Abstract
Blocking HIV-1 cell entry has long been a major goal of anti-HIV drug development. Here, we report a successful design of two highly potent chimeric HIV entry inhibitors composed of one CCR5-targeting RANTES (regulated on activation normal T cell expressed and secreted) variant (5P12-RANTES or 5P14-RANTES (Gaertner, H., Cerini, F., Escola, J. M., Kuenzi, G., Melotti, A., Offord, R., Rossitto-Borlat, I., Nedellec, R., Salkowitz, J., Gorochov, G., Mosier, D., and Hartley, O. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17706–17711)) linked to a gp41 fusion inhibitor, C37. Chimeric inhibitors 5P12-linker-C37 and 5P14-linker-C37 showed extremely high antiviral potency in single cycle and replication-competent viral assays against R5-tropic viruses, with IC50 values as low as 0.004 nm. This inhibition was somewhat strain-dependent and was up to 100-fold better than the RANTES variant alone or in combination with unlinked C37. The chimeric inhibitors also fully retained the antiviral activity of C37 against X4-tropic viruses, and this inhibition can be further enhanced significantly if the target cell co-expresses CCR5 receptor. On human peripheral blood mononuclear cells, the inhibitors showed very strong inhibition against R5-tropic Ba-L strain and X4-tropic IIIB strain, with IC50 values as low as 0.015 and 0.44 nm, which are 45- and 16-fold better than the parent inhibitors, respectively. A clear delivery mechanism requiring a covalent linkage between the two segments of the chimera was observed and characterized. Furthermore, the two chimeric inhibitors are fully recombinant and are easily produced at low cost. These attributes make them excellent candidates for anti-HIV microbicides. The results of this study also suggest a potent approach for optimizing existing HIV entry inhibitors or designing new inhibitors.
Keywords: AIDS, Chemokines, HIV, Protein Chimeras, Virus Entry, 5P12-RANTES, CCR5, Entry Inhibitor, RANTES, gp41
Introduction
Approximately 33 million people are living with HIV, and millions more are infected each year.2 There is currently no vaccine, and treatments usually involve inhibiting viral activity post-infection by inhibiting the HIV protease or reverse transcriptase. More recently, therapies that target other parts of the viral life cycle have been approved, including an HIV integrase inhibitor (3).
One of the most promising areas in the fight against HIV/AIDS has been the development of entry inhibitors, which generally bind to either the viral surface or the human cell surface to stop HIV before it can enter a cell. The HIV surface protein gp120 first makes contact with the human cell surface protein CD4, which causes a conformational rearrangement in gp120, allowing the protein to then bind its co-receptor on the cell surface (either the chemokine receptor CCR5 or CXCR4). During this process, the HIV protein gp41 is exposed, and its fusion peptide enters the cell surface. Toward the end of the infection process, the C-terminal helical trimer folds over to contact the N-terminal trimer of gp41, forming a 6-helix bundle that likely pulls the membranes of the two entities in closer proximity to assist fusion of the virus to the cell (4, 5). Recently, it has been reported that some of these events may occur in the endosome (6).
Inhibition of HIV entry can be achieved by blocking one or more of the events that lead to infection. Proteins, particularly lectins that bind to gp120, have been shown to be effective inhibitors (7–9). Fusion inhibitors, such as peptides that bind to gp41, can stop the 6-helix bundle formation (4). In particular, the so-called C-peptides that are derived from the C terminus of gp41 effectively bind to the N terminus of gp41 to inhibit infection. One of these peptides, T-20, has been approved for clinical use (10–12). Another strategy to inhibit HIV infection involves binding the co-receptor on the human cell surface, particularly CCR5 (1, 13–20). Natural ligands for CCR5, namely chemokines MIP-1β, MIP-1α, and RANTES,3 were found to be able to block HIV infection (21). It was later shown that variants of these chemokines, particularly RANTES, could lead to even stronger inhibition (1, 13–16, 20). Despite the effectiveness of these entry inhibition strategies, many of them have serious drawbacks. Although the recent RANTES variants are extremely potent, they work by binding CCR5, so are only effective against R5-tropic virus (1). Although the C-peptides are effective against most strains of HIV, their potency is limited to nanomolar levels, and the virus can evolve to reduce the ability of the peptide to bind gp41 (4, 10–12, 22).
Considering the stepwise nature of the HIV entry process, there are certain time windows in which multiple targets are simultaneously susceptible to inhibition. Binding of co-receptor inhibitors and fusion inhibitors to their targets can both be achieved after the exposure of gp41 and before gp120 interacts with its co-receptor. Evidence suggests that co-receptor binding is a key factor in the kinetic properties of fusion and that lowered co-receptor density or weakened co-receptor-gp120 binding slows down gp41-mediated cell fusion and prolongs the time window of the intermediate states of gp41 for fusion inhibitors to bind (23, 24). We reasoned that a properly engineered chimeric molecule containing one co-receptor inhibitor and one fusion inhibitor can block HIV cell entry at both steps more potently and could overcome the drawbacks of either individual component.
We chose to use CCR5 ligand RANTES variants 5P12-RANTES and 5P14-RANTES as the co-receptor inhibitor portion of our chimeric protein. These variants were recently developed by Gaertner and co-workers (1, 13) and are among the most potent R5 entry inhibitors yet reported, with HIV inhibition at mid-picomolar levels in in vitro assays. In addition to their high potency, they are small proteins that are able to be made recombinantly and are easy to produce at low cost. For the fusion inhibitor part of the chimeric protein, we chose the C-peptide C37. C37 and the nearly identical C34 are well characterized C-peptides and are highly effective at nanomolar concentrations in vitro (4, 12, 22, 25, 26). It has also been shown that covalently linking C34 to a range of unrelated proteins did not diminish their anti-HIV activity, and in one case the linked C-peptide showed a longer in vivo lifetime (27, 28).
We report here the success of this strategy. Our chimeric inhibitors 5P12-linker-C37 and 5P14-linker-C37 show much higher anti-HIV potency than the parent inhibitors 5P12-RANTES, 5P14-RANTES, and C37 against R5-tropic HIV strains, leading to HIV inhibition at low picomolar levels. The chimeric proteins also retain the anti-HIV activity of C37 against X4-tropic strains. This X4-tropic anti-HIV potency can be further enhanced up to 6,000-fold on certain target cell lines expressing both CCR5 and CXCR4 co-receptors. A clear mechanism of C37 delivery by the RANTES variants is observed and characterized, which is a key to the greatly enhanced activity of our chimeric inhibitors.
EXPERIMENTAL PROCEDURES
Protein Preparation
The genes for 5P12-RANTES, 5P12-linker-C37, 5P14-RANTES, 5P14-linker-C37, and P2-linker-C37 were made using standard thermocycling methods. Oligonucleotides were purchased from Bioneer Corp. (Alameda, CA). Mutations to the 5P12-linker-C37 were made using the QuikChange site-directed mutagenesis method (Stratagene, La Jolla, CA). These genes were expressed along with an N-terminal small ubiquitin-like modifier tag in the pET SUMO expression vector from Invitrogen. The vectors were transformed into BL21(DE3) and grown in 1 liter of 15N minimal medium using 15NH4Cl as the only nitrogen source. Protein production was induced with 1 mm isopropyl β-d-thiogalactopyranoside when the absorbance at 600 nm reached 0.7. The cells were incubated with shaking for 20 h at 22 °C after induction and then harvested by centrifugation. The cell pellet was resuspended in cracking buffer (500 mm NaCl, 20 mm Tris, pH 8.0) with 10 mm benzamidine and French pressed twice at 16,000 p.s.i. The solution was centrifuged at 20,000 × g for 30 min. The supernatant was discarded, and the pellet was resuspended in 10 ml of unfolding buffer (5 m guanidinium HCl, 3 mm EDTA, 50 mm Tris, 50 mm NaCl, pH 8.0) with 10 mm β-mercaptoethanol. The resuspended solution was incubated at room temperature for 2 h with stirring followed by a centrifugation at 20,000 × g for 60 min. The supernatant containing the denatured protein was added dropwise into 90 ml of folding buffer (50 mm NaCl, 20 mm Tris, pH 8.0) with 10 mm β-mercaptoethanol. The solution was incubated overnight at 4 °C, and the precipitants were then removed by centrifugation at 20,000 × g for 60 min. The supernatant was dialyzed in 4 liters of dialysis buffer (50 mm NaCl, 20 mm Tris, pH 8.0) with slow stirring, and the buffer was changed after 6 h. After dialysis, the solution was centrifuged again, and the supernatant containing the refolded protein was passed through a nickel chelating column (GE Healthcare) and eluted with imidazole in 500 mm NaCl, 50 mm Tris buffer, pH 8.0. The purified proteins were dialyzed in 4 liters of 50 mm NaCl, 20 mm Tris buffer, pH 8.0, to remove imidazole. To cleave the SUMO tag, recombinant yeast ULP1 protease was added, and the solution was incubated overnight at 4 °C. (ULP1 protease was produced and purified in our laboratory as briefly described. ULP1 was expressed in LB medium using a pET-28b, vector and the cells were collected and French pressed. The ULP1 protease from the supernatant was purified using a nickel chelating column.) Precipitated matter was removed by centrifugation at 20,000 × g for 30 min, and the product was separated from the SUMO tag using an acetonitrile gradient on a C4 reversed phase chromatography column (Vydac, Hesperia, CA) on an Akta purification system (GE Healthcare) and then lyophilized by the Labconco freeze-dry system (Labconco Corp., Kansas City, MO). In our hands, we were able to obtain a yield of 5 mg of pure protein from 1 liter of Escherichia coli preparation. For proteins containing a 20-amino acid linker, the protocol was modified to include an extra step of centrifugation to remove unfolded protein before adding TFA and acetonitrile for the final C4 column purification step. For the C37 peptide, the N-acetylated and C-amidated fusion inhibitor C37 was purchased from GenScript (Piscataway, NJ).
NMR Spectroscopy
All NMR data were acquired at 25 °C on a four-channel 600 MHz Bruker Avance III spectrometer equipped with a GRASP II gradient accessory and a TCI cryoprobe, which has an actively shielded Z-gradient coil. NMR samples were prepared by adding 15N-labeled lyophilized proteins into 20 mm sodium phosphate buffer, pH 2.5, with 5% D2O. The chemical shift was referenced relative to internal 2,2-dimethyl-2-silapentane-5-sulfonic acid (29). The data were processed using NmrPipe (30) and analyzed using PIPP (31). For two-dimensional HSQC spectra, sweep width = 6982.631 Hz (1H) and 1700.030 Hz (15N), with 512* points in 1H and 128* points in 15N.
Cell Lines and Viruses
HeLa-ADA and HeLa-P5L cells were kindly provided by Dr. M. Alizon and Dr. Anne Brelot (Cochin Institute, Paris, France) (32). HeLa-TZM-bl, HL2/3, and Magi-CXCR4 cells were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health; the HeLa-TZM-bl cell line was from Dr. John C. Kappes, Dr. Xiaoyun Wu, and Tranzyme Inc. (33–36); HL2/3 was from Dr. Barbara K. Felber and Dr. George N. Pavlakis (37); and Magi-CXCR4 was donated by Dr. Michael Emerman (38). 293FT cells were kindly provided by Dr. Jennifer Manilay and were originally obtained from Invitrogen. Viruses used in replication-competent viral assays and PBMC assays, including HIV-1 Ba-L, ADA, and IIIB, were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health; HIV-1 Ba-L was from Dr. Suzanne Gartner, Dr. Mikulas Popovic, and Dr. Robert Gallo (39); HIV-1 ADA-M was from Dr. Howard Gendelman (40–43); and HIV-1 IIIB was from Dr. Robert C. Gallo (44–46).
Cell-Cell Fusion Assay
HIV-1 cell-cell fusion assays were carried out as described previously (32). Briefly, 104/well target cells (HeLa-P5L for R5-tropic fusion assay, HeLa-TZM-bl, and Magi-CXCR4 for X4-tropic fusion assay) were seeded in a 96-well plate. After 12 h of incubation, the medium was replaced with 50 μl per well of fresh RPMI 1640 medium (DMEM for the X4 assay). Different concentrations of inhibitors were added and mixed well. 104/well effector cells (HeLa-ADA for R5 assay and HL2/3 for X4 assay) in 50 μl of medium were then added to each well. The cells were incubated at 37 °C for 24 h to allow fusion. Cells were lysed with 20 μl of 0.5% Nonidet P-40 (US Biological) in PBS, pH 7.4, for 30 min, and then 30 μl of PBS with 8 mm substrate chlorophenol red-β-d-galactopyranoside (Calbiochem), 20 mm KCl, and 10 mm β-mercaptoethanol (Sigma) was added to each well. The absorbance signal at wavelength 570 and 630 nm were measured, and the 570:630 ratio for each well was calculated. Data were analyzed using KaleidaGraph (Synergy Software, Reading, PA).
Single-round Viral Infection Assay
Plasmids used to generate the pseudotyped viral particles were all obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, National Institutes of Health; plasmid pNL4-3.Luc.R−E− (with deletion of the envelope and vpr genes) and envelope plasmids pSV-ADA and pSV-JRFL were from Dr. Nathaniel Landau (47); pHEF-VSVG was from Lung-Ji Chang (48). pCAGGS-SF162-gp160 was from Leonidas Stamatatos and Dr. Cecilia Cheng-Mayer (49–51); HXB2-env was from Dr. Kathleen Page and Dr. Dan Littman (52); HIV-1 clone Ba-L.01 was from Dr. John Mascola (53); pSVIII-91US005.11 was from Dr. Beatrice Hahn (54); and 6535, clone 3 (SVPB5), was from Dr. David Montefiori, Dr. Feng Gao, and Dr. Ming Li (55). To make the pseudo-typed viral particles, 293FT cells were co-transfected with the pNL4-3.Luc.R−E− plasmid and an envelope plasmid using the Profection Mammalian Transfection System (Promega). 48 h post-transfection, the supernatant were collected, centrifuged, and filtered with a 0.45-μm syringe filter. The viral stocks were stored at −80 °C. For the infection assay, 104 cells per well (HeLa-TZM-bl cell for both the R5 and X4-tropic assays and, Magi-CXCR4 cells for the control “X4 only” assays) were seeded in a 96-well plate. The next day, after changing the medium, different concentration of inhibitors were added to the wells and mixed, and then the virus particles were added. The final volume was adjusted to 100 μl per well. After incubation for 3 days (the medium was changed once), the cells were lysed, and the substrate chlorophenol red-β-d-galactopyranoside was added (as described above). The absorbance signals at wavelengths 570 and 630 nm were measured, and the 570:630 ratio for each well was calculated. The data were plotted using Microsoft Excel, and the IC50 value was determined using a linear equation fitted between two data points surrounding 50% inhibition. For presentation purposes, data shown in the figures were plotted and fitted as curves using a four-parameter logistic equation in Kaleidagraph (Synergy Software).
Replication-competent Viral Assay and PBMC Assay
The replication competent viral assays were performed as described previously (56). TZM-bl cells were used for both the CCR5 and CXCR4 tropic assays, and the HeLa-CXCR4 cells were used for the control X4 only assays. The PBMCs were freshly isolated and used in viral assays as described previously (56, 57). Each assay was conducted in parallel with control compounds AMD3100 (CXCR4 inhibitor; positive control inhibitor for IIIB and negative control inhibitor for Ba-L and ADA) and TAK779 (CCR5 inhibitor; positive control inhibitor for Ba-L and ADA and negative control inhibitor for IIIB) (data not shown). Cytotoxicity of the inhibitors was assayed using the CellTiter 96 AQueous One Solution cell proliferation assay (Promega) (56). All tested compounds showed no toxicity at the highest tested concentration (100 nm). Data were plotted and presented as described for the single-round viral assays.
Receptor Density Comparison Using Flow Cytometry
The CCR5 receptor expression levels on HeLa-TZM-bl cells and HeLa-P5L cells were compared using flow cytometry. The cells were incubated with FITC-conjugated anti-CCR5 antibody (clone 2D7, BD Biosciences), and the fluorescence values were determined using a FACSAria cytometer (BD Biosciences). Flow cytometric data were analyzed using FlowJo software (TreeStar).
