Abstract
DNA polymerase and DNA helicase are essential components of DNA replication. The helicase unwinds duplex DNA to provide single-stranded templates for DNA synthesis by the DNA polymerase. In bacteriophage T7, movement of either the DNA helicase or the DNA polymerase alone terminates upon encountering a nick in duplex DNA. Using a minicircular DNA, we show that the helicase·polymerase complex can bypass a nick, albeit at reduced efficiency of 7%, on the non-template strand to continue rolling circle DNA synthesis. A gap in the non-template strand cannot be bypassed. The efficiency of bypass synthesis depends on the DNA sequence downstream of the nick. A nick on the template strand cannot be bypassed. Addition of T7 single-stranded DNA-binding protein to the complex stimulates nick bypass 2-fold. We propose that the association of helicase with the polymerase prevents dissociation of the helicase upon encountering a nick, allowing the helicase to continue unwinding of the duplex downstream of the nick.
Keywords: DNA Damage, DNA Helicase, DNA Polymerase, DNA Replication, DNA-protein Interaction, Protein-protein Interactions
Introduction
The efficient and precise replication of DNA genomes relies on the interplay of the multiple proteins constituting the replisome. The interaction of the replicative helicase with the DNA polymerase is one of the most critical among the several protein-protein interactions within the replisome. Replicative DNA polymerases catalyze a nucleophilic attack of the 3′-hydroxyl of the primer on the α-phosphate of a dNTP aligned to the single-stranded template. With few exceptions, the single-stranded template arises from unwinding of the DNA duplex by DNA helicase at the expense of NTP hydrolysis (1).
Synergy between the helicase and polymerase has been observed in a variety of replication systems, including Escherichia coli, bacteriophage T4- and T7-infected cells, and mitochondria (2–9). In general, association of the two proteins occurs directly or through auxiliary proteins. The association of helicase with polymerase enhances the processivity of polymerase and the unwinding rate of helicase significantly, suggesting that either protein in the complex can be reciprocally regulated (1–10).
Bacteriophage T7 has evolved an efficient mechanism for DNA replication. Only four proteins, the T7 gene 5 DNA polymerase (gp5),2 gene 4 helicase-primase (gp4), gene 2.5 ssDNA-binding protein (gp2.5) and E. coli thioredoxin (trx), the processivity factor, are needed to reconstitute a functional replisome (11). In the T7 replication system the helicase and primase occupy distinct domains within the C- and N-terminal halves, respectively, of gp4. This direct association, without aid of accessory proteins, provides a model to study the interaction and synergy between these key components in DNA replication (1). gp5 has both polymerase and 3′→5′ exonuclease activities. gp5 by itself is a non-processive polymerase, catalyzing the incorporation of <50 nucleotides per binding event (12). In phage-infected cells gp5 is found in a tight one-to-one complex (gp5·trx) with its processivity factor, E. coli trx. This association increases the processivity to several hundred nucleotides per binding event (8, 12, 13).
The helicase activity of phage T7 is located in the C-terminal half of gp4. gp4 assembles as a hexamer and uses the energy of hydrolysis of dTTP to translocate unidirectionally 5′→3′ on ssDNA and to unwind duplex DNA (14, 15). Gene 4 encodes two co-linear 63- and 56-kDa proteins (16). The full-length 63-kDa gp4, having both the C-terminal helicase domain and the N-terminal primase domain, has both helicase and primase activity. The 56-kDa gp4 is translated from an internal ribosome-binding site and lacks the N-terminal Cys4 zinc-binding domain indispensable for primer synthesis (17). Despite being deficient in primer synthesis, the 56-kDa gp4 retains the ability to transfer the primer to the polymerase (18). Oligomerization of gp4 involves N-terminal residues within the helicase and the linker connecting the primase and helicase (19–21). Consequently, the 56-kDa gp4 forms hexamers, a requirement for helicase activity, because the ssDNA, upon which the helicase translocates, passes through the central core of the hexamer (22). Because both the 56-kDa gp4 and 63-kDa gp4 have the same helicase domain and robust DNA-unwinding activity, both proteins support strand-displacement DNA synthesis mediated by T7 DNA polymerase (23–25).
T7 helicase has at least three modes for binding to the gp5·trx complex. One mode involves an interaction between the acidic C-terminal tail of gp4 and a basic patch on gp5 located near the region where the template strand exits the active site. This electrostatic interaction is essential to establish the initial helicase-polymerase interaction at the replication fork (26). A second electrostatic mode involves an interaction between the acidic C-terminal tail of gp4 and two basic loops in the thioredoxin-binding domain of gp5 (27). A tighter binding mode between helicase and polymerase occurs when gp5·trx is bound to a DNA in a polymerization mode (27). The structural basis for this tight binding during DNA synthesis is not known. These interactions not only provide gp5·trx immediate access to the single-stranded template generated by helicase, but also increase the processivity of polymerase from several hundred bases to >17 kb as determined by single molecule experiments (28). The helicase also benefits from these interactions. A 10-fold increase in the rate of unwinding is observed when DNA synthesis accompanies helicase unwinding (9).
During genome replication, replication forks often encounter DNA damage such as lesions or strand interruptions that, if not removed by specific repair mechanisms, can inactivate the replisome (29–31). Single phosphodiester bond interruptions (nicks) are likely to be the most frequent DNA damage a replisome will encounter. They can be introduced by endonucleases, recombination events, and the discontinuous nature of lagging-strand synthesis that leaves nicks between Okazaki fragments (32). In addition, the repair of other types of DNA damage, for example the excision repair of thymine dimers, also produces nicks (33). If not removed by DNA ligase, a nick poses a challenge to the replisome, especially the helicase, because most helicases require a fork structure for their unwinding activity. A nick will terminate the replication fork and abort helicase function (29, 32).
In the current study we have examined the ability of T7 gp5·trx and 56-kDa gp4 to replicate duplex DNA containing either nicks or gaps at various positions within the two strands. We have used the 56-kDa gp4, because it is even more efficient in mediating strand-displacement synthesis, and it is devoid of primase activity. We have used a minicircular DNA consisting of 70 bp of duplex DNA and a single-stranded 5′-tail (33). This chemically synthesized minicircle thus has a preformed fork onto which the proteins can assemble. The small size of the minicircle allows for the determination of the stoichiometry of the components. We show that a T7 helicase·polymerase complex can bypass a nick on the non-template strand, another advantage derived from the T7 helicase-polymerase interaction.
EXPERIMENTAL PROCEDURES
Materials
Oligonucleotides were obtained from Integrated DNA Technology. T4 polynucleotide kinase, T4 DNA ligase, terminal transferase, and dNTPs were purchased from New England Biolabs. Radiolabeled nucleotides were purchased from PerkinElmer.
Preparation of DNA Substrates and Proteins
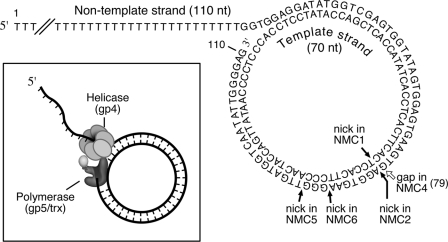
Oligonucleotides were purified using a denaturing gel before use. DNA sequences were derived from a minicircular substrate described previously (33) (Fig. 1). A 70-nt circular template strand DNA (Fig. 1) was prepared as described previously (33). Radioactive labeling of DNA at the 5′-end was performed using T4 polynucleotide kinase and [γ-32P]ATP. Minicircular DNAs were constructed by annealing the non-template strand (NTS) DNA fragments to the circular template strand (TS) (Fig. 1) followed by purification of all minicircular DNAs on a non-denaturing gel. Substrate NMC1 with a nick on the template strand was constructed by annealing the 110-nt NTS to the 70-nt linear TS (5′-pCACTTC…TCACCT (Fig. 1)). Nicks on the non-template strand were introduced by annealing two fragments of the designated 5′-phosphorylated NTS to the circular TS (Fig. 1). DNA with a 1-nt gap on the non-template strand (NMC4) was created by annealing two fragments of NTS (1–77 and 80–110) to the circular template, then a ddTMP was incorporated at the position of the 78th nucleotide in the non-template strand (Fig. 1) using terminal transferase. The resulting gap at position 79 cannot be filled by T7 DNA polymerase, because it cannot initiate synthesis from the dideoxy-terminated fragment. gp5·trx, 56-kDa gp4, and gp2.5 were purified as described previously (11, 24, 34).
FIGURE 1.
Minicircular DNA. The minicircular DNA contains a 110-nt non-template strand and a circular 70-nt template strand. The positions of the nicks are indicated by black arrows. The position of the gap is indicated by a white arrow. The inset shows the scheme of the assembly of the T7 helicase/DNA polymerase complex and the subsequent strand-displacement synthesis. NMC, nicked minicircle.
Strand-displacement DNA Synthesis
The reaction (10 μl) contained 40 mm Tris-HCl, pH 7.5, 10 mm DTT, 10 mm MgCl2, 50 mm potassium glutamate, 0.6 mm dNTP, and 20 nm of the indicated DNA. The reaction was initiated by adding 50 nm gp5·trx or 50 nm each of gp5·trx and helicase. After incubation for 10 min at 37 °C, the reaction mixture was loaded onto either a 10% non-denaturing or denaturing polyacrylamide gel, and the DNA products were separated by electrophoresis. After the gel was dried, radioactivity from the individual DNA bands was measured using a Fuji BAS 1000 Bioimaging analyzer.
RESULTS
Strand-displacement DNA Synthesis by T7 DNA Polymerase and Helicase on a Minicircle
A minicircular DNA, depicted in Fig. 1, offers several advantages for examining leading strand DNA synthesis (33). In the minicircle the 5′-tail of the non-template strand provides a loading site for T7 helicase, whereas the 3′-end serves as a primer for gp5·trx to initiate synthesis. The absence of a 3′ terminus on the template strand eliminates hydrolysis of this strand by the exonuclease activity of T7 DNA polymerase. The helicase·polymerase complex mediates strand-displacement DNA synthesis by a rolling circle mechanism, producing DNA products longer than 17 kb (33) (Fig. 1).
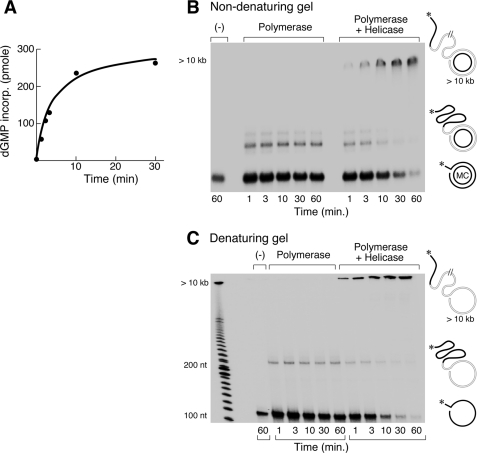
In the experiment shown in Fig. 2A T7 DNA polymerase and DNA helicase mediate strand-displacement synthesis on the minicircle depicted in Fig. 1. In order to monitor synthesis the 5′ terminus of the non-template strand is labeled with 32P. Extensive rolling circle DNA synthesis occurs to yield products that exceed the resolving capacity of the gel (Fig. 2, B and C). The products synthesized by gp5·trx in the absence and presence of gp4 were analyzed by electrophoresis through non-denaturing (Fig. 2B) and denaturing (Fig. 2C) gels. Interestingly, gp5·trx, in the absence of DNA helicase, displays limited strand-displacement DNA synthesis (Fig. 2B). However, the reaction is aborted after an extension of the strand by ∼90 nt as revealed by the denaturing gel (Fig. 2C). On another minicircle construct of similar length, but containing an A/T-rich sequence, the polymerase extended the strand ∼40 nt (data not shown). The limited strand-displacement DNA synthesis catalyzed by the polymerase alone is most likely affected by the structural features of the different DNAs (see “Discussion”). In the presence of T7 DNA helicase, gp5·trx mediated extensive strand-displacement synthesis to produce molecules whose length exceeded the resolving capacity of the gel (>10 kb).
FIGURE 2.
Strand-displacement DNA synthesis by T7 helicase·polymerase on minicircular DNA. A, time course of strand-displacement DNA synthesis mediated by T7 helicase·polymerase. Reaction containing [3H]dGTP (10 cpm/pmol) was stopped by adding EDTA to a final concentration of 20 mm at the indicated time points, and incorporation of [3H]dGMP into DNA was measured. B and C, gel analysis of time course of strand-displacement DNA synthesis by T7 polymerase or polymerase/helicase complex. Reactions were the same as in A except that the 5′ terminus of the non-template strand was labeled with 32P. Products were separated on a non-denaturing (B) or denaturing (C) 10% TBE (89 mm Tris, 89 mm boric acid, 2 mm EDTA (pH 8.0)) polyacrylamide gel.
T7 Helicase/Polymerase Cannot Bypass a Nick on the Template Strand
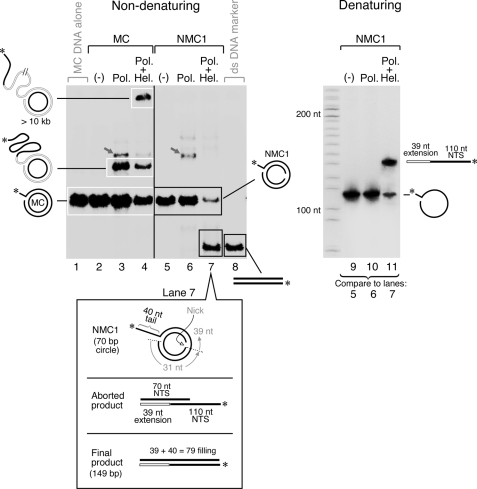
One advantage of using the minicircle is that it has the ability to modify either of the two strands by chemical synthesis of the appropriate oligonucleotides; then the minicircle is assembled by annealing the designed DNA fragments. We have introduced nicks into each of the two strands and examined their affect on strand-displacement synthesis mediated by the helicase·polymerase complex. A nick in DNA is defined as a single phosphodiester bond interruption, and in our experiments they contain a 5′-phosphoryl and a 3′-hydroxyl end group. We first introduced a nick into the template strand (position is indicated by an arrow in Fig. 1). Product formation was monitored as in Fig. 2 by labeling the 5′ terminus of the non-template strand with 32P. The helicase·polymerase complex did not mediate extensive rolling circle DNA synthesis on this minicircle containing a nick (Fig. 3, lane 7) as it did with the intact minicircle (Fig. 3, lane 4), indicating that the nick aborts synthesis. Extensive synthesis is indicated by the slow migrating species near the top of the non-denaturing gel. Instead, most of the DNA was converted into linear dsDNA of ∼150 bp (Fig. 3, lane 7) resulting from strand-displacement DNA synthesis that aborted at the nick. Indeed, a 5′-radiolabeled dsDNA marker of 150 bp has a mobility identical to the product of aborted synthesis (Fig. 3, lane 8). The limited strand-displacement synthesis seen with gp5·trx alone (Fig. 3, lane 3) was not observed in the presence of a nick in the template strand (Fig. 3, lane 6). A minor band marked by a gray arrow in Fig. 3 is most likely a mobility-shift band resulting from protein-DNA interactions. This minor band is observed on a non-denaturing gel (Fig. 2B) but not a denaturing gel (Fig. 2C). This minor band disappears after phenol extraction of the samples (data not shown). Analysis of the product on a denaturing gel confirmed that there is no extension of NMC1 by the polymerase alone (Fig. 3, lane 10). The analysis also revealed the nature of the abortive product mediated by the helicase·polymerase complex (Fig. 3, lane 11).
FIGURE 3.
Effect of a nick in the template strand on strand-displacement synthesis. Intact minicircular DNA (lanes 2–4) or minicircular DNA containing a nick in the template strand (lanes 5–7) were examined for their ability to support rolling circle DNA synthesis mediated by gp5·trx and helicase. Each DNA was labeled with 32P at the 5′ terminus of the non-template strand. Lane 1 contains minicircular DNA only, and lane 8 is a 5′-radiolabeled dsDNA marker of 150 bp. A gray arrow signifies the band probably resulting from a protein·DNA complex. Lanes 9–11 represent analyses of samples shown in lanes 5–7 on a 10% TBE denaturing gel containing 7 m urea.
T7 Helicase/Polymerase Can Bypass a Nick on the Non-template Strand
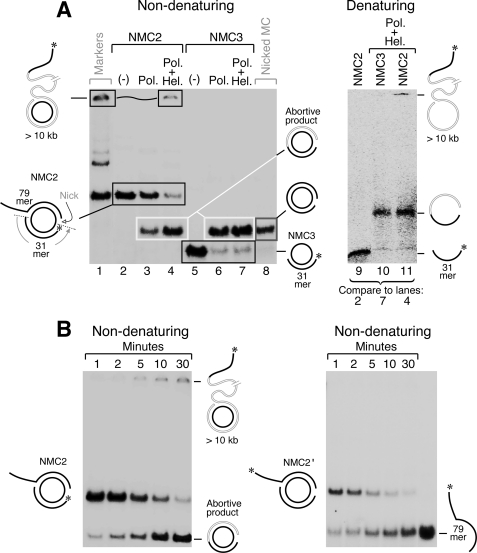
A nick was also introduced into the non-template strand by annealing two oligonucleotides (a 5′-32P-labeled 31-mer and a 79-mer) to the circular template strand as shown in Fig. 4A, resulting in the 5′-P at the nick labeled with 32P (NMC2). In the absence of helicase, T7 gp5·trx extends the 31-mer by strand-displacement DNA synthesis of the 79-mer NTS fragment. Displacement of the 79-mer from the circular template strand results in a nicked minicircle (Fig. 4A, lane 3). This reaction is significantly enhanced by the presence of helicase (Fig. 4A, lane 4). As anticipated, nearly all DNA synthesis is aborted at the nick, because T7 gp5·trx cannot initiate synthesis at a nick and helicase cannot load onto DNA at a nick (35, 36). However, a small amount of high molecular weight DNA was observed (Fig. 4A, lane 4), indicating that the helicase·polymerase complex bypasses some nicks.
FIGURE 4.
T7 helicase·polymerase can bypass a nick located on the non-template strand. A, minicircular DNA with a nick in the non-template strand designated NMC2 (lanes 2–4) or the same minicircular DNA missing the non-template strand upstream of the nick designated NMC3 (lanes 5–7) were examined for their ability to support rolling circle DNA synthesis mediated by T7 DNA polymerase and helicase. The NMC2 and NMC3 DNA constructs are depicted. The 5′-phosphate at each nick was labeled with 32P. Lane 1 shows the position of the intact minicircle and the fully extended DNA product, and lane 8 is a radiolabeled fully double-stranded minicircle DNA with a nick in the non-template strand. Lanes 9–11 represent analyses of samples shown in lanes 2, 4, and 7 on a 10% TBE denaturing gel containing 7 m urea. B, time course of DNA synthesis by T7 helicase·polymerase complex on the minicircular DNA with a nick in the non-template strand. The 5′-phosphate at the nick (left panel) or on the single-stranded tail (right panel) was labeled with 32P. Reaction conditions were the same as that for lane 4 in A except that the reaction was stopped at various time points.
To confirm that the nick bypass requires complex formation between T7 polymerase and helicase, another DNA construct (NMC3), lacking the 79-mer fragment, was prepared (Fig. 4A). T7 gp5·trx initiates synthesis at the 3′ terminus of the annealed fragment to produce a nicked minicircle (Fig. 4A, lane 6). The presence of helicase in the reaction did not allow gp5·trx to bypass the nick (Fig. 4A, lane 7). This result shows that T7 gp5·trx cannot bypass a nick unless it is in complex with the helicase; the presence of helicase in solution does not fulfill this function. The existence of the 79-mer NTS in NMC2 serves as a track for helicase and polymerase to function together in their leading-strand DNA synthesis mode before encountering the nick. To confirm that the high molecular weight band consists of extended DNA and is not a consequence of protein-DNA interactions, reaction products were also separated on a denaturing gel (Fig. 4A, lanes 9–11). The results are consistent with those obtained on the non-denaturing gel. The nick bypass efficiency as defined by the percentage of high molecular weight DNA (top band) out of total products (high molecular weight DNA plus aborted product (lower band)) was 6.7% as determined from lane 4 in Fig. 4A.
To further confirm the origin of the long extended product and the efficiency of nick bypass, the time course of DNA synthesis by T7 helicase·polymerase on minicircles containing a nick was examined. In addition to the nicked circle (NMC2) described above where the 32P label is located at the nick we also examined the same minicircular nicked DNA with the label at the 5′ terminus of the displaced tail of the non-template strand (NMC2′ depicted in Fig. 4B). Again a small amount of 32P-labeled high molecular weight product was observed when the label was present at the nick, but none was observed when the label was present at the 5′-end of the single-stranded tail (Fig. 4B). These result show that no high molecular weight DNA arose from synthesis initiating at the 3′ terminus of the nick or that ligation of the nick in the original minicircle had occurred.
Effect of Local DNA Structure on Nick Bypass
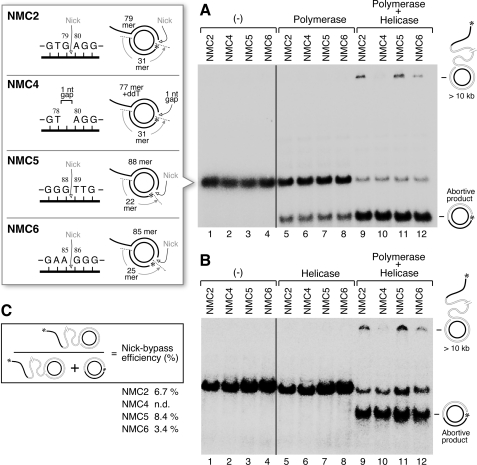
The effect of DNA structure on the ability of helicase·polymerase to bypass the nick was examined using three minicircles containing a nick in the non-template strand (Fig. 5). One minicircle (NMC4) containing a 1-nt gap instead of a nick in the non-template strand was constructed as described under “Experimental Procedures.” In the second construct (NMC5) three G:C base pairs were placed adjacent to the nick at the 3′ terminus of the fragment. In the third construct (NMC6) three G:C base pairs are also located adjacent to the nick but at the 5′ terminus of the fragment.
FIGURE 5.
Effect of local DNA structure on the ability of helicase·polymerase to bypass a nick. Four minicircle DNAs, depicted in the figure, were constructed and examined for the ability of helicase·polymerase to bypass interruptions in the non-template strand. The first (NMC2) contains a nick in the non-template strand and is the same as that used in Fig. 3. The second (NMC4) has a gap upstream of the nick with the same position as that in NMC2. The third (NMC5) and fourth (NMC6) each have a nick in the non-template strand with three G:C base pairs on opposite sides of the nick. In NMC5 the three G:C pairs are located at the 3′ side of the nick and in NMC6 at the 5′ side. The 5′-phosphate located at each interruption was labeled with 32P. These DNAs (20 nm) were each incubated in the absence of enzymes (A and B, lanes 1–4), with 50 nm DNA polymerase (A, lanes 5–8), with 50 nm helicase (B, lanes 5–8), or with 50 nm helicase plus 50 nm DNA polymerase (A and B, lanes 9–12) at 37 °C for 10 min in a reaction as described. The reaction mixtures were separated on a non-denaturing 10% TBE polyacrylamide gel. C, bypass efficiency was determined by the ratio of the intensity of the top band (fully extended strand-displacement DNA product due to nick bypass) to the sum of the intensity of the top and bottom (aborted product) bands.
The ability of these three constructs to support bypass DNA synthesis relative to the nicked circle (NMC2) described in the last section is shown in Fig. 5. T7 helicase·polymerase can bypass the nicks adjacent to the G:C base pairs on either side of the nick, but the complex cannot bypass the 1-nt gap. The efficiency of bypass synthesis is 8.4% when the G:C is base paired at the 3′ terminus and 3.4% when at the 5′ terminus. We also examined the ability of T7 DNA helicase alone to continue unwinding of DNA containing a nick using these minicircles. Gene 4 DNA helicase alone was unable to unwind any of the minicircles used in the current study (Fig. 5B, lanes 5–8). It was previously shown that T7 DNA helicase requires both a 5′- and a 3′-ssDNA tail for proper loading onto DNA (35, 37).
T7 Gene 2.5 ssDNA-binding Protein Enhances Nick Bypass
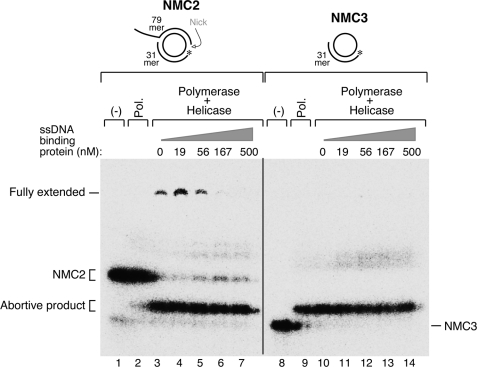
A minimal T7 replisome consists of gp5, gp4, gp2.5, and E. coli thioredoxin (1, 10). The T7 gene 2.5 ssDNA-binding protein is essential for coordinated leading- and lagging-strand DNA synthesis (33). It also enables T7 helicase·polymerase to initiate strand-displacement synthesis from a nick in duplex DNA (36). Furthermore, it facilitates the annealing of complementary strands of DNA through homologous base pairing (38).
Although the mechanism by which gp2.5 mediates these events remains elusive, its DNA-binding and known interaction with DNA polymerase are thought to be essential (1, 33, 36). Using two minicircles, one with a nick in the non-template strand (NMC2) and one consisting of an oligonucleotide annealed to the single-stranded minicircle (NMC3), we examined the effect of gp2.5 on nick bypass by helicase·polymerase (Fig. 6). The latter DNA has 39 nt of exposed single strand DNA, whereas the nicked duplex minicircle has only the 5′-single-stranded tail of 40 nt. With the nicked minicircle the addition of low concentrations (19 nm) of gp2.5 increased the efficiency of nick bypass 2-fold (from 6.7% to 15.1%, comparing lane 3 and lane 4 in Fig. 6). However, further increasing the concentration of gp2.5 did not improve nick bypass but eventually inhibited nick bypass (Fig. 6, lanes 5–7). No product of nick bypass was observed with the circle annealed to the oligonucleotide confirming that strand-displacement DNA synthesis occurs only with an ongoing helicase·polymerase complex.
FIGURE 6.
Effect of T7 ssDNA-binding protein on bypassing a nick. Minicircle DNA, depicted in the figure, with a nick in the non-template strand (NMC2, left panel) or the same minicircle lacking the oligonucleotide bearing the 5′-tail (NMC3, right panel) were examined for their ability to support rolling circle DNA synthesis mediated by T7 DNA polymerase and helicase in the presence of the indicated amount of T7 gene 2.5 ssDNA-binding protein.
DISCUSSION
The response of DNA replication machinery to genomic lesions such as nicks in DNA is an interesting and important issue. Whereas studies have been carried out on the consequence of a nick in DNA on the progress of DNA polymerase, less attention has been directed to the DNA helicase, an essential component of all replisomes. Indeed, the helicase may well be the first protein in the replisome to encounter a nick. If the DNA helicase, translocating 5′ to 3′ on the lagging strand, ceases unwinding upon encountering a nick, it is likely that both leading- and lagging-strand DNA synthesis will be aborted. Neither Thermus aquaticus DnaB helicase (39) nor T7 helicase (35) alone can continue unwinding DNA past a nick in vitro. Both of these helicases require a single-stranded tail for loading onto DNA prior to unwinding the duplex. In addition to the 5′-tail a 3′-single-stranded tail is required to exclude that strand from the central core of the helicase (39). If the nick is located in the strand that is not occupied by the helicase, then the absence of a 3′-ssDNA tail will stop the helicase or, alternatively, allow the helicase to accommodate both strands within its central core and move along the duplex. If the nick is in the strand upon which the helicase is translocating, then it is likely that the helicase will terminate unwinding as it departs from the end of the interrupted strand. The latter need not be the case if the binding of multiple nucleotides within the central cavity is sufficient to maintain contact with the DNA. However, the situation within the replisome is quite different due to other interactions with the helicase. In particular the high affinity interaction of the polymerase with the helicase has marked effects on the replication process. Not only does DNA synthesis by the polymerase enhance unwinding (9, 10), but the interaction of the two proteins also dramatically increases the processivity (28). In the current study we show that the interaction between T7 helicase and polymerase provides the helicase with another advantage: the potential to bypass a nick on the DNA strand on which it is translocating. Recent work also showed that the T4 replisome is capable of bypassing an abasic lesion located in the leading strand which, a reaction that would pose a challenge for the individual proteins (40).
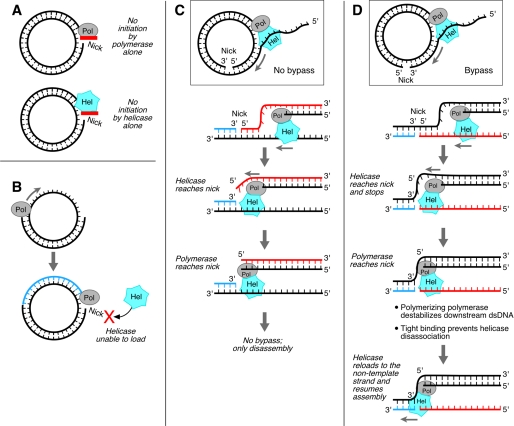
What is the mechanism underlying the bypass of a nick by the polymerase/helicase complex? Neither T7 helicase nor T7 DNA polymerase alone can bypass a nick. Helicase requires contacts with both strands of the duplex, encircling the 5′-strand and excluding the 3′-strand from the central hole formed by the hexamer ring (39). A nick does not allow for these contacts (Fig. 7A). However, when the helicase is associated with a polymerizing polymerase, the tight binding (KD = 90 nm (26)) between the two could allow the helicase to remain bound to the template rather than to dissociate, thus providing an opportunity for the helicase to contact the 5′-end of the DNA at the nick. In this model the intrinsic and transient destabilization (breathing) of the downstream DNA duplex can be captured by the helicase to initiate unwinding. The T7 helicase also can bind to the T7 polymerase via its acidic C-terminal tail (Fig. 7B). However, because the helicase is not in an unwinding mode, the unwinding and polymerizing are not coupled and the two proteins are not tightly bound (27). This weak binding of the two proteins does not allow the helicase to bypass the nick. If the nick is located in the template strand the complex fails to bypass the nick (Fig. 7C). Most likely the structure generated when gp5·trx encounters the nick precludes it from moving to the next nucleotide in the template. The helicase should encounter the nick before the polymerase during leading strand synthesis. This makes the situation different for nick bypass depending on the position of the nick. In the case of a nick in the template, when polymerase reaches the nick, the helicase has already halted movement or has encircled the dsDNA. In either condition the helicase loses contact with the template. As a result the opportunity for polymerase to contact the DNA terminus on the other side of the nick is also lost. However, if the nick is in the non-template strand (Fig. 7D), the helicase might first stop, but when the polymerase reaches the nick it will attempt to continue DNA synthesis, because the template is continuous. Such a polymerizing polymerase and its tight binding to helicase provide the helicase with the opportunity to contact the DNA terminus downstream of the nick. Another benefit derived from the contact of the helicase with the polymerase is that the polymerase may provide additional destabilization energy to the downstream DNA (41). Consistent with this model, the stability of DNA duplex downstream of the nick does affect the efficiency of bypass. Three adjacent G:C pairs downstream of the nick decrease bypass efficiency by approximately a half. In contrast, G:C pairs upstream of the nick do not show a significant effect. In this scenario the nick is stabilized within the central core of the helicase to allow the complex to pass over this site. The inefficiency of bypass synthesis (7%) suggests that the stabilization of the nick within the helicase is weak, a conclusion supported by the inability of the complex to bypass a gap with a single nucleotide missing.
FIGURE 7.
T7 helicase·polymerase encountering a nick. A, T7 helicase and/or gp5·trx cannot initiate DNA unwinding or polymerization of nucleotides at a nick. The helicase needs an ssDNA tail to load while polymerase needs ssDNA as template. B, a polymerizing DNA polymerase cannot bypass a nick in the presence of exogenous helicase. The pre-assembly of helicase·polymerase in the leading-strand synthesis mode is necessary for nick bypass. C, an ongoing helicase·polymerase complex cannot bypass a nick on the template strand. When polymerase reaches the nick, the helicase that encountered the nick earlier has already stopped or has started translocation on the dsDNA. In either condition the polymerase loses the opportunity to contact the DNA terminus on the other side of the nick. D, an ongoing helicase·polymerase complex can bypass a nick on the non-template strand. Helicase unwinding might at first be terminated at the nick, but when polymerase reaches the nick it may attempt to continue DNA synthesis, because the template is continuous and the polymerase is capable of at least destabilizing the dsDNA downstream of the nick. These events would favor the loading of the helicase onto the non-template strand downstream of the nick to re-establish the replication fork. The tight association of helicase with polymerase increases the chance of helicase contacting the DNA terminus downstream of the nick. gp2.5 may facilitate the binding.
In our experiments the efficiency of bypassing a nick mediated by the helicase·polymerase complex during DNA replication is low (∼7%). Further assistance could come from other components in the T7 replisome. For example, gene 2.5 single-stranded DNA-binding protein can improve the efficiency of nick bypass by 15%. Interestingly, the improvement was observed only at concentrations of gp2.5 comparable to other components of the replisome. The requirement for a low concentration of gp2.5 to obtain enhanced bypass synthesis suggests a role in the stabilization of the helicase·polymerase complex at the nick through protein interactions (Fig. 7D). Higher concentrations of gp2.5 could lead to destabilization of the helicase·polymerase complex, because the C termini of gp2.5 and helicase potentially compete for the same binding site on the polymerase (1, 27, 28).
Another surprising finding is that T7 gp5·trx alone can mediate strand-displacement synthesis for ∼90 nucleotides (40 on another A/T-rich minicircle) on the intact minicircular DNA (Fig. 2, B and C). Previous studies had found that such synthesis by gp5·trx alone was limited to 5 nt or less (9, 42). When a nick is present on the template strand, there is no strand-displacement synthesis catalyzed by the polymerase alone (Fig. 3, lanes 6 and 10). The molecular basis for this difference in the extent of strand-displacement synthesis is not known. However, a number of factors are likely to affect the extent of strand-displacement synthesis by T7 DNA polymerase. Other DNA polymerases such as E. coli DNA polymerase I and phi 29 DNA polymerase can catalyze extensive strand-displacement synthesis on duplex DNA (43, 44). T7 DNA polymerase also has the intrinsic ability to catalyze strand-displacement synthesis. However, this activity is not robust and is susceptible to the conformation of the DNA. The different extents of strand-displacement synthesis we observe on various DNAs could be due to differences in the torsional tension of the minicircular DNAs. In these small molecules differences in base composition could either enhance or decrease the efficiency of removing torsional stress during DNA synthesis on the circular molecule. In addition, products from strand-displacement synthesis on minicircular templates contain repeated sequence every 70 nucleotides allowing for intra- or intermolecular (5′-CAATATTG-3′) base pairing. The resulting secondary structure would prevent further progression of the polymerase, resulting in discrete products. The abundance of consecutive guanines in the products raises the possibility of forming a stable G-quartet structure to pause replication. Such inhibitