1995) Science 268, 873–876 [DOI] [PubMed] [Google Scholar]
- 13. Traynelis S. F., Burgess M. F., Zheng F., Lyuboslavsky P., Powers J. L. (1998) J. Neurosci. 18, 6163–6175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Westphal R. S., Tavalin S. J., Lin J. W., Alto N. M., Fraser I. D., Langeberg L. K., Sheng M., Scott J. D. (1999) Science 285, 93–96 [DOI] [PubMed] [Google Scholar]
- 15. Ehlers M. D., Zhang S., Bernhadt J. P., Huganir R. L. (1996) Cell 84, 745–755 [DOI] [PubMed] [Google Scholar]
- 16. Ehlers M. D., Fung E. T., O'Brien R. J., Huganir R. L. (1998) J. Neurosci. 18, 720–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jeffrey R. A., Ch'ng T. H., O'Dell T. J., Martin K. C. (2009) J. Neurosci. 29, 15613–15620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Standley S., Roche K. W., McCallum J., Sans N., Wenthold R. J. (2000) Neuron 28, 887–898 [DOI] [PubMed] [Google Scholar]
- 19. Scott D. B., Blanpied T. A., Swanson G. T., Zhang C., Ehlers M. D. (2001) J. Neurosci. 21, 3063–3072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hawkins L. M., Prybylowski K., Chang K., Moussan C., Stephenson F. A., Wenthold R. J. (2004) J. Biol. Chem. 279, 28903–28910 [DOI] [PubMed] [Google Scholar]
- 21. Okabe S., Miwa A., Okado H. (1999) J. Neurosci. 19, 7781–7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Rao A., Craig A. M. (1997) Neuron 19, 801–812 [DOI] [PubMed] [Google Scholar]
- 23. Pérez-Otaño I., Ehlers M. D. (2005) Trends Neurosci. 28, 229–238 [DOI] [PubMed] [Google Scholar]
- 24. Crump F. T., Dillman K. S., Craig A. M. (2001) J. Neurosci. 21, 5079–5088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fong D. K., Rao A., Crump F. T., Craig A. M. (2002) J. Neurosci. 22, 2153–2164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tingley W. G., Roche K. W., Thompson A. K., Huganir R. L. (1993) Nature 364, 70–73 [DOI] [PubMed] [Google Scholar]
- 27. Tingley W. G., Ehlers M. D., Kameyama K., Doherty C., Ptak J. B., Riley C. T., Huganir R. L. (1997) J. Biol. Chem. 272, 5157–5166 [DOI] [PubMed] [Google Scholar]
- 28. Ehlers M. D., Tingley W. G., Huganir R. L. (1995) Science 269, 1734–1737 [DOI] [PubMed] [Google Scholar]
- 29. Forrest D., Yuzaki M., Soares H. D., Ng L., Luk D. C., Sheng M., Stewart C. L., Morgan J. I., Connor J. A., Curran T. (1994) Neuron 13, 325–338 [DOI] [PubMed] [Google Scholar]
- 30. Goslin K., Asmussen H., Banker G. (1998) in Culturing Nerve Cells (Banker G., Goslin K. eds) pp. 339–370, 2nd Ed., MIT Press, Cambridge [Google Scholar]
- 31. Kaech S., Banker G. (2006) Nat. Protoc. 1, 2406–2415 [DOI] [PubMed] [Google Scholar]
- 32. Rubinson D. A., Dillon C. P., Kwiatkowski A. V., Sievers C., Yang L., Kopinja J., Rooney D. L., Ihrig M. M., McManus M. T., Gertler F. B., Scott M. L., Van Parijs L. (2003) Nat. Genet. 33, 401–406 [DOI] [PubMed] [Google Scholar]
- 33. Dull T., Zufferey R., Kelly M., Mandel R. J., Nguyen M., Trono D., Naldini L. (1998) J. Virol. 72, 8463–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Manadas B., Santos A. R., Szabadfi K., Gomes J. R., Garbis S. D., Fountoulakis M., Duarte C. B. (2009) J. Proteome Res. 8, 4536–4552 [DOI] [PubMed] [Google Scholar]
- 35. Caldeira M. V., Melo C. V., Pereira D. B., Carvalho R. F., Carvalho A. L., Duarte C. B. (2007) Mol. Cell Neurosci. 35, 208–219 [DOI] [PubMed] [Google Scholar]
- 36. Gomes A. R., Ferreira J. S., Paternain A. V., Lerma J., Duarte C. B., Carvalho A. L. (2008) Mol. Cell Neurosci. 37, 323–334 [DOI] [PubMed] [Google Scholar]
- 37. Rao A., Kim E., Sheng M., Craig A. M. (1998) J. Neurosci. 18, 1217–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zukin R. S., Bennett M. V. (1995) Trends Neurosci. 18, 306–313 [DOI] [PubMed] [Google Scholar]
- 39. Liao D., Scannevin R. H., Huganir R. (2001) J. Neurosci. 21, 6008–6017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Blahos J., 2nd, Wenthold R. J. (1996) J. Biol. Chem. 271, 15669–15674 [DOI] [PubMed] [Google Scholar]
- 41. Hall R. A., Soderling T. R. (1997) J. Biol. Chem. 272, 4135–4140 [DOI] [PubMed] [Google Scholar]
- 42. Barria A., Malinow R. (2002) Neuron 35, 345–353 [DOI] [PubMed] [Google Scholar]
- 43. Huh K. H., Wenthold R. J. (1999) J. Biol. Chem. 274, 151–157 [DOI] [PubMed] [Google Scholar]
- 44. Leonard A. S., Hell J. W. (1997) J. Biol. Chem. 272, 12107–12115 [DOI] [PubMed] [Google Scholar]
- 45. Chung H. J., Huang Y. H., Lau L. F., Huganir R. L. (2004) J. Neurosci. 24, 10248–10259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sanz-Clemente A., Matta J. A., Isaac J. T., Roche K. W. (2010) Neuron 67, 984–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lan J. Y., Skeberdis V. A., Jover T., Grooms S. Y., Lin Y., Araneda R. C., Zheng X., Bennett M. V., Zukin R. S. (2001) Nat. Neurosci. 4, 382–390 [DOI] [PubMed] [Google Scholar]
- 48. Lau C. G., Takayasu Y., Rodenas-Ruano A., Paternain A. V., Lerma J., Bennett M. V., Zukin R. S. (2010) J. Neurosci. 30, 242–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bradley J., Carter S. R., Rao V. R., Wang J., Finkbeiner S. (2006) J. Neurosci. 26, 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Petralia R. S., Wang Y. X., Hua F., Yi Z., Zhou A., Ge L., Stephenson F. A., Wenthold R. J. (2010) Neuroscience 167, 68–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Al-Hallaq R. A., Yasuda R. P., Wolfe B. B. (2001) J. Neurochem. 77, 110–119 [DOI] [PubMed] [Google Scholar]
- 52. Sheng M., Cummings J., Roldan L. A., Jan Y. N., Jan L. Y. (1994) Nature 368, 144–147 [DOI] [PubMed] [Google Scholar]
- 53. Zhang J., Diamond J. S. (2009) J. Neurosci. 29, 4274–4286 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.