Abstract
Krüppel-like transcription factors (KLFs) have elicited significant attention because of their regulation of essential biochemical pathways and, more recently, because of their fundamental role in the mechanisms of human diseases. Neonatal diabetes mellitus is a monogenic disorder with primary alterations in insulin secretion. We here describe a key biochemical mechanism that underlies neonatal diabetes mellitus insulin biosynthesis impairment, namely a homozygous mutation within the insulin gene (INS) promoter, c.-331C>G, which affects a novel KLF-binding site. The combination of careful expression profiling, electromobility shift assays, reporter experiments, and chromatin immunoprecipitation demonstrates that, among 16 different KLF proteins tested, KLF11 is the most reliable activator of this site. Congruently, the c.-331C>G INS mutation fails to bind KLF11, thus inhibiting activation by this transcription factor. Klf11−/− mice recapitulate the disruption in insulin production and blood levels observed in patients. Thus, these data demonstrate an important role for KLF11 in the regulation of INS transcription via the novel c.-331 KLF site. Lastly, our screening data raised the possibility that other members of the KLF family may also regulate this promoter under distinct, yet unidentified, cellular contexts. Collectively, this study underscores a key role for KLF proteins in biochemical mechanisms of human diseases, in particular, early infancy onset diabetes mellitus.
Keywords: Diabetes, Genetic Diseases, Glucose Metabolism, Pancreatic Islet, Transcription Regulation, KLF
Introduction
KLF7 proteins constitute a group of 16 different proteins that are structurally characterized by the presence of an Sp1-like three-zinc finger domain at their C terminus and a variable transcriptional regulatory domain at their N terminus (1, 2). This family of proteins has elicited significant attention because of several important features. By regulating gene expression, at the cellular level, these proteins can modulate cell proliferation, apoptosis, senescence, cell adhesion, angiogenesis, and metabolism (3, 4). Emerging evidence demonstrates that KLF genes can regulate cardiovascular development, bone homeostasis, brain function, pancreatic homeostasis, and liver morphogenesis and metabolic health (5). These data have led to a nascent area in biochemistry, namely addressing to what extent KLF proteins are involved in novel mechanisms of human diseases. Although their role in cancer pathways has been partially characterized, the field is rapidly expanding to address their mechanisms in altering human health. Most recently, a study associating mutations in a KLF1-binding site with anemia has been reported (6). These data have laid the foundation for unique avenues of investigation into the biochemistry of KLF proteins across the spectrum of human diseases.
Congruent with these new lines of discovery, the current study primarily deals with the biochemical characterization of a novel KLF-mediated transcriptional pathway that regulates the insulin gene (INS). When disrupted, this pathway leads to neonatal diabetes mellitus (NDM), a monogenic and nonautoimmune form of diabetes in early infancy. The discovery is important, because KLF-mediated pathways have been previously associated with diabetes in human genetic studies. The first study on the role of KLF proteins in diabetes revealed that missense mutations in KLF11 cause a form of MODY (7). Additionally, it has been demonstrated that KLF11 may contribute to MODY by directly regulating the promoter of PDX1 (8). Candidate gene studies have suggested that single-nucleotide polymorphisms in KLF11 and KLF7 loci may also associate with type 2 diabetes risk (9, 10). Lastly, KLF14, a gene identified by our laboratory, was recently found as a key link to this disease via genome-wide association studies (11). However, mechanistic information on how these pathways trigger disease is currently lacking. We report the characterization of a novel, functional KLF cis-regulatory region located at the c.-331 position within the human INS promoter that activates INS expression. Mutations at this site have been recently described in NDM patients (12), although its characterization as a KLF binding site, its transcriptional activity, and the contribution of chromatin modifying enzymes to this function have not been reported. Following genetic confirmation in an independent NDM cohort study and the discovery of the c.-331 C>G INS promoter mutation, a family-wide screening of the 16 distinct KLF proteins demonstrates that KLF11 is a consistent activator of this promoter via a p300-mediated mechanism. ChIP assays demonstrate that KLF11 binds to the conserved c.-331 KLF binding site in the rat Ins2 gene, demonstrating that this site is occupied by KLF11 in vivo. The c.-331 C>G INS promoter mutation present in NDM disrupts KLF11 binding to this site, hindering INS transcription. In addition, the Klf11−/− mouse recapitulates alterations in insulin biosynthesis. Collectively, alterations in KLF11-mediated transcription by three different mechanisms, namely KLF11 mutations involved in MODY (7), as well as the two described here, the c.-331 mutant INS promoter involved in NDM and Klf11 deletion in mice, strongly support a role for KLF11 protein in insulin biosynthesis and diabetes development to a greater extent than previously anticipated. Furthermore, our screening data suggest that, in addition to KLF11, a small subset of other KLF proteins could regulate this promoter under distinct cellular contexts that remain to be defined. More broadly, this study sheds light on a promising avenue of biochemical research, namely how alterations of novel KLF-mediated pathways may underlie the mechanisms of important and common human diseases.
EXPERIMENTAL PROCEDURES
Molecular Genetic Analysis of the Human INS Promoter
Mutations were screened in six NDM patients from consanguineous parents (clinical characteristics are shown in supplemental Table S1) who were referred to the French Network for the Study of NDM (13). All of the patients were negative for any missense mutations in KCNJ11, ABCC8, and INS or for abnormalities in the chromosome 6q24. Control individuals were 355 and 747 nondiabetic adults from France and Turkey, respectively. Local ethics committees approved the study, and written parental consent was obtained for genetic testing of subjects. The human INS promoter (from nucleotide c.-448) was amplified by PCR (conditions available upon request to authors) and sequenced using an automated 3730xl DNA Analyzer (Applied Biosystems). Electrophoregram reads were assembled and compared with the reference sequence NM_000207.2, using the SeqScape software (version v2.5; Applied Biosystems). Nucleotide numbers and variant locations are displayed by base numbers counting from the ATG translation initiation codon according to the Human Genome Variation Society nomenclature.
KLF Expression Analyses in Human Pancreatic Islets and β Cells
For microarray analysis, human islets of Langerhans (n = 3) and sorted β cells (n = 3) were isolated from adult brain-dead donors in accordance with French regulations and with local institutional ethical committee approval, as previously described (14, 15). RNAs from these samples were processed using the TotalPrep RNA amplification kit (Ambion Illumina). cDNAs were fluorescently labeled and hybridized to the HumanHT-12_v4_BeadChip array according to the manufacturer's instructions (Illumina). Fluorescence intensities were controlled, quantified, and analyzed using BeadStudio (Illumina). Each cDNA sample was hybridized twice to the microarray. Expression was confirmed by standard PCR from two independent cDNA samples of both human islets and β cells, using the FastStart Taq (Roche Applied Science).
Cell Culture, Transfection, and Luciferase Assay
The INS promoter reporter construct was a kind gift of Danielle Melloul (Hadassah University Hospital, Jerusalem, Israel) (16). The c.-331C>G mutated INS promoter was generated using QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's recommendations and verified by sequencing. Rat INS-1 and mouse β-TC-3 β cells were cultured as previously described (17, 18). The cell lines (∼80% confluent) were electroporated in 0.4-cm cuvettes (2 × 106 cells) at 350 V (INS-1) or 275 V (β-TC-3) for one 10-ms pulse with 5 μg of the wild-type (WT) or mutated human INS promoter reporter plasmid using an ECM 830 square wave electroporator (BTX Harvard Apparatus). Where indicated, the cells were co-transfected with 5 μg of empty vector or each human KLF construct. In experiments with dominant negative (DN) p300, 3 μg of the WT INS promoter reporter plasmid was co-transfected with 3 μg of parental control vector or KLF11 and 6 μg of parental control vector or DN-p300 (8). Forty-eight hours after transfection, the cells were lysed, and reporter activity was read using the Luciferase assay system (Promega) and a 20/20 luminometer (Turner Designs), according to the manufacturer's protocol. The data in relative light units were normalized to control and shown as the means ± S.E. of the means. The experiments were performed in triplicate, three independent times. Statistics was performed using Student's t tests.
EMSA
Annealed oligonucleotides corresponding to the c.-331 KLF binding site from the INS promoter (5′-ATCTGCCGACCCCCCCACCCCAGGCCCTAATG-3′), its human c.-331C>G mutation (5′-ATCTGCCGACCCCCCGACCCCAGGCCCTAATG-3′), and the corresponding rat Ins2 promoter (5′-TGGCCATCTGCTGATCCACCCTTAATGGGACA-3′) were end-labeled with [γ-32P]ATP using T4 polynucleotide kinase, as indicated by the manufacturer (Promega). Recombinant fusion protein expression was induced in BL21 cells (Stratagene) and purified by using glutathione-Sepharose 4B affinity chromatography in accordance with the manufacturer's instructions (Amersham Biosciences). Subsequently, EMSA was performed as previously described (19). Where indicated, the following were added to the binding reactions: 2 μg of anti-GST antibody, 2 μg of anti-mIgG antibody, or 250-fold molar excess unlabeled WT hINS probe.
ChIP Assay
ChIP assay was performed as previously described (20). Briefly, INS-1 cells were transduced with adenovirus containing polyhistidine-tagged KLF11 or an empty vector control at a multiplicity of infection of 10 virus particles per cell. A 143-bp region of the rIns2 promoter was amplified by PCR, using specific primers to encompass the potential KLF binding site (forward, 5′-TAGCACCAGGCAAGTGTTTG-3′; reverse, 5′-GGGGTTACTGAATCCCCACT-3′) and visualized by 2.5% agarose gel.
Generation of Klf11−/− Mice
The Klf11 knock-out model was generated at the University of Washington, Seattle, following standard homologous recombination techniques to inactivate the endogenous Klf11 gene in embryonic stem cells, generating chimeras, and isolating colony founders carrying the knock-out of this gene (21). This animal was originally generated in a mixed background and subsequently transferred to the Mayo Animal Facilities where it was crossed back into a pure C57BL/6 background for more than 20 generations to produce the inbred strain used in this study. In all of the experiments, Klf11−/− animals were compared with Klf11+/+ littermates.
Blood Glucose and Serum Insulin Measurements and Oral Glucose Tolerance Test in Mice
Blood glucose measurements were obtained from tail vein bleeding using OneTouch Ultra glucometer (Lifescan; Johnson & Johnson). Serum harvested from cardiac puncture was centrifuged at 500 × g at 4 °C for 20 min and stored at −20 °C for assays. Serum insulin levels were measured with insulin ELISA kit using mouse insulin standards (Crystal Chem Inc.). For the oral glucose tolerance test (OGTT), the mice were fasted overnight and were given d-glucose solution at 2 g/kg of body weight by gavage. Serial blood glucose and serum insulin measurements (0, 20, 60, and 120 min) were obtained from tail vein bleeding. Blood glucose and serum insulin were measured as stated above and comparable with previous studies (22, 23). Statistics were performed using an analysis of variance model.
Immunohistochemistry
Insulin levels by immunohistochemistry were examined in six Klf11−/− mice and their corresponding gender-, age-, and weight-matched littermates. Analysis in each mouse was performed in islets from different regions of the pancreas and utilizing 10 sections, sampling different depths within the organ. Sections were incubated with anti-insulin antibody (Sigma) for 1 h at room temperature. The detection reaction was performed as previously described (20).
RESULTS
Genetic Screening Identifies a Mutation within the INS Promoter in NDM Patients That Impairs INS Transcription
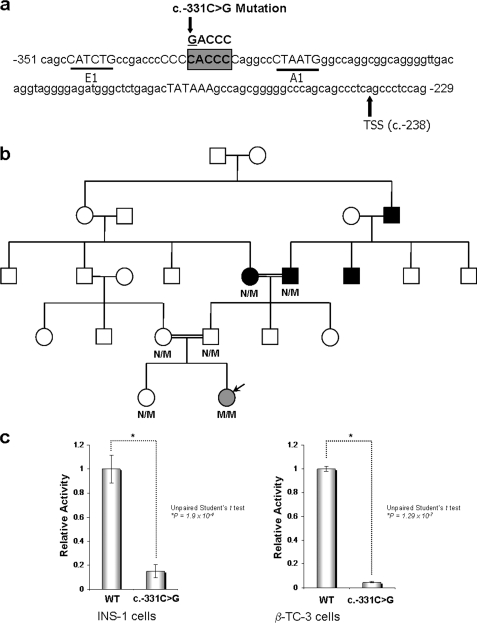
We searched for novel mechanisms underlying the regulation of the human INS gene as revealed from genetic studies of the INS promoter. We screened for genetic alterations that may target the proximal INS promoter in patients with NDM (supplemental Table S1) from six consanguineous families who were negative for any missense mutations in KCNJ11, ABCC8, and INS, or chromosome 6q24 abnormalities (i.e. the most frequent causes of NDM (24–26)). Through this approach, we found only one homozygous mutation (c.-331C>G) within the INS promoter in two NDM patients from consanguineous Turkish families, which was not present in 1,002 control individuals (two different control cohorts of 355 and 747 nondiabetic adults from French and Turkish origins, respectively). The c.-331C>G mutation is located between the E1 and A1 elements (27–29), which is a highly conserved region across many mammalian species (Fig. 1a). The existence of this mutation was also confirmed in an independent cohort of NDM patients (12), thus fulfilling the Medical Genetics gold standard of independent validation, lending high reliability to the existence and importance of this mutation. An extended genotype-phenotype correlation of patients affected by this mutation as well as the mechanism underlying its effect on insulin transcription remained to be determined. Consequently, we provide a detailed comparative description of the clinical genetic features of NDM in patients carrying the c.-331 mutation and clinical characteristics of members from one of the families (Fig. 1b and supplemental Tables S1 and S2). Briefly, the two individuals carrying the homozygous c.-331C>G INS promoter mutation displayed severe intrauterine growth retardation and alteration in insulin biosynthesis that requires supplementation with this hormone (0.3–0.4 units/kg/day) at birth. In the proband-1, discontinuation of insulin therapy triggered an abnormal OGTT, showing a delayed peak of both plasma glucose and serum insulin levels (supplemental Table S2). A recent study reported that isolated rat β cells treated with glibenclamide exhibited higher rates of proinsulin synthesis (30). Unfortunately, attempts to increase insulin levels by administration of sulfonylurea did not restore the metabolic balance because of a failure in normalizing the levels of this hormone. Ultimately, the proband-1 requires bolus insulin therapy (0.08 units/kg/day), further reflecting its defect in insulin biosynthesis. Therefore, we sought to investigate the functional behavior of the c.-331C>G mutated human INS promoter using reporter assays. Interestingly, INS-1 cells demonstrate up to an 85% reduction (± 5.2%) in promoter activity upon mutation, whereas β-TC-3 cells have reduced activity up to 96.4% (± 0.9%), thus recapitulating the insulin defect observed in the mutated NDM patients (Fig. 1c). These experiments also revealed that the severe disruption in transcriptional activity caused by the c.-331C>G mutation is typical of alterations at the level of basal transcription, which, as demonstrated by our laboratory in studies on KLF13-mediated regulation of the CYP1A1 promoter, also compromise activated transcription (31). Collectively, the information provided here on the c.-331C>G mutation is of significant relevance to biochemistry and medicine, because these results provide a more complete genotype-phenotype correlation, which is critical to the objectives of Online Mendelian Inheritance in Man for further research in this area. More importantly, it represents an optimal model for functional studies aimed at describing novel mechanisms that underlie forms of diabetes that result from INS promoter dysfunction. Subsequently, a variety of methods were used to identify how this mutation impairs insulin transcription at the biochemical level.
FIGURE 1.
Mutation and functional analysis of the INS promoter in NDM. a, diagram shows the human INS promoter sequence surrounding the c.-331 site (c.-229 to c.-351). The previously described cis-regulatory elements, E1 and A1, are in capital letters and underlined. The TATA box is also in capital letters, and the transcription start site (TSS) is indicated with an arrow (c.-238). The CACCC box, which is recognized by the KLF protein family is in capital letters and highlighted in a gray box with the c.-331C>G mutation of this element shown above the sequence. The numbering is described relative to the translational start site (c.1) according to Human Genome Variation Society guidelines. b, pedigree of Family 1. Proband-1-CE is indicated by a black arrow. c, loss of INS promoter activity with the c.-331C>G mutation in both INS-1 and β-TC-3 cells. Luciferase-based reporter assays were performed using the WT and c.-331C>G mutant INS promoters. The mean, standard error, and p values are shown for three replicates performed independently at least three times.
The c.-331C>G Mutation Disrupts a Novel KLF-binding Site That Activates INS Transcription in a p300-dependent Manner
The c.-331C>G mutation maps to a CACCC sequence motif. DNA binding data from SELEX and mutational analysis of GC/GT boxes performed by our laboratory demonstrate that the CACCC sequence conforms perfectly to a KLF binding site (7, 8). Because the c.-331C>G mutation strongly decreases human INS promoter activity (Fig. 1c), we hypothesized that this dysfunction is caused by impaired binding of a KLF activator protein, yet to be defined. KLF transcription factors form a group of 16 proteins that bind with similar selectivity, although unpredictable affinity, to the CACCC box (32). To identify which KLF(s) bind and regulate the c.-331 CACCC site but not the mutated GACCC site in NDM patients, we designed a four-tier biochemical screening approach.
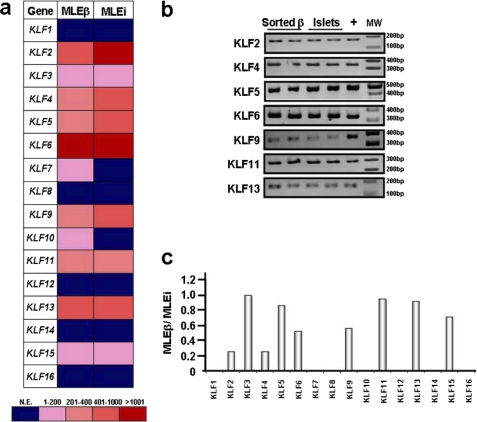
First, we performed a careful genetic-based survey of which KLF genes are expressed in human islets and β cells to identify the first candidate regulators of the c.-331 CACCC site, as NDM is due to severe defects of insulin secretion by the pancreatic β cell (24–26). For this purpose, we designed a KLF family-wide expression microarray (Illumina technology). The median levels of expression (MLEs) in three samples of islets (MLEi) or FACS-assisted sorted β cells (MLEβ) were determined for each KLF gene. We found that among all KLF genes, KLF1, KLF7, KLF8, KLF10, KLF12, KLF14, and KLF16 are not expressed in both human islets and sorted β cells. In contrast, KLF2, KLF4, KLF5, KLF6, KLF9, KLF11, and KLF13 are highly expressed in both human islets and sorted β cells (>250 MLE; Fig. 2a). These data were also confirmed by RT-PCR (Fig. 2b). In sorted human β cells, some expression/enrichment values remained the same as in the whole islets, suggesting concentrations of distinct KLF mRNA in this specific cell population, whereas others decreased, suggesting that another cell type within the islet may be enriched with those KLFs. This phenomenon is better captured by the MLEβ/MLEi ratio. Fig. 2c shows that distinct KLF genes were enriched by our FACS-sorted human β cell population versus the islet, as indicated by a MLEβ/MLEi ratio close to 1 (> 0.85) for KLF3 (0.99), KLF5 (0.86), KLF11 (0.95), and KLF13 (0.92), which demonstrates that most of the expression signal detected in islets for these genes was then recovered in the sorted β cells. On the other hand, KLF2 (0.26), KLF4 (0.26), KLF6 (0.52), KLF9 (0.56), and KLF15 (0.72) had a MLEβ/MLEi ratio of less than 0.85, indicating that the expression in the entire islet cannot be accounted for by its expression in sorted β cells. However, it is important to note that in the case of KLF3 and KLF15, the relative abundance of transcript was not significant in either islets (MLEi) or sorted β cells (MLEβ) (KLF3: MLEi = 48.9, MLEβ = 48.5; KLF15: MLEi = 20.2, MLEβ = 14.5). All of these values, as well as their relative enrichment in either islet or β cells, as defined by the percentage of genes that fall lower in expression levels than each KLF protein, are provided in supplemental Table S3. This careful array experiment demonstrates that pancreatic islet β cells are significantly enriched in specific KLF proteins that likely influence the biology of these cells by driving a complex cascade of gene expression. In addition, this constitutes the most comprehensive expression analysis of KLF genes in β cells thus far reported and, as the first step of our screening, was efficient in ruling out nine KLF proteins (KLF1, KLF3, KLF7, KLF8, KLF10, KLF12, KLF14, KLF15, and KLF16) as candidates for the regulation of the c.-331 CACCC site and warranted further testing of the remaining KLF family members via well characterized biochemical assays.
FIGURE 2.
Expression profiling of KLF genes reveals enrichment of distinct KLF proteins in human islets and β cells. Illumina-based KLF gene family-wide array profiling was used to determine the expression profile in whole human islets and FACS-assisted sorted human β cells. a, heat map indicating the relative value of expression of each KLF gene in a particular sample (see supplemental Table S3 for numerical data). Color scale of values is shown below. b, expression of KLF proteins found to be expressed by Illumina array was confirmed by RT-PCR in two cDNA samples of each, sorted human β cells and human islets. Positive control (+) is total human cDNA. MW, molecular weight marker. c, histogram showing the relative enrichment of KLF genes specifically in human β cells within the islet as defined by the ratio between the median level of expression in sorted β cells compared with whole islets (MLEβ/MLEi). KLF3, KLF5, KLF11, and KLF13 had values close to 1 (>0.85), indicating that most of the expression signal detected in islets for these genes was recovered in the sorted β cells. Note, however, that the abundance of KLF3 transcript was not significant in either whole islets or β cells, as shown in Fig. 2A.
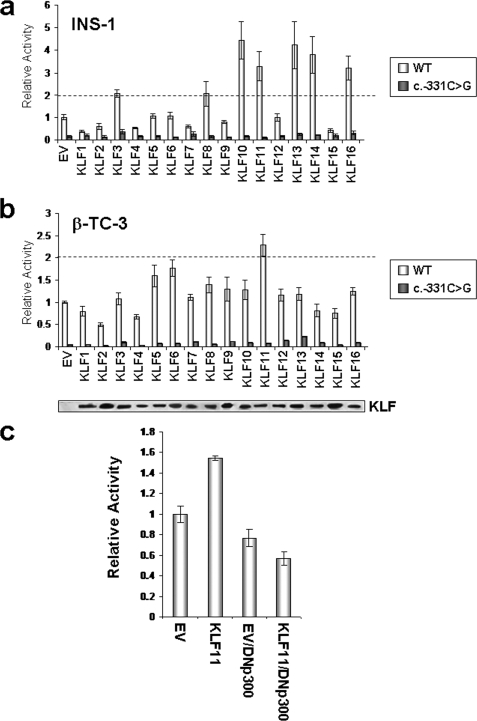
Next, we performed reporter experiments to assess which KLF proteins activate the wild-type but not the mutant INS promoter. Because the investigation of human INS gene regulation is problematic, because human β cell lines are unavailable, we performed this reporter-based screening step using two independently derived rodent lines, rat INS-1 (Fig. 3a) and mouse β-TC-3 (Fig. 3b) β cells. For the proper evaluation of these experiments, we statistically defined the a priori criteria that proteins that could only regulate the transfected human INS promoter in two different, well established β cell models constitute the best candidates for transcriptional regulators of the c.-331 CACCC site and thus justify additional biochemical characterizations. Fig. 3a demonstrates that the KLF family members, KLF10 (4.5-fold ± 0.17), KLF11 (3.3-fold ± 0.11), KLF13 (4.2-fold ± 0.25), KLF14 (3.8-fold ± 0.22), and KLF16 (3.2-fold ± 0.31) significantly increased wild-type INS promoter activity in INS-1 cells, but this activity was abolished by the c.-331C>G mutation. However, according to our array data (Fig. 2), KLF10, KLF14, and KLF16 are not readily expressed in β cells. Fig. 3b shows that in β-TC-3 cells, KLF11 activated this promoter 2.3-fold (± 0.08), whereas KLF5 and KLF6 had less effect (mean 1.6- and 1.8-fold, respectively). Because KLF5 and KLF6 had no activity in INS-1 cells, following our rigorous a priori evaluation criteria, they were eliminated as candidates.
FIGURE 3.
KLF11 activates the human INS promoter but not the c.-331C>G mutant promoter. a, rat INS-1 β cells were co-transfected with either the WT or c.-331C>G mutant INS promoter reporter construct and each of the KLF family members (KLF1–KLF16) or empty vector (EV) control. Luciferase activity was measured, and the means and standard errors (as indicated by the error bars) were determined for each experimental condition from triplicates of three independent experiments. The dashed line represents the 2-fold cut-off value for significant activation above empty vector control. b, KLF11 was the only KLF family member to increase WT INS promoter activity more than 2-fold in β-TC-3 cells, but this activity was abolished with the c.-331C>G mutant promoter. His- and FLAG-tagged KLF protein expression was confirmed by Western blot (lower panel). c, KLF11 requires functional p300 to activate the INS promoter. INS-1 cells were co-transfected with the WT INS promoter and either empty vector control or KLF11 along with an empty vector control or a DN-p300. Note that plasmid DNA amounts were adjusted to accommodate either control vector or DN-p300; thus KLF11 activation of the WT INS promoter was adjusted to 1.6-fold above empty vector control. The mean promoter activity ± standard error is shown relative to WT INS promoter transfected with control empty vector.
Notably, however, KLF11 consistently activated the INS promoter in both cell lines and is highly present in both human islets and β cells. Thus, KLF11 became our focus as a regulator of the c.-331 CACCC site. Mechanistically, Fig. 3c demonstrates that KLF11 requires the functional histone acetyltransferase, p300, to activate the human INS promoter in the INS-1 β cell line, thus linking regulation of the c.-331 mutation to a distinct chromatin-mediated pathway. KLF11 increased activity of the wild-type INS promoter, but co-transfection with DN-p300 abolished this activity (74% of empty vector control with DN-p300 ± 6.7%), supporting a significant role for this histone acetyltransferase in normal β cell function and insulin secretion. Other histone acetyltransferases such as p300/CREB binding protein-associated factor and CREB binding protein did not modify the activating function of KLF11 (data not shown). This result is important, because p300 mechanisms and acetylation of histones have been shown by ChIP to be dominant in INS expression (33–35).
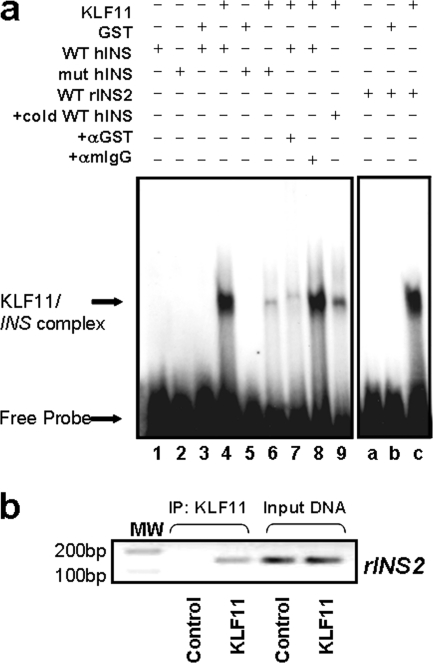
In our third screening step, we assessed whether the c.-331C>G mutation impairs KLF11-mediated INS activation. By EMSA, we investigated whether KLF11 binds the c.-331 CACCC site in both human INS and rat Ins2 promoters. More importantly, we determined whether the human c.-331C>G mutation disrupts this binding. We analyzed the ability of KLF11 to bind to the c.-331 wild-type site with a 32-mer probe containing the CACCC box. The extended length of the EMSA probe was able to better represent the context within the human INS promoter than shorter sequences (corresponding to c.-346 to c.-315 nucleotides; Fig. 4a). We also assessed the ability of KLF11 to bind to a similar probe representing the NDM c.-331C>G mutation, which alters the core CACCC box to GACCC. Fig. 4a demonstrates that KLF11 binds to the wild-type INS promoter binding site (lane 4). This binding, however, was significantly disrupted using the c.-331C>G NDM mutant sequence (lane 6). The binding activity of KLF11 to this sequence was found to be specific by supershift using antibodies (lanes 7 and 8). Competition assays demonstrated that the KLF11 binding activity was competed with unlabeled wild-type probe (lane 9). A probe corresponding to the same conserved region of the rat Ins2 promoter also demonstrated that KLF11 indeed forms a complex with this site in vitro (lanes a–c). Together, these studies demonstrate that KLF11 specifically binds to the c.-331 CACCC site of the human INS and rat Ins2 promoters. Thus, these data characterize a novel biochemical role for KLF11, namely to bind and regulate the WT c.-331 CACCC site of the INS promoter and reveal that this binding is disrupted by the c.-331C>G mutation found in NDM.
FIGURE 4.
KLF11 binds to the c.-331 novel KLF binding site, which is disrupted by the c.-331C>G NDM mutation. a, EMSA was performed using either the wild-type human INS CACCC promoter site (WT hINS; lanes 1, 3, 4, and 7–9) or the c.-331C>G mutation (mut hINS; lanes 2, 5, and 6) with GST protein (lanes 3 and 5), recombinant KLF11 (KLF11; lanes 4 and 6–9), or probe alone (lanes 1 and 2). The specific complexes that form between KLF11 and the probe are indicated on the left. Although the anti-GST antibody shifted the KLF11-WT hINS complex (lane 7), the same amount of an anti-mIgG antibody did not (lane 8). Notably, KLF11 did not shift the c.-331C>G mutant hINS probe (lane 6). Furthermore, the addition of an excess of unlabeled WT hINS probe competed for the binding (lane 9). Finally, the corresponding region of the rat Ins2 promoter was utilized as a probe for gel shift assays (WT rIns2, lanes a–c) with control GST protein alone (lane b), KLF11 (KLF11; lane c), or probe alone (lane a), which demonstrated that KLF11 indeed forms a complex with this region of the rat Ins2 promoter in vitro (lane c). b, KLF11 occupies the CACCC box of the rat Ins2 promoter in vivo. The region of the Ins2 promoter fragment containing the conserved CACCC box was amplified by PCR after ChIP from KLF11-infected cells but not the empty vector-infected sample immunoprecipitated (IP) with the same anti-His antibody (negative control), demonstrating that this region of the rIns2 promoter is a target of KLF11 in INS-1 β cells. Positive amplification of PCR products is shown in the input DNA lanes, demonstrating that this region of the rIns2 promoter is present in all samples before immunoprecipitation. Anti-mouse IgG was used as an additional negative control (data not shown). MW, molecular weight marker.
Finally, we performed ChIP assays to determine whether the region of the evolutionarily conserved CACCC site was occupied in vivo by KLF11. The conservation of this site in the rat Ins2 promoter allowed the utilization of ChIP assays in INS-1 cells to assess whether KLF11 binds to the endogenous promoter in vivo. Fig. 4b shows that indeed this KLF site within the Ins2 promoter is an actual target of KLF11. It is noteworthy that in this study, we utilized epitope-tagged KLF11 protein because, despite extensive testing and experimentation, a specific KLF11 antibody that is useful for ChIP assays remains to be developed (36). In conclusion, our results demonstrate that disruption in KLF11-mediated activation of the c.-331 CACCC site within the human INS promoter compromises a p300-dependent chromatin remodeling pathway that regulates basal INS transcriptional activity, which may also impact its inducible expression and contribute to the pathogenesis of NDM. Lastly, our results do not rule out the possibility that other KLF proteins, distinct from KLF11, may mediate this function under different cell biological contexts (e.g. activation of different signaling pathways).
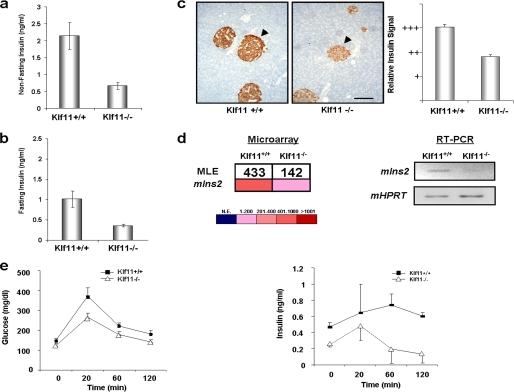
Mouse Modeling Experiments Demonstrate That Disruption of Klf11 Function Impairs Insulin Biosynthesis
The participation of KLF11 in the regulation of insulin biosynthesis in vivo was further supported by mouse genetic experiments utilizing a Klf11−/− mouse in a genetically pure C57BL/6 background, as a functional model to study globally disrupted KLF11-mediated transcription. Interestingly, knock-out of Klf11 in mice results in decreased insulin levels in serum from nonfasting (2.16 ± 0.40 versus 0.68 ± 0.10 ng/ml, p < 0.05, n = 8) and fasting (1.01 ± 0.20 versus 0.29 ± 0.05 ng/ml, p < 0.05, n = 8) animals (Fig. 5, a and b, respectively). Based upon our biochemical analysis of transcriptional control of the c.-331 KLF site through KLF11 binding, we hypothesized that reduced insulin levels in Klf11−/− mice may be due to a failure to fully activate the murine Ins2 promoter that contains the conserved KLF box. Congruent with the blood biochemistry data, a reduction in insulin production was initially evidenced by semi-quantitative immunohistochemistry, which showed concomitant attenuation of the insulin signal in Klf11−/− islet β cells (Fig. 5c). For a more quantitative assessment, we measured the expression of the Ins2 mRNA levels in Klf11−/− mice using, for internal data consistency, the same microarray technique employed to measure the expression of KLF genes in human islet. Indeed, the levels of insulin in the Klf11−/− pancreas were 66% of Klf11+/+ littermates values (p = 0.002). This result was also validated by RT-PCR (Fig. 5d). In agreement with decreased insulin, these mice showed abnormal responses to OGTT. Peak glucose levels occurred at 20 min in both Klf11−/− and control groups; however, Klf11−/− mice had 40% lower values than Klf11+/+ littermates (371.2 ± 41.6 versus 265.9 ± 19.8, p < 0.05; Fig. 5e, left panel). At 60 and 120 min, blood glucose levels in Klf11−/− mice remained lower than in Klf11+/+ littermates (178.6 ± 14.8 versus 220.9 ± 17.7 at 60 min and 141.8 ± 11.7 versus 182 ± 16 mg/dl; p < 0.05). In addition, serial plasma insulin levels were checked at each time point of the OGTT. There was a 2-fold increase in insulin levels compared with baseline at the 20-min time point (0.24 ± 0.02 to 0.48 ± 0.17 ng/ml) in Klf11−/− mice versus only a 0.3-fold increase in Klf11+/+ littermates (0.47 ± 0.05 to 0.64 ± 0.35 ng/ml p < 0.05; Fig. 5e, right panel). This rapid increase in insulin in Klf11−/− mice did not persist either at 60 or 120 min. As in many other studies that tried to model diabetes-associated alterations in mice, including Hnf1a, Gck, and Glut2 (37–40), Klf11−/− animals did not ultimately develop diabetes, likely because of concomitant alterations of metabolic pathways in peripheral tissues (41, 42). This notion is supported by our gene expression profiling of these mice, which demonstrates that Klf11−/− mice display abnormal expression of several metabolic enzymes in adipose and muscle tissue (supplemental Fig. S1), where KLF11 plays a key physiological role (43). This type of effect is likely due to the fact that the disruption of Klf11-mediated transcription is global in Klf11−/− mice, rather than localized as with the c.-331C>G INS mutation. Nevertheless, the disruption of insulin homeostasis is the first key phenotypic abnormality ever described for the Klf11−/− mice. Together with the rest of our comprehensive biochemical experiments, these mouse genetic experiments support a key role for the KLF11/p300-mediated pathway in the regulation of INS transcription.
FIGURE 5.
Klf11−/− mice display defects in insulin biosynthesis and blood levels. a, nonfasting insulin levels in Klf11−/− animals were significantly lower when compared with Klf11+/+ littermates. b, insulin levels were also lower in Klf11−/− animals than in Klf11+/+ littermates after overnight fasting. c, immunohistochemistry shows decreased insulin immunoreactivity within Klf11−/− islets, on the right as compared with Klf11+/+ littermates, on the left. Representative images are shown, and semiquantitative evaluation is depicted as a bar graph on the right. Scale bar, 200 μm. d, Ins2 mRNA levels were significantly lower in pancreata from Klf11−/− mice compared with Klf11+/+ littermate values by microarray (left panel) and RT-PCR (right panel). e, OGTT in Klf11−/− mice show peak glucose levels at 20 min in both groups, although values in Klf11−/− mice were lower than in Klf11+/+ littermates (left panel). Lower values were also observed at 60 and 120 min. Serial plasma insulin levels corresponding to each time point of OGTT (right panel) are also shown.
DISCUSSION
Recent reports indicate that KLF proteins are involved in mediating several biochemical mechanisms that lead to human diseases when disrupted (5). Thus, the search for KLF-mediated mechanisms is fueling a new and promising area of research in both biological chemistry and medicine. The current study deals with the characterization of a mutation in a novel KLF binding site that regulates insulin biosynthesis, leading to NDM. The patients affected by this mutation experience typical time-dependent fluctuations in their insulin and glucose levels, suggesting that under certain circumstances, the lack of insulin biosynthesis may be compensated, most likely at the level of peripheral resistance. Unfortunately, however, this mutation ultimately leads to severe disease with intrauterine growth retardation and postnatal morbidity. Therefore, identifying the mechanisms underlying the regulation of both the wild-type and mutated INS promoters is of paramount importance to uncover the role of KLFs in pancreatic β cell biochemistry. These data will significantly expand the repertoire of mechanisms underlying the development of diabetes.
Our investigations were encouraged by the observation that the c.-331C>G mutation has been identified in two independent cohorts of human subjects. The high reliability that repetition of genetic data provides led us to design a KLF family-wide screening for identifying potential KLF protein(s) that can bind this site and activate the wild-type INS promoter but not the mutant counterpart. In an unbiased approach used previously in our laboratory (44, 45), we screened all of the 16 different KLF proteins for their ability to perform this function. These experiments revealed that KLF11 is the most consistent activator of the locus. Although our study focused on KLF11 because of our experimental exclusion criteria of other candidates, it does not eliminate other KLF proteins in different biological contexts.
These data should be discussed in lieu of previous biochemical and diabetes genetic studies, which confirmed that some of the KLF proteins, reported here as potential regulators of the INS promoter, are indeed diabetes susceptibility genes (7–11). Therefore, previous reports, the data of the current studies, and investigations using high resolution genetic and biochemical techniques underscore the emerging importance of KLF proteins in the etiopathogenesis of nonautoimmune diabetes.
As mentioned, impairment in KLF11-mediated mechanisms, because of KLF11 mutations (7) or mutation in the c.-331 KLF site of INS promoter as described here, leads to impairment in INS transcription in MODY and NDM. Using genetically engineered mice, we sought to partially model the impairment in KLF11-mediated processes by deleting this gene through homologous recombination. Notably, this animal recapitulates the defect in insulin biosynthesis observed in human situations that are characterized by alterations in the biochemical function of KLF11. This result is highly informative because the mechanism of Klf11 disruption used in this mouse model causes alterations in insulin biochemistry similar to what is observed in humans although via different mechanisms, namely the functional disruption of this protein as observed in MODY versus the lack of its transactivating DNA binding function on the INS promoter in NDM, respectively. Therefore, the existence of three different mechanisms of disruption of KLF11-mediated transcriptional regulation that result in impaired insulin biosynthesis in both human and mice underscores the importance of this transcription factor in this key biochemical process.
Complementary observations reported here indicate that, in addition to KLF11, a reduced subset of KLF proteins may be able to also regulate insulin biosynthesis. For instance, these other KLF proteins may regulate promoters under basal or signaling-induced regulated conditions. Thus, it is currently difficult to predict under what biological context these other KLF factors could regulate INS expression. In this regard, characterizing the role of these other KLF genes in diabetes and the cellular contexts in which these phenomena occur will require significant efforts that are beyond the scope of this article. Nevertheless, the discovery of additional KLF proteins as candidates for regulation of insulin biosynthesis offers significant value added by providing the background for fueling future investigations in this important field of medical genetics.
An important focus of further discussion is how our mouse model experiments hold congruency with the biochemical and genetic data reported here. Fortunately, this animal has provided the ultimate test for a role of KLF11 in insulin homeostasis. Indeed, Klf11−/− animals display lower levels of pancreatic insulin than control littermates, as documented by several techniques. However, because insulin levels are not abolished, it is likely that the other transcription factors involved in insulin biosynthesis can compensate for the loss of Klf11, although not completely. Although these data support the overarching hypothesis of this study, we recognize that the most appropriate model would be mutation of the c.-331 KLF site to recapitulate the abnormal promoter using elegant, yet complex, kick-in technology. Therefore, future studies involving the generation of these mice will be even more informative on the mechanisms underlying the c.-331C>G mutation in NDM. Nevertheless, the important data provided by our mouse model support that KLF disruption results in impaired insulin homeostasis, which is congruent with the main observations reported for KLF11 in MODY and NDM in humans.
In summary, we have identified the c.-331 CACCC box in the INS promoter as a novel, bona fide KLF binding site. We provide evidence that supports the role for KLF11 in the regulation of this site and predict that impairment in its transcriptional function leads to a disruption in insulin biosynthesis. The importance of KLF11 in the regulation of insulin biosynthesis is further illustrated by our mouse model experiments. Additionally, we identify a small subset of KLF proteins that may also regulate insulin biosynthesis that serve as the catalyst for defining the ultimate role of KLF proteins in diabetes. Collectively, these results expand the known repertoire of key biochemical functions controlled by KLF proteins and provide a well characterized example of their critical participation in human disease.
Supplementary Material
Acknowledgments
We are grateful to the patients and their families for participation in the study. We thank Didier Chevenne and Munevver Turkmen for involvement in the clinical data. We acknowledge the Mayo Clinic Advanced Genomic Technology Center Microarray Shared Resource for mouse microarray experiments. We also thank Marion Rouault and Céline Liger for their technical support. We thank Adrienne Grzenda, Dr. William Faubion, and Dr. Angela Mathison for critically reading this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DK52913 (to R. U.) and DK050203 (to R. S.). This work was also supported by Agence Nationale de la Recherche-Maladies Rares (ANR-MRAR) Research Program Grant ANR-07-MRAR-000 (to M. P.), Transnational European Research Grant on Rare Diseases ERANET-09-RARE-005 (to M. V. and M. P.), and European Union Integrated Project EuroDia LSHM-CT-2006-518153 in Framework Programme 6 of the European Community (to P. F.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Fig. S1.
- KLF
- Krüppel-like transcription factor
- NDM
- neonatal diabetes mellitus
- MODY
- maturity onset diabetes of the young
- DN
- dominant negative
- OGTT
- oral glucose tolerance test
- MLE
- median level of expression
- MLEi
- MLE in three samples of islets
- MLEβ
- MLE in FACS-assisted sorted β cells.
REFERENCES
- 1. Lomberk G., Urrutia R. (2005) Biochem. J. 392, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaczynski J., Cook T., Urrutia R. (2003) Genome Biol. 4, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Black A. R., Black J. D., Azizkhan-Clifford J. (2001) J. Cell. Physiol. 188, 143–160 [DOI] [PubMed] [Google Scholar]
- 4. Bureau C., Hanoun N., Torrisani J., Vinel J. P., Buscail L., Cordelier P. (2009) Curr. Genomics 10, 353–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McConnell B. B., Yang V. W. (2010) Physiol. Rev. 90, 1337–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bieker J. J. (2010) Nat. Genet. 42, 733–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Neve B., Fernandez-Zapico M. E., Ashkenazi-Katalan V., Dina C., Hamid Y. H., Joly E., Vaillant E., Benmezroua Y., Durand E., Bakaher N., Delannoy V., Vaxillaire M., Cook T., Dallinga-Thie G. M., Jansen H., Charles M. A., Clément K., Galan P., Hercberg S., Helbecque N., Charpentier G., Prentki M., Hansen T., Pedersen O., Urrutia R., Melloul D., Froguel P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4807–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fernandez-Zapico M. E., van Velkinburgh J. C., Gutiérrez-Aguilar R., Neve B., Froguel P., Urrutia R., Stein R. (2009) J. Biol. Chem. 284, 36482–36490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kanazawa A., Kawamura Y., Sekine A., Iida A., Tsunoda T., Kashiwagi A., Tanaka Y., Babazono T., Matsuda M., Kawai K., Iiizumi T., Fujioka T., Imanishi M., Kaku K., Iwamoto Y., Kawamori R., Kikkawa R., Nakamura Y., Maeda S. (2005) Diabetologia 48, 1315–1322 [DOI] [PubMed] [Google Scholar]
- 10. Gutiérrez-Aguilar R., Froguel P., Hamid Y. H., Benmezroua Y., Jørgensen T., Borch-Johnsen K., Hansen T., Pedersen O., Neve B. (2008) J. Clin. Endocrinol. Metab. 93, 3128–3135 [DOI] [PubMed] [Google Scholar]
- 11. Voight B. F., Scott L. J., Steinthorsdottir V., Morris A. P., Dina C., Welch R. P., Zeggini E., Huth C., Aulchenko Y. S., Thorleifsson G., McCulloch L. J., Ferreira T., Grallert H., Amin N., Wu G., Willer C. J., Raychaudhuri S., McCarroll S. A., Langenberg C., Hofmann O. M., Dupuis J., Qi L., Segrè A. V., van Hoek M., Navarro P., Ardlie K., Balkau B., Benediktsson R., Bennett A. J., Blagieva R., Boerwinkle E., Bonnycastle L. L., Boström K. B., Bravenboer B., Bumpstead S., Burtt N. P., Charpentier G., Chines P. S., Cornelis M., Couper D. J., Crawford G., Doney A. S., Elliott K. S., Elliott A. L., Erdos M. R., Fox C. S., Franklin C. S., Ganser M., Gieger C., Grarup N., Green T., Griffin S., Groves C. J., Guiducci C., Hadjadj S., Hassanali N., Herder C., Isomaa B., Jackson A. U., Johnson P. R., Jorgensen T., Kao W. H., Klopp N., Kong A., Kraft P., Kuusisto J., Lauritzen T., Li M., Lieverse A., Lindgren C. M., Lyssenko V., Marre M., Meitinger T., Midthjell K., Morken M. A., Narisu N., Nilsson P., Owen K. R., Payne F., Perry J. R., Petersen A. K., Platou C., Proenca C., Prokopenko I., Rathmann W., Rayner N. W., Robertson N. R., Rocheleau G., Roden M., Sampson M. J., Saxena R., Shields B. M., Shrader P., Sigurdsson G., Sparsø T., Strassburger K., Stringham H. M., Sun Q., Swift A. J., Thorand B., Tichet J., Tuomi T., van Dam R. M., van Haeften T. W., van Herpt T., van Vliet-Ostaptchouk J. V., Walters G. B., Weedon M. N., Wijmenga C., Witteman J., Bergman R. N., Cauchi S., Collins F. S., Gloyn A. L., Gyllensten U., Hansen T., Hide W. A., Hitman G. A., Hofman A., Hunter D. J., Hveem K., Laakso M., Mohlke K. L., Morris A. D., Palmer C. N., Pramstaller P. P., Rudan I., Sijbrands E., Stein L. D., Tuomilehto J., Uitterlinden A., Walker M., Wareham N. J., Watanabe R. M., Abecasis G. R., Boehm B. O., Campbell H., Daly M. J., Hattersley A. T., Hu F. B., Meigs J. B., Pankow J. S., Pedersen O., Wichmann H. E., Barroso I., Florez J. C., Frayling T. M., Groop L., Sladek R., Thorsteinsdottir U., Wilson J. F., Illig T., Froguel P., van Duijn C. M., Stefansson K., Altshuler D., Boehnke M., McCarthy M. I. (2010) Nat. Genet. 42, 579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Garin I., Edghill E. L., Akerman I., Rubio-Cabezas O., Rica I., Locke J. M., Maestro M. A., Alshaikh A., Bundak R., del Castillo G., Deeb A., Deiss D., Fernandez J. M., Godbole K., Hussain K., O'Connell M., Klupa T., Kolouskova S., Mohsin F., Perlman K., Sumnik Z., Rial J. M., Ugarte E., Vasanthi T., Johnstone K., Flanagan S. E., Martínez R., Castaño C., Patch A. M., Fernández-Rebollo E., Raile K., Morgan N., Harries L. W., Castaño L., Ellard S., Ferrer J., Perez de Nanclares G., Hattersley A. T. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 3105–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Polak M., Dechaume A., Cavé H., Nimri R., Crosnier H., Sulmont V., de Kerdanet M., Scharfmann R., Lebenthal Y., Froguel P., Vaxillaire M. (2008) Diabetes 57, 1115–1119 [DOI] [PubMed] [Google Scholar]
- 14. Lukowiak B., Vandewalle B., Riachy R., Kerr-Conte J., Gmyr V., Belaich S., Lefebvre J., Pattou F. (2001) J. Histochem. Cytochem. 49, 519–528 [DOI] [PubMed] [Google Scholar]
- 15. Bouatia-Naji N., Bonnefond A., Cavalcanti-Proença C., Sparsø T., Holmkvist J., Marchand M., Delplanque J., Lobbens S., Rocheleau G., Durand E., De Graeve F., Chèvre J. C., Borch-Johnsen K., Hartikainen A. L., Ruokonen A., Tichet J., Marre M., Weill J., Heude B., Tauber M., Lemaire K., Schuit F., Elliott P., Jørgensen T., Charpentier G., Hadjadj S., Cauchi S., Vaxillaire M., Sladek R., Visvikis-Siest S., Balkau B., Lévy-Marchal C., Pattou F., Meyre D., Blakemore A. I., Jarvelin M. R., Walley A. J., Hansen T., Dina C., Pedersen O., Froguel P. (2009) Nat. Genet. 41, 89–94 [DOI] [PubMed] [Google Scholar]
- 16. Marshak S., Totary H., Cerasi E., Melloul D. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 15057–15062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Asfari M., Janjic D., Meda P., Li G., Halban P. A., Wollheim C. B. (1992) Endocrinology 130, 167–178 [DOI] [PubMed] [Google Scholar]
- 18. Gerrish K., Cissell M. A., Stein R. (2001) J. Biol. Chem. 276, 47775–47784 [DOI] [PubMed] [Google Scholar]
- 19. Cook T., Gebelein B., Mesa K., Mladek A., Urrutia R. (1998) J. Biol. Chem. 273, 25929–25936 [DOI] [PubMed] [Google Scholar]
- 20. Fernandez-Zapico M. E., Mladek A., Ellenrieder V., Folch-Puy E., Miller L., Urrutia R. (2003) EMBO J. 22, 4748–4758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Song C. Z., Gavriilidis G., Asano H., Stamatoyannopoulos G. (2005) Blood Cells Mol. Dis. 34, 53–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iwabu M., Yamauchi T., Okada-Iwabu M., Sato K., Nakagawa T., Funata M., Yamaguchi M., Namiki S., Nakayama R., Tabata M., Ogata H., Kubota N., Takamoto I., Hayashi Y. K., Yamauchi N., Waki H., Fukayama M., Nishino I., Tokuyama K., Ueki K., Oike Y., Ishii S., Hirose K., Shimizu T., Touhara K., Kadowaki T. (2010) Nature 464, 1313–1319 [DOI] [PubMed] [Google Scholar]
- 23. Miyawaki K., Yamada Y., Yano H., Niwa H., Ban N., Ihara Y., Kubota A., Fujimoto S., Kajikawa M., Kuroe A., Tsuda K., Hashimoto H., Yamashita T., Jomori T., Tashiro F., Miyazaki J., Seino Y. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14843–14847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Aguilar-Bryan L., Bryan J. (2008) Endocr. Rev. 29, 265–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vaxillaire M. D. P., Bonnefond A., Froguel P. (2009) Pediatr. Endocrinol. Rev. 6, 405–417 [PubMed] [Google Scholar]
- 26. Greeley S. A., Tucker S. E., Naylor R. N., Bell G. I., Philipson L. H. (2010) Trends Endocrinol. Metab. 21, 464–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Melloul D., Marshak S., Cerasi E. (2002) Diabetologia 45, 309–326 [DOI] [PubMed] [Google Scholar]
- 28. Naya F. J., Stellrecht C. M., Tsai M. J. (1995) Genes Dev. 9, 1009–1019 [DOI] [PubMed] [Google Scholar]
- 29. Whelan J., Poon D., Weil P. A., Stein R. (1989) Mol. Cell. Biol. 9, 3253–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ling Z., Wang Q., Stangé G., In't Veld P., Pipeleers D. (2006) Diabetes 55, 78–85 [PubMed] [Google Scholar]
- 31. Kaczynski J. A., Conley A. A., Fernandez Zapico M., Delgado S. M., Zhang J. S., Urrutia R. (2002) Biochem. J. 366, 873–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pearson R., Fleetwood J., Eaton S., Crossley M., Bao S. (2008) Int. J. Biochem. Cell Biol. 40, 1996–2001 [DOI] [PubMed] [Google Scholar]
- 33. Qiu Y., Guo M., Huang S., Stein R. (2002) Mol. Cell. Biol. 22, 412–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Qiu Y., Sharma A., Stein R. (1998) Mol. Cell. Biol. 18, 2957–2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mosley A. L., Corbett J. A., Ozcan S. (2004) Mol. Endocrinol. 18, 2279–2290 [DOI] [PubMed] [Google Scholar]
- 36. Nagai R., Friedman S. L., Kasuga M. (eds) (2009) The Biology of Krüppel-like Factors, pp. 33–49, Springer, Tokyo [Google Scholar]
- 37. Bady I., Marty N., Dallaporta M., Emery M., Gyger J., Tarussio D., Foretz M., Thorens B. (2006) Diabetes 55, 988–995 [DOI] [PubMed] [Google Scholar]
- 38. Leturque A., Brot-Laroche E., Le Gall M. (2009) Am. J. Physiol. Endocrinol. Metab. 296, E985–E992 [DOI] [PubMed] [Google Scholar]
- 39. Pontoglio M., Sreenan S., Roe M., Pugh W., Ostrega D., Doyen A., Pick A. J., Baldwin A., Velho G., Froguel P., Levisetti M., Bonner-Weir S., Bell G. I., Yaniv M., Polonsky K. S. (1998) J. Clin. Invest. 101, 2215–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Terauchi Y., Takamoto I., Kubota N., Matsui J., Suzuki R., Komeda K., Hara A., Toyoda Y., Miwa I., Aizawa S., Tsutsumi S., Tsubamoto Y., Hashimoto S., Eto K., Nakamura A., Noda M., Tobe K., Aburatani H., Nagai R., Kadowaki T. (2007) J. Clin. Invest. 117, 246–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Alevizos I., Misra J., Bullen J., Basso G., Kelleher J., Mantzoros C., Stephanopoulos G. (2007) Cell Cycle 6, 1631–1638 [DOI] [PubMed] [Google Scholar]
- 42. Hsiao G., Chapman J., Ofrecio J. M., Wilkes J., Resnik J. L., Thapar D., Subramaniam S., Sears D. D. (2011) Am. J. Physiol. Endocrinol. Metab. 300, E164–E174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yamamoto K., Sakaguchi M., Medina R. J., Niida A., Sakaguchi Y., Miyazaki M., Kataoka K., Huh N. H. (2010) Biochem. Biophys. Res. Commun. 400, 175–180 [DOI] [PubMed] [Google Scholar]
- 44. Truty M. J., Lomberk G., Fernandez-Zapico M. E., Urrutia R. (2009) J. Biol. Chem. 284, 6291–6300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fernandez-Zapico M. E., Lomberk G. A., Tsuji S., Demars C. J., Bardsley M. R., Lin Y. H., Almada L., Han J. J., Mukhopadhyay D., Ordog T., Buttar N. S., Urrutia R. (2011) Biochem. J. 435, 529–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.