RESULTS
Design of the Chimeric Inhibitors
5P12-RANTES and 5P14-RANTES are variants of the chemokine RANTES developed by Gaertner et al. (1), each with 10 different amino acid mutations at the N terminus. Unlike natural RANTES, which is an agonist to CCR5, 5P12 triggers neither receptor sequestration nor cell signaling, whereas 5P14 causes receptor sequestration but not signaling (1). Lack of receptor signaling activity is a valuable property in an anti-HIV agent because immunologic activation could lead to more susceptibility to HIV infection. The flexible N terminus of both 5P12 and 5P14 are critical for their receptor-related anti-HIV function (1), but the C-terminal amino acids, which typically form an α-helix in the chemokine structure, are likely not functionally important. So we chose to link the C37 peptide to the C terminus of 5P12 and 5P14, leaving the N terminus intact. The C termini of 5P12-RANTES and 5P14-RANTES were covalently linked to the N terminus of C37 peptide using 10-amino acid flexible glycine/serine linkers “GGGGSGGGGS.” We denoted the engineered chimeric proteins 5P12-linker-C37 and 5P14-linker-C37. All the individual RANTES variants and chimeric inhibitors were expressed in E. coli and purified to no less than 95% purity as determined by SDS-PAGE. Proteins were tested by NMR to determine the structural integrity. 1H-15N correlation spectra revealed that all proteins are folded (Fig. 1). 5P12-linker-C37 and 5P14-linker-C37 exhibited the same peak placement as free 5P12 and 5P14, respectively, except for extra peaks in the unstructured region of the spectrum, which are likely caused by the linker and the C37 peptide, which is known to be unstructured in the absence of its binding partner (26). These data indicate that linking C37 to a RANTES variant does not compromise the native structure of the RANTES variant.
FIGURE 1.
1H-15N two-dimensional HSQC spectra of the chimeric proteins and the parent RANTES variants to verify the structural integrity of the proteins. A, overlay of 5P12-linker-C37 (red peaks) with 5P12-RANTES (black peaks). B, overlay of 5P14-linker-C37 (red peaks) with 5P14-RANTES (black peaks).
Antiviral Potencies of 5P12-Linker-C37 and 5P14-Linker-C37 against R5-Tropic HIV Viruses
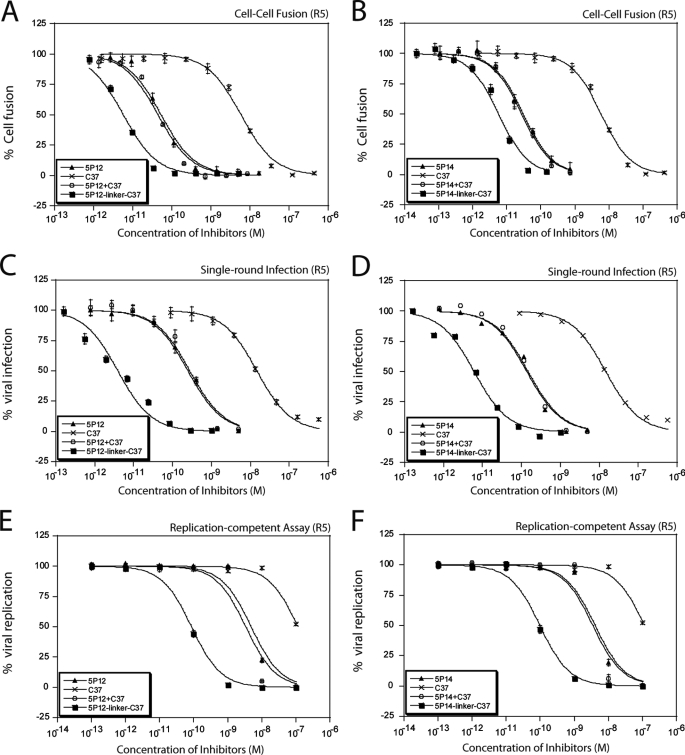
In vitro assays, including cell-cell fusion assays, single-cycle viral infection assays, replication-competent viral assays, and PBMC assays were conducted to evaluate the antiviral potencies of the chimeric inhibitors (Fig. 2). Control compounds, including RANTES variants alone, C37 alone, and a mixture of RANTES variants with C37 (1:1 ratio, unlinked), were tested in parallel with the chimeric inhibitors.
FIGURE 2.
Antiviral activities of the chimeric inhibitors against R5-tropic virus. Chimeric inhibitors showed higher anti-HIV potency than the control groups (RANTES variants alone, C37 alone, or 1:1 mixture of the RANTES variants and C37). A and B, cell-cell fusion assay using effector cells expressing ADA viral envelopes. C and D, single-cycle viral infection assay (Ba-L strain pseudoviral particles infecting TZM-bl cells). E and F, replication-competent viral assay (Ba-L strain virus infecting TZM-bl cells). Data shown are typical results of single assays done in triplicate. Error bars represent standard deviations of the data. Refer to Tables 1 and 2 and supplemental Tables S1 and S3 for number of repeats.
In R5-tropic cell-cell fusion assays, consistent with previously published data (1), 5P12-RANTES and 5P14-RANTES showed anti-HIV IC50 values of 50 and 30 pm, respectively, and the IC50 of C37 was in the low nanomolar range, 2 orders of magnitude higher. As expected, because of the large differences of the RANTES variants and C37 in antiviral potencies, simply mixing 5P12 or 5P14 with C37 in 1:1 ratio exhibited similar potency as 5P12 or 5P14 alone. But the chimeric inhibitors exhibited antiviral potencies stronger than either of the components alone or the unlinked combination of the two (Fig. 2, A and B, and supplemental Table S1). These findings suggest that the enhancement is not due to simply adding two inhibitors together but rather an intramolecular mechanism of the covalently linked inhibitors.
Further testing with multiple strains of R5 virus in single-cycle viral infection assays in TZM-bl cells revealed similar results (Table 1 and Fig. 2, C and D). For all six strains tested, 5P12-linker-C37 and 5P14-linker-C37 exhibited up to 100-fold greater potency compared with 5P12 and 5P14 alone or compared with a 1:1 mixture of them with C37 (Table 1 and supplemental Table S3). It was also found that the potency enhancement is strain-dependent. For virus strains that are particularly sensitive to C37, such as Ba-L, 5P12-linker-C37 and 5P14-linker-C37 showed 70- and 23-fold potency enhancement over 5P12 and 5P14, respectively. But for virus strains that are less sensitive to C37, such as 6535, 5P12-linker-C37 was only 2.5-fold better than 5P12, although the 5P14-linker-C37 showed no enhancement over 5P14. These data indicate that the linked C-peptide is critical for the potency enhancement of the chimeric inhibitor and suggest that the effectiveness of the fusion inhibitor part of the chimera against a specific viral strain determines the magnitude of the relative potency enhancement over the RANTES variant alone (supplemental Fig. S1).
TABLE 1.
Anti-HIV activities of the chimeric inhibitors in single-cycle viral assay
Results are average IC50 ± S.D. (nm) from four or more independent experiments in triplicate. 5P12 + C37 and 5P14 + C37 groups showed similar R5 antiviral activity to 5P12 and 5P14 alone and similar X4 antiviral activity to C37 (supplemental Table S3).
| HIV virus | Tropism | 5P12-linker-C37 | 5P12 | C37 | 5P14-linker-C37 | 5P14 |
|---|---|---|---|---|---|---|
| Ba-L | R5 | 0.004 ± 0.001 | 0.29 ± 0.09 | 15 ± 0.6 | 0.007 ± 0.001 | 0.16 ± 0.01 |
| SF162 | R5 | 0.006 ± 0.001 | 0.59 ± 0.09 | 38 ± 13 | 0.017 ± 0.005 | 0.22 ± 0.06 |
| ADA | R5 | 0.025 ± 0.004 | 0.47 ± 0.09 | 44 ± 14 | 0.037 ± 0.002 | 0.18 ± 0.03 |
| JRFL | R5 | 0.015 ± 0.001 | 0.51 ± 0.01 | 49 ± 7.4 | 0.02 ± 0.006 | 0.14 ± 0.03 |
| US005 | R5 | 0.03 ± 0.006 | 0.20 ± 0.02 | 59 ± 16 | 0.025 ± 0.006 | 0.09 ± 0.03 |
| 6535 | R5 | 0.22 ± 0.05 | 0.55 ± 0.07 | 261 ± 57 | 0.08 ± 0.02 | 0.10 ± 0.03 |
| HXB2 (Magi-X4) | X4 | 18 ± 7.4 | >500 | 9.6 ± 1.3 | 14 ± 5.0 | >500 |
| HXB2 (TZM-bl) | X4 | 0.001 ± 0.0003 | >500 | 6.1 ± 0.3 | 0.001 ± 0.0001 | >500 |
| VSV-G | Control | >500 | >500 | >500 | >500 | >500 |
A series of assays with replication-competent virus were also carried out. CCR5-tropic HIV-1 ADA and HIV-1 Ba-L strains were used to infect TZM-bl cells at different concentrations of inhibitors. Compared with the results of the single-round viral infection assay, all inhibitors were less effective in inhibiting viral replication. For example, 5P12-RANTES and 5P14-RANTES had nanomolar rather than mid-picomolar inhibition against tested strains. But the chimeric inhibitors consistently showed stronger inhibition, with 5P12-linker-C37 showing up to 157-fold enhancement over 5P12 alone and with 5P14-linker-C37 showing up to 56-fold better inhibition than 5P14 alone (Table 2 and Fig. 2, E and F).
TABLE 2.
Anti-HIV activities of the chimeric inhibitors in replication-competent viral assay
Results are average IC50 ± the uncertainty of the average (half the difference) (nm) from two independent experiments in triplicate. ± 0 indicates the two experiments yielded identical IC50 values.
| HIV virus | Tropism | 5P12-linker-C37 | 5P12 | C37 | 5P14-linker-C37 | 5P14 |
|---|---|---|---|---|---|---|
| Ba-L | R5 | 0.08 ± 0 | 12.61 ± 8.19 | >100 | 0.08 ± 0.01 | 4.52 ± 0.64 |
| ADA | R5 | 0.65 ± 0.08 | 28.5 ± 5.9 | >100 | 0.63 ± 0.06 | 21.8 ± 0.1 |
| IIIB (HeLa-X4) | X4 | 10.1 ± 0.1 | >500 | 10.4 ± 0.2 | 9.23 ± 1.77 | >500 |
| IIIB (TZM) | X4 | 0.05 ± 0 | > 00 | 77.1 ± 21.1 | 0.04 ± 0 | >500 |
As described above, the chimeric inhibitors consistently showed better inhibition than the parent compounds against HIV in engineered cell lines, which may be different from natural human cells in properties such as receptor expression level. To determine the success of these chimeric inhibitors on primary human cells and to get an estimation of their potency against HIV on its natural targets, the inhibitors were tested on human PBMCs. The PBMC results confirmed the previous findings, with 5P12-linker-C37 being 45-fold better than 5P12 alone and 5P14-linker-C37 being 26-fold better than 5P14 alone against the Ba-L strain (Table 3). Further testing on primary strains confirmed the higher potency of 5P12-linker-C37 compared with 5P12, although 5P14-linker-C37 appeared to have similar activity as 5P14 (Table 3).
TABLE 3.
Anti-HIV activities of the chimeric inhibitors in PBMC assay
| HIV virus | Ba-L (R5)a | 91US001 (R5)b | 91US004 (R5)b | IIIB (X4)a | CMU02 (X4)b | 92UG001 (X4)b | 92UG001 (X4R5)b |
|---|---|---|---|---|---|---|---|
| 5P12-linker-C37 | 0.015 ± 0.005 | 0.02 | 0.04 | 0.44 ± 0.02 | 0.07 | 0.57 | 18.1 |
| 5P12 | 0.675 ± 0.5 | 0.18 | 0.21 | >100 | >100 | >100 | >100 |
| C37 | 13.85 ± 8.3 | 28.6 | 49.4 | 7.0 ± 2.5 | 18.2 | 39 | 58.7 |
| 5P14-linker-C37 | 0.015 ± 0.005 | 0.09 | 0.27 | 3.1 ± 2.3 | >100 | >100 | >100 |
| 5P14 | 0.395 ± 0.26 | 0.07 | 0.12 | >100 | 0.04 | 42.4 | 9.47 |
a Results are average IC50 ± the uncertainty of the average (half the difference) (nm) from two independent experiments in triplicate.
b Results are IC50 (nm) from one independent assay in triplicate.
Antiviral Potencies of 5P12-Linker-C37 and 5P14-Linker-C37 against X4-Tropic HIV Viruses
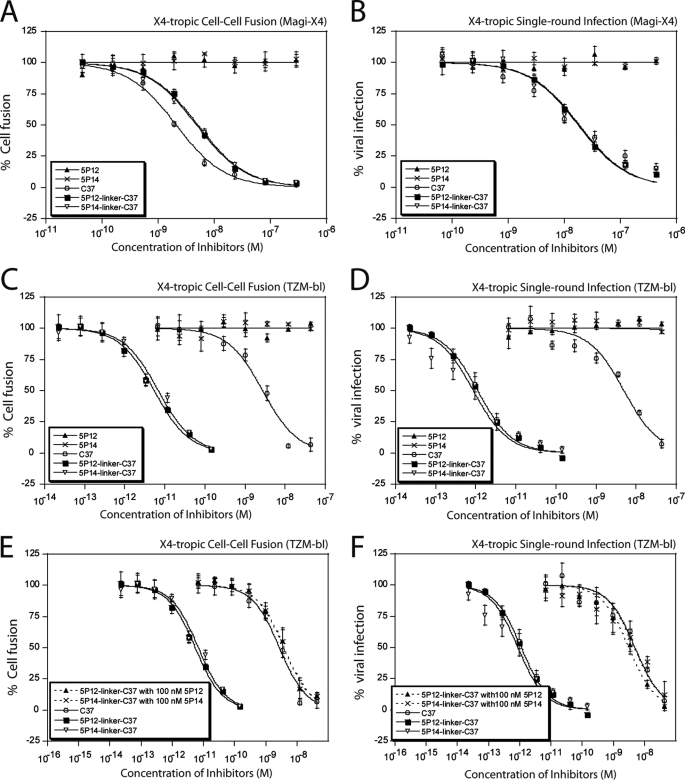
5P12 and 5P14 work by binding CCR5 and are therefore only effective against R5-tropic virus. As expected, they showed no inhibition against X4 envelopes, either in cell-cell fusion assays or in viral assays using X4-tropic virus against Magi-X4 cells (Table 1 and Fig. 3, A and B). In contrast, the peptide C37 is active against both R5- and X4-tropic virus due to its ability to bind gp41, and this peptide exhibits nanomolar level of inhibition potency in X4 fusion and viral assays. The designed chimeric inhibitors 5P12-linker-C37 and 5P14-linker-C37 also show anti-X4 activities due to the action of the C37 segment of the molecule, as shown in Fig. 3, A and B. These chimeric inhibitors exhibit IC50 values nearly identical to C37, demonstrating the effectiveness even when the other segment (the RANTES variant) is not utilized for the inhibition.
FIGURE 3.
Antiviral activities of the chimeric inhibitors against X4-tropic virus. The chimeric inhibitors retain the antiviral activity of C37 in X4 assays. A, Magi-X4 cell (expresses only CXCR4, but not CCR5, on the surface)-based cell-cell fusion assays. B, Magi-X4 cell-based single-cycle viral infection assays. The antiviral activity of chimeric inhibitors against X4 virus is greatly enhanced if the cells co-express CCR5 receptors. C, TZM-bl cell (expresses both CCR5 and CXCR4 on the surface)-based fusion assays. D, TZM-bl-based single-cycle viral infection assays. This enhancement of the chimeric protein requires binding to CCR5. When the CCR5 receptors on the cell surface are occupied by preincubation with CCR5-binding protein, the chimeric inhibitors showed no enhancement over C37. E, TZM-bl cell-based fusion assays. F, TZM-bl cell-based single-cycle viral infection assays. The cells were preincubated with 100 nm 5P12 or 5P14. Data shown are typical results of single assays done in triplicate. Error bars represent standard deviations of the data. Refer to Table 1 and 2 and supplemental Tables S2 and S3 for number of repeats.
More striking results were observed when performing assays with X4-tropic virus on the TZM-bl cell line, which expresses both CXCR4 and CCR5 receptors. Although the IC50 value of C37 alone remains the same as on the Magi-X4 cell line, the antiviral potency of the chimeric inhibitors increased 400- and 6,000-fold compared with C37 in cell fusion assays and in single-cycle viral assays against the HXB2 strain, respectively (Table 1, supplemental Table S2, and Fig. 3, C and D). Parent inhibitor controls 5P12-RANTES and 5P14-RANTES showed no inhibition, indicating that CCR5 binding by the RANTES variants does not inhibit X4-tropic viral entry, as expected. Furthermore, potency enhancement was not observed when 5P12 or 5P14 was mixed with C37 in a 1:1 ratio, which led to the same activity as C37 alone. The higher potency of the chimeric proteins against X4 virus under these conditions suggests that the strong enhancement was due to an intramolecular mechanism involving both components of the chimeric protein. This enhancement likely involves the RANTES variants being bound to CCR5 receptors, placing the C37 part of the chimera in the proper position to bind its target gp41.
Given the fact that CCR5 and CXCR4 receptors form hetero-oligomers on the cell surface (58, 59), it is very likely that by binding to the CCR5 receptors, the RANTES variants in the chimeric protein could specifically deliver C37 to its virus target, which is presumably using the nearby CXCR4 as a co-receptor, to achieve the strong enhancement of X4 inhibition potency. To provide evidence for this, we carried out experiments in which the CCR5 receptors were blocked prior to adding X4-tropic virus and the chimeric inhibitors. In both fusion and single-cycle viral assays, target cells were preincubated with 100 nm 5P12 or 5P14 before 5P12-linker-C37 or 5P14-linker-C37 was added. Because the CCR5 receptors were blocked by an excessive amount of RANTES variants and the chimeric inhibitors could not bind to CCR5 receptors, their X4 inhibition potency reverted to that of C37 alone (Fig. 3, E and F).
Replication-competent viral assays were also conducted to confirm the antiviral activities of the chimeric inhibitors against X4-tropic virus replication. Similar results were observed as from the single cycle assays, and although the chimeric inhibitors 5P12-linker-C37 and 5P14-linker-C37 showed the same IC50 values as C37 on HeLa-X4 cells (which express only CXCR4 receptors on the surface), they exhibited 1500–1900-fold increased potency over C37 against the X4-tropic HIV-1 IIIB strain on TZM-bl cells, which co-expressed CCR5 and CXCR4 receptors (Table 2). Altogether, the data show that chimeric inhibitors fully retain the anti-X4 activity of C37 and that this anti-X4 potency can be further enhanced when the target cells co-express CCR5 on the surface.
Viral assays against PBMCs, some of which also express both CCR5 and CXCR4 receptors on the surface, were then carried out to evaluate the X4-tropic antiviral potencies of these chimeric inhibitors on natural human cells. As expected, 5P12 and 5P14 alone did not show any inhibition against the X4-tropic HIV-1 IIIB strain. The peptide C37 alone showed an IC50 of 7 nm. The chimeric inhibitors, however, showed inhibition that was up to 16-fold better than C37 alone as judged by IC50, most likely due to the co-expression of CCR5 receptors on some of the cells (Table 3). This result showed that the chimeric inhibitors were extremely potent against X4-tropic viruses even on natural human cells. Further testing on clinical X4-tropic strains CMU02 and 92UG029 exhibited 260- and 68-fold improvement by 5P12-linker-C37 over C37. 5P14-linker-C37 also showed greatly enhanced activity over C37 for strain CMU02, although not for 92UG029 (Table 3).
The chimeric inhibitors were also tested against a clinical dual-tropic strain, 92UG001. In this case, the RANTES variants 5P12 and 5P14 alone showed no activity, although C37 had an IC50 of 59 nm. Both 5P12-linker-C37 and 5P14-linker-C37 showed severalfold increased potency over C37 alone (Table 3).
Mechanism of the Chimeric Inhibitors
In vitro assays, including R5- and X4-tropic cell fusion assays, single-cycle viral infection assays, replication-competent viral assays, and PBMC assays indicate the success of the strategy to covalently link RANTES variants with a C-peptide. These experiments also provide evidence suggesting the high potency is probably due to the excellent inhibition of the RANTES variants, along with their ability to specifically deliver C37 to its gp41 target. Therefore, experiments were designed to further characterize the mechanism of action of the chimeric inhibitors, first focusing on the relative importance of the C37 segment.
A series of mutations were made to 5P12-linker-C37 (Fig. 4, Table 4, and supplemental Table S4), and the corresponding effects were evaluated with cell fusion and single-cycle viral assays. It has previously been shown that the mutation of Ile to Asp in position 642 in the C-peptide causes a 10,000-fold drop of anti-HIV activity, almost completely abolishing its function, whereas the Ile mutation to Asp at position 656 causes a moderate 80-fold decrease of activity (23). Therefore, mutations to Asp were made in the 642th and 656th positions of C37 in 5P12-linker-C37. To test whether these mutations reduce the activity of the C37 segment of the chimeric protein, we tested the mutants with Magi-X4 cell-based fusion and viral assays. 5P12-linker-C37I642D completely lost its ability to inhibit X4-tropic virus at lower than 500 nm concentration, whereas 5P12-linker-C37I656D showed 10- and 3-fold decrease in activity in fusion and viral assays, respectively, compared with the wild type chimera (Fig. 4A, Table 4, and supplemental Table S4). These mutations that weaken the potency of the C-peptide also reduced the overall effectiveness of the chimeric protein against R5-tropic viruses. The antiviral potencies of both 5P12-linker-C37I642D and 5P12-linker-C37I656D were much lower compared with 5P12-linker-C37, inhibiting similarly to free 5P12 (Fig. 4C, Table 4, and supplemental Table S4). A decrease in antiviral potency was observed in X4-tropic viral assays on TZM-bl cells that contain both CCR5 and CXCR4 on the surface (Fig. 4E and Table 4).
FIGURE 4.
Mechanism of action of the chimeric inhibitors. A, mutations on the C37 segment of 5P12-linker-C37 cause reduced or loss of activity against X4-tropic virus, whereas substitution of 5P12 with a different N terminus (that of P2-RANTES) has no effect against X4-tropic virus. The X4-tropic antiviral potency was determined on Magi-X4 cells (which express only CXCR4, but not CCR5, on the surface) against HXB2 strain pseudotyped virus particles. C, mutations on either the RANTES variant segment or the C37 segment of 5P12-linker-C37 cause reduced activity against R5-tropic virus. The R5-tropic antiviral potency was determined using TZM-bl cells (which express both CCR5 and CXCR4 on the surface) against Ba-L strain pseudotyped virus particles. E, when the cells co-express CCR5, mutations on either the RANTES variant segment or the C37 segment of 5P12-linker-C37 cause reduced activity against X4-tropic virus. The X4-tropic antiviral potency was determined on TZM cells (which express both CCR5 and CXCR4 on the surface) against HXB2 strain pseudotyped virus particles. B, changing the original 10-amino acid linker to a shorter 3-amino acid linker or a longer 20-amino acid linker does not affect the native activity of C37 against X4-tropic virus. The X4-tropic antiviral potency was determined on Magi-X4 cells (which express only CXCR4, but not CCR5, on the surface) against HXB2 strain pseudotyped virus particles. D and F, 5P12–3AA-C37 shows reduced antiviral activity in both R5-tropic single-cycle viral assays and TZM-bl cell-based X4-tropic single-cycle viral assays, whereas 5P12–20AA-C37 shows very similar activity to 5P12-linker-C37. The R5-tropic antiviral potency was determined using TZM-bl cells (which express both CCR5 and CXCR4 on the surface) against Ba-L strain pseudotyped virus particles (D). The TZM-bl cell based X4-tropic antiviral potency was determined on TZM cells against HXB2 strain pseudotyped virus particles (F). Data shown are typical results of single assays done in triplicate. Error bars represent standard deviations of the data. Refer to Table 3 and supplemental Table S4 for number of repeats.
TABLE 4.
Anti-HIV activities of 5P12-linker-C37 mutations in single-cycle viral assay
Results are average IC50 ± S.D. (nm) from four or more independent experiments in triplicate.
| HIV virus | Tropism | 5P12-linker-C37 | Mutation in RANTES |
Mutation in C37 |
Change of linker length |
||
|---|---|---|---|---|---|---|---|
| P2-RANTES-linker-C37 | 5P12-linker-C37I642D | 5P12-linker-C37I656D | 5P12-GGS-C37 | 5P12-(GGGGS)4-C37 | |||
| Ba-L | R5 | 0.004 ± 0.001 | 0.82 ± 0.09 | 0.24 ± 0.04 | 0.20 ± 0.02 | 0.06 ± 0.001 | 0.002 ± 0.001 |
| SF162 | R5 | 0.006 ± 0.001 | 0.80 ± 0.02 | 0.75 ± 0.36 | 0.27 ± 0.07 | 0.1 ± 0.02 | 0.004 ± 0.0005 |
| ADA | R5 | 0.025 ± 0.004 | 0.61 ± 0.05 | 0.29 ± 0.02 | 0.15 ± 0.02 | 0.1 ± 0.003 | 0.016 ± 0.004 |
| HXB2 (Magi-X4) | X4 | 18 ± 7.4 | 17 ± 2.4 | >500 | 60 ± 20 | 27 ± 3.1 | 11 ± 6.4 |
| HXB2 (TZM) | X4 | 0.001 ± 0.0003 | 0.28 ± 0.07 | >500 | 0.10 ± 0.02 | 0.28 ± 0.05 | 0.002 ± 0.001 |
| VSV-G | Control | >500 | >500 | >500 | >500 | >500 | >500 |
The RANTES portion of 5P12-linker-C37 was also mutated to determine the role of the RANTES segment in the whole chimeric protein. In particular, the N terminus (the first 10 amino acids) of the potent 5P12 was changed to form another RANTES variant, “P2-RANTES.” P2-RANTES is also an R5 ligand but with lower antiviral potency against R5 virus (nanomolar level of inhibition in cell fusion and pseudotyped viral infection) (13). Substitution of the RANTES part of the linker protein did not affect the C-peptide portion of the chimera, as evidenced by the similar activity of P2-linker-C37 as 5P12-linker-C37 and C37 in Magi-X4 cell-based X4-tropic assays where only the C37 portion would be expected to be active (Table 4, supplemental Table S4, and Fig. 4A). But this variant did show decreased activity in R5 and TZM-bl X4 assays (Table 4 and Fig. 4, C and E). Therefore, the RANTES portion of the chimeric protein is also critical for the enhanced activity. These data indicate that both parts of the linker protein are necessary and that they both have to be functioning to show an enhancement of potency.
Having demonstrated the necessity of both portions of the chimera, we hypothesized that the observed enhancement of the chimeric inhibitors was likely due to the specific delivery of C37 to the nearby gp41 target by the RANTES variant as it binds to the CCR5 co-receptor. To probe this possibility, we tested the effects of different linker length on the overall activity of the linker protein. Mutant chimeric inhibitors with the 3-amino acid linker “GGS” and 20-amino acid linker “(GGGGS)4” were made to compare with our 5P12-linker-C37 that has a flexible 10-amino acid linker (GGGGS)2. NMR experiments were done to confirm the structural integrity of the mutants (data not shown). The spectra showed that the 20-amino acid long linker led to a significant portion of unfolded protein using our regular purification method, so we modified the protocol to obtain pure and fully folded chimeric protein with the 20-amino acid long linker. Finally, two-dimensional HSQC spectra verified the structural integrity of all mutant chimeric proteins and showed that the change of linker length did not affect the structure of 5P12 (data not shown). As a control, X4-tropic single round virus assays using Magi-X4 cells (containing no CCR5) confirmed that different linker length did not affect the antiviral function of the C37 portion of these chimeras (Table 4 and Fig. 4B). This was expected, because we have already shown that under these conditions the C-peptide is the only component involved in inhibition, whereas the RANTES variant is not active against X4-tropic strains. In contrast, R5-tropic viral assays with three different strains showed that the shorter linker 5P12-GGS-C37 has the lowest antiviral potency, 15-fold lower compared with that 5P12-linker-C37. The moiety with the longer linker 5P12-(GGGGS)4-C37 showed almost the same activity as 5P12-linker-C37 (Table 4 and Fig. 4D). Similar results were observed in X4-tropic assays using TZM-bl (containing both surface R5 and X4) as target cells, where 5P12-GGS-C37 showed the lowest ability to inhibit and where 5P12-linker-C37 had similar activity to 5P12-(GGGGS)4-C37 (Table 4 and Fig. 4F).
Because the individual activities of 5P12 and C37 were not affected, the change in antiviral activity in these altered chimeras was likely caused by the change in linker length. The data suggest that the linker must be long enough to allow both parts of the linker protein to be functional and support the hypothesis that the enhancement of potency in the chimeric inhibitor is due to the specific delivery of C37 to the nearby gp41 target by the RANTES variant as it binds to CCR5.
DISCUSSION
In this study, a potent strategy to inhibit HIV by targeting multiple steps of HIV cell entry is described. It was reasoned that during the HIV entry process, there are time windows when co-receptor inhibition and gp41 fusion inhibition can be achieved simultaneously. Based on this hypothesis, we designed chimeric inhibitors that contain both a co-receptor inhibitor and a fusion inhibitor, and the two components were linked together using a flexible glycine/serine linker. The chimeric inhibitors 5P12-linker-C37 and 5P14-linker-C37 exhibited antiviral potency higher than either of the individual components alone or in combination. They were able to inhibit R5-tropic HIV at low picomolar levels in all the in vitro assays and therefore are among the most potent entry inhibitors yet reported. The chimeric inhibitors also fully retained the anti-X4 activity of C37, and this anti-X4 potency can be further enhanced when the cells co-express CCR5 on the surface. The chimeric inhibitors therefore overcome the major drawbacks of the parent co-receptor inhibitors 5P12 and 5P14, which lack activity against X4-tropic virus. Another advantage is that by blocking HIV entry at two steps, the chimeric inhibitors are less likely to be evaded by virus through mutations.
A few potent protein-based chimeric HIV entry inhibitors have been previously reported (60–62). Most relevant to this study is a similar strategy recently reported using an antibody to CCR5 covalently linked to two T-2635 fusion inhibitors (60). This molecule, called BFFI, also blocked HIV at both the co-receptor binding step and the 6-helix bundle formation step and showed very strong antiviral activity. However, this BFFI only exhibited 2-fold potency enhancement over the parent CCR5 mAb in PBMC assays against R5-tropic viruses, despite the large enhancement shown in the TZM-bl cell-based in vitro assays. BFFI also failed to inhibit X4-tropic viruses on cell lines expressing only CXCR4 receptors or on PBMCs. Against X4-tropic virus, BFFI was only active when the cells co-expressed large amounts of CCR5 along with CXCR4. This is probably because the large mAb sterically blocked the effective binding of the fusion inhibitor to its target (28). Another major drawback of BFFI was that it is produced in mammalian cells, making large scale production of this inhibitor less feasible because of expense. Our chimeric inhibitors, however, are straightforward and inexpensive to produce in E. coli, highly active against R5 viruses, active against X4-tropic viruses regardless of the presence of CCR5 receptors on the surface of the target cell, and are extremely potent on PBMCs.
The overall effectiveness of the chimeric inhibitors relies heavily on the following two major factors aside from the innate effectiveness of the components: viral susceptibility to the C37 peptide and CCR5 receptor density on the target cell. As shown in Table 1, we tested the effectiveness of the chimeric inhibitors on six different single-cycle R5 viruses. The viruses showed variable sensitivity to C37, with C37 inhibition IC50 ranging from 15 to 261 nm, while showing a quite similar sensitivity to 5P12 or 5P14 alone. The relative potency enhancement of the chimeric inhibitors over the parent RANTES variants similarly varied from 1- to 100-fold and was largely in proportion to the susceptibility of the virus to C37; generally, the more sensitive the virus to C37, the more potency enhancement of the chimeric inhibitor over the RANTES variants alone against that virus (supplemental Fig. S1).
The effectiveness of the chimeras also depend on receptor density on the target cells, which is also true for other inhibitors (2). Lower receptor density leads to more sensitivity to inhibition. In this study, R5-tropic fusion assays were carried out on two cell lines with differing amounts of CCR5 on the surface, and the results showed that the lower the CCR5 density, the more potent the chimeric inhibitor (supplemental Fig. S2 and supplemental Table S5).
Extensive in vitro viral assays and mutagenesis studies were carried out to investigate the mechanism of the chimeric inhibitors. In R5-tropic viral assays, the chimeric inhibitors showed up to a 100-fold potency enhancement over the parent RANTES variants, whereas a simple mixture of the RANTES variants and C37 showed no enhancement, indicating that C37 enhanced the R5 antiviral potency of the RANTES variants, and the mechanism involves both components being covalently linked. Similar conclusions can be drawn from the results of an X4-tropic viral assay on TZM-bl cells, which contain both CCR5 and CXCR4 on their surface. Although the only active part of the chimeric inhibitor against X4-tropic virus is C37, as much as 6000-fold enhancement of potency was observed. This effect disappeared when the CCR5 receptors were blocked, suggesting that the RANTES variant binds CCR5 and specifically delivers the C37 portion of the chimera to gp41. Mutagenesis on either part of the chimeric protein showed that both parts are essential and that they are likely functioning at the same time. Change of linker length also provided valuable information about the spatial requirements of this intramolecular mechanism.
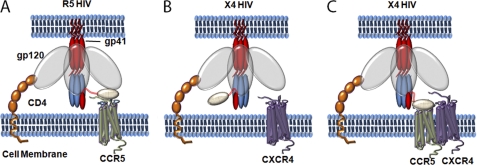
Based on these findings, we propose the following model to explain the mechanism of our chimeric inhibitors on both R5 and X4-tropic viruses. The inhibitors likely inhibit R5-tropic virus by binding to both the CCR5 co-receptor and the gp41 N-terminal trimer of hairpin simultaneously or near simultaneously. By binding to the co-receptor, the chimeric inhibitors could specifically deliver C37 near to its target on gp41 and potentially increase the local concentration of C37 on the cell surface (Fig. 5A). When inhibiting X4 virus on cells containing only CXCR4 receptors, the chimeric inhibitors behave essentially as C37 alone by binding only to gp41 (Fig. 5B). When the cells express both CCR5 and CXCR4, the chimeric inhibitors can deliver C37 to its target by binding to a CCR5 receptor that is presumably in proximity to a CXCR4 that is being used as a co-receptor for infection due to the known hetero-oligomerization of CCR5 with CXCR4 (58, 59). By specific delivery of C37 to its target and possibly increasing the local concentration of C37 on the cell surface, the chimeric inhibitors block HIV more efficiently than C37 alone (Fig. 5C).
FIGURE 5.
Model of action of the chimeric inhibitors. A, during the process of R5-tropic viral entry, the chimeric inhibitor can bind to the CCR5 receptor and block the co-receptor-gp120 interaction and at the same time deliver the C37 fusion inhibitor to the nearby gp41 targets. In this way, the chimeric inhibitor blocks R5-tropic HIV entry at both steps more effectively. B, during the process of X4-tropic viral entry, only the C37 part of the chimeric inhibitor is active, and the chimeric inhibitor functions as a fusion inhibitor by binding to the N-terminal trimer-of-hairpins of gp41. C, when the target cells of the X4-tropic virus co-express both CCR5 and CXCR4 receptors, the chimeric inhibitors can inhibit viral entry more efficiently. Because CCR5 and CXCR4 form hetero-oligomers on the cell surface, the chimeric inhibitors can bind to CCR5 and deliver the C37 peptide to the nearby X4 infection site. By specific delivery of C37 to its target and possibly increasing the local concentration of C37 on the cell surface, the chimeric inhibitors block HIV more efficiently than C37 alone.
We report here the success of a strategy to covalently link potent CCR5-binding proteins with a gp41-binding C-peptide. The chimeric inhibitors exhibited extremely high antiviral potency and were able to inhibit R5, X4, and dual-tropic viruses including clinical strains. Because the inhibitors block HIV at two steps, they are likely more resistant to viral mutations. Also, as fully recombinant inhibitors, they are inexpensive and relatively easy to produce. Overall, these inhibitors are excellent candidates for HIV microbicides. This study could also provide insight for a general approach for optimizing existing HIV entry inhibitors or designing new inhibitors.
Supplementary Material
Acknowledgments
We thank Dr. David Gravano, Dr. Ioannis Kagiampakis, Dr. Yongguang Gao, Dr. Hongjun Jin, and Dr. Yonggang Chang for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant AI079777.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2 and Tables S1–S5.
UNAIDS, 2010 Report on the Global AIDS Epidemic, WHO/UNAIDS.
- RANTES
- regulated on activation normal T cell expressed and secreted
- 5P12
- 5P12-RANTES
- 5P14
- 5P14-RANTES
- 5P12-linker-C37
- 5P12-RANTES covalently linked with gp41-binding peptide C37 via a 10-amino-acid peptide GGGGSGGGGS
- 5P14-linker-C37
- 5P14-RANTES covalently linked with gp41-binding peptide C37 via a 10-amino acid peptide GGGGSGGGGS
- HSQC
- heteronuclear single quantum coherence
- PBMC
- peripheral blood mononuclear cells.
REFERENCES
- 1. Gaertner H., Cerini F., Escola J. M., Kuenzi G., Melotti A., Offord R., Rossitto-Borlat I., Nedellec R., Salkowitz J., Gorochov G., Mosier D., Hartley O. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17706–17711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Heredia A., Gilliam B., DeVico A., Le N., Bamba D., Flinko R., Lewis G., Gallo R. C., Redfield R. R. (2007) AIDS 21, 1317–1322 [DOI] [PubMed] [Google Scholar]
- 3. Pace P., Rowley M. (2008) Curr. Opin. Drug Discovery Dev. 11, 471–479 [PubMed] [Google Scholar]
- 4. Root M. J., Steger H. K. (2004) Curr. Pharm. Des. 10, 1805–1825 [DOI] [PubMed] [Google Scholar]
- 5. Eckert D. M., Kim P. S. (2001) Annu. Rev. Biochem. 70, 777–810 [DOI] [PubMed] [Google Scholar]
- 6. Miyauchi K., Kim Y., Latinovic O., Morozov V., Melikyan G. B. (2009) Cell 137, 433–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang H. G., Williams R. E., Lin P. F. (2004) Curr. Pharm. Des. 10, 1785–1793 [DOI] [PubMed] [Google Scholar]
- 8. Emau P., Tian B., O'keefe B. R., Mori T., McMahon J. B., Palmer K. E., Jiang Y., Bekele G., Tsai C. C. (2007) J. Med. Primatol. 36, 244–253 [DOI] [PubMed] [Google Scholar]
- 9. Mori T., O'Keefe B. R., Sowder R. C., 2nd, Bringans S., Gardella R., Berg S., Cochran P., Turpin J. A., Buckheit R. W., Jr., McMahon J. B., Boyd M. R. (2005) J. Biol. Chem. 280, 9345–9353 [DOI] [PubMed] [Google Scholar]
- 10. Champagne K., Shishido A., Root M. J. (2009) J. Biol. Chem. 284, 3619–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu S., Lu H., Niu J., Xu Y., Wu S., Jiang S. (2005) J. Biol. Chem. 280, 11259–11273 [DOI] [PubMed] [Google Scholar]
- 12. Poveda E., Briz V., Soriano V. (2005) AIDS Rev. 7, 139–147 [PubMed] [Google Scholar]
- 13. Hartley O., Dorgham K., Perez-Bercoff D., Cerini F., Heimann A., Gaertner H., Offord R. E., Pancino G., Debré P., Gorochov G. (2003) J. Virol. 77, 6637–6644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hartley O., Offord R. E. (2005) Curr. Protein Pept. Sci. 6, 207–219 [DOI] [PubMed] [Google Scholar]
- 15. Hartley O., Gaertner H., Wilken J., Thompson D., Fish R., Ramos A., Pastore C., Dufour B., Cerini F., Melotti A., Heveker N., Picard L., Alizon M., Mosier D., Kent S., Offord R. (2004) Proc. Natl Acad. Sci. U.S.A. 101, 16460–16465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kawamura T., Bruse S. E., Abraha A., Sugaya M., Hartley O., Offord R. E., Arts E. J., Zimmerman P. A., Blauvelt A., Bruce S. E. (2004) J. Virol. 78, 7602–7609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lederman M. M., Veazey R. S., Offord R., Mosier D. E., Dufour J., Mefford M., Piatak M., Jr., Lifson J. D., Salkowitz J. R., Rodriguez B., Blauvelt A., Hartley O. (2004) Science 306, 485–487 [DOI] [PubMed] [Google Scholar]
- 18. Pastore C., Picchio G. R., Galimi F., Fish R., Hartley O., Offord R. E., Mosier D. E. (2003) Antimicrob. Agents Chemother. 47, 509–517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuhmann S. E., Hartley O. (2008) Annu. Rev. Pharmacol. Toxicol. 48, 425–461 [DOI] [PubMed] [Google Scholar]
- 20. Gaertner H., Lebeau O., Borlat I., Cerini F., Dufour B., Kuenzi G., Melotti A., Fish R. J., Offord R., Springael J. Y., Parmentier M., Hartley O. (2008) Protein Eng. Des. Sel. 21, 65–72 [DOI] [PubMed] [Google Scholar]
- 21. Cocchi F., DeVico A. L., Garzino-Demo A., Arya S. K., Gallo R. C., Lusso P. (1995) Science 270, 1811–1815 [DOI] [PubMed] [Google Scholar]
- 22. Root M. J., Kay M. S., Kim P. S. (2001) Science 291, 884–888 [DOI] [PubMed] [Google Scholar]
- 23. Kahle K. M., Steger H. K., Root M. J. (2009) PLoS Pathog. 5, e1000674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Furuta R. A., Wild C. T., Weng Y., Weiss C. D. (1998) Nat. Struct. Biol. 5, 276–279 [DOI] [PubMed] [Google Scholar]
- 25. Chan D. C., Chutkowski C. T., Kim P. S. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 15613–15617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu M., Kim P. S. (1997) J. Biomol. Struct. Dyn. 15, 465–471 [DOI] [PubMed] [Google Scholar]
- 27. Stoddart C. A., Nault G., Galkina S. A., Thibaudeau K., Bakis P., Bousquet-Gagnon N., Robitaille M., Bellomo M., Paradis V., Liscourt P., Lobach A., Rivard M. E., Ptak R. G., Mankowski M. K., Bridon D., Quraishi O. (2008) J. Biol. Chem. 283, 34045–34052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hamburger A. E., Kim S., Welch B. D., Kay M. S. (2005) J. Biol. Chem. 280, 12567–12572 [DOI] [PubMed] [Google Scholar]
- 29. Wishart D. S., Bigam C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. (1995) J. Biomol. NMR 6, 135–140 [DOI] [PubMed] [Google Scholar]
- 30. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 31. Garrett D. S., Powers R., Gronenborn A. M., Clore G. M. (1991) J. Magn. Reson. 95, 214–220 [DOI] [PubMed] [Google Scholar]
- 32. Pleskoff O., Tréboute C., Brelot A., Heveker N., Seman M., Alizon M. (1997) Science 276, 1874–1878 [DOI] [PubMed] [Google Scholar]
- 33. Takeuchi Y., McClure M. O., Pizzato M. (2008) J. Virol. 82, 12585–12588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wei X., Decker J. M., Liu H., Zhang Z., Arani R. B., Kilby J. M., Saag M. S., Wu X., Shaw G. M., Kappes J. C. (2002) Antimicrob. Agents Chemother. 46, 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Derdeyn C. A., Decker J. M., Sfakianos J. N., Wu X., O'Brien W. A., Ratner L., Kappes J. C., Shaw G. M., Hunter E. (2000) J. Virol. 74, 8358–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Platt E. J., Wehrly K., Kuhmann S. E., Chesebro B., Kabat D. (1998) J. Virol. 72, 2855–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ciminale V., Felber B. K., Campbell M., Pavlakis G. N. (1990) AIDS Res. Hum. Retroviruses 6, 1281–1287 [DOI] [PubMed] [Google Scholar]
- 38. Vodicka M. A., Goh W. C., Wu L. I., Rogel M. E., Bartz S. R., Schweickart V. L., Raport C. J., Emerman M. (1997) Virology 233, 193–198 [DOI] [PubMed] [Google Scholar]
- 39. Gartner S., Markovits P., Markovitz D. M., Kaplan M. H., Gallo R. C., Popovic M. (1986) Science 233, 215–219 [DOI] [PubMed] [Google Scholar]
- 40. Gendelman H. E., Baca L. M., Kubrak C. A., Genis P., Burrous S., Friedman R. M., Jacobs D., Meltzer M. S. (1992) J. Immunol. 148, 422–429 [PubMed] [Google Scholar]
- 41. Westervelt P., Gendelman H. E., Ratner L. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 3097–3101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gendelman H. E., Orenstein J. M., Baca L. M., Weiser B., Burger H., Kalter D. C., Meltzer M. S. (1989) AIDS 3, 475–495 [DOI] [PubMed] [Google Scholar]
- 43. Gendelman H. E., Orenstein J. M., Martin M. A., Ferrua C., Mitra R., Phipps T., Wahl L. A., Lane H. C., Fauci A. S., Burke D. S., et al. (1988) J. Exp. Med. 167, 1428–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Popovic M., Read-Connole E., Gallo R. C. (1984) Lancet 2, 1472–1473 [DOI] [PubMed] [Google Scholar]
- 45. Popovic M., Sarngadharan M. G., Read E., Gallo R. C. (1984) Science 224, 497–500 [DOI] [PubMed] [Google Scholar]
- 46. Ratner L., Haseltine W., Patarca R., Livak K. J., Starcich B., Josephs S. F., Doran E. R., Rafalski J. A., Whitehorn E. A., Baumeister K., et al. (1985) Nature 313, 277–284 [DOI] [PubMed] [Google Scholar]
- 47. Connor R. I., Chen B. K., Choe S., Landau N. R. (1995) Virology 206, 935–944 [DOI] [PubMed] [Google Scholar]
- 48. Chang L. J., Urlacher V., Iwakuma T., Cui Y., Zucali J. (1999) Gene Ther. 6, 715–728 [DOI] [PubMed] [Google Scholar]
- 49. Cheng-Mayer C., Liu R., Landau N. R., Stamatatos L. (1997) J. Virol. 71, 1657–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Stamatatos L., Wiskerchen M., Cheng-Mayer C. (1998) AIDS Res. Hum. Retroviruses 14, 1129–1139 [DOI] [PubMed] [Google Scholar]
- 51. Stamatatos L., Lim M., Cheng-Mayer C. (2000) AIDS Res. Hum. Retroviruses 16, 981–994 [DOI] [PubMed] [Google Scholar]
- 52. Page K. A., Landau N. R., Littman D. R. (1990) J. Virol. 64, 5270–5276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li Y., Svehla K., Mathy N. L., Voss G., Mascola J. R., Wyatt R. (2006) J. Virol. 80, 1414–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gao F., Morrison S. G., Robertson D. L., Thornton C. L., Craig S., Karlsson G., Sodroski J., Morgado M., Galvao-Castro B., von Briesen H., Beddows S., Weber J., Sharp P. M., Shaw G. M., Hahn B. H. (1996) J. Virol. 70, 1651–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li M., Gao F., Mascola J. R., Stamatatos L., Polonis V. R., Koutsoukos M., Voss G., Goepfert P., Gilbert P., Greene K. M., Bilska M., Kothe D. L., Salazar-Gonzalez J. F., Wei X., Decker J. M., Hahn B. H., Montefiori D. C. (2005) J. Virol. 79, 10108–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lackman-Smith C., Osterling C., Luckenbaugh K., Mankowski M., Snyder B., Lewis G., Paull J., Profy A., Ptak R. G., Buckheit R. W., Jr., Watson K. M., Cummins J. E., Jr., Sanders-Beer B. E. (2008) Antimicrob. Agents Chemother. 52, 1768–1781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ptak R. G., Gallay P. A., Jochmans D., Halestrap A. P., Ruegg U. T., Pallansch L. A., Bobardt M. D., de Béthune M. P., Neyts J., De Clercq E., Dumont J. M., Scalfaro P., Besseghir K., Wenger R. M., Rosenwirth B. (2008) Antimicrob. Agents Chemother. 52, 1302–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Babcock G. J., Farzan M., Sodroski J. (2003) J. Biol. Chem. 278, 3378–3385 [DOI] [PubMed] [Google Scholar]
- 59. Percherancier Y., Berchiche Y. A., Slight I., Volkmer-Engert R., Tamamura H., Fujii N., Bouvier M., Heveker N. (2005) J. Biol. Chem. 280, 9895–9903 [DOI] [PubMed] [Google Scholar]
- 60. Kopetzki E., Jekle A., Ji C., Rao E., Zhang J., Fischer S., Cammack N., Sankuratri S., Heilek G. (2008) Virol. J. 5, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kagiampakis I., Gharibi A., Mankowski M. K., Snyder B. A., Ptak R. G., Alatas K., LiWang P. J. (2011) Antimicrob. Agents Chemother. 55, 264–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ji C., Kopetzki E., Jekle A., Stubenrauch K. G., Liu X., Zhang J., Rao E., Schlothauer T., Fischer S., Cammack N., Heilek G., Ries S., Sankuratri S. (2009) J. Biol. Chem. 284, 5175–5185 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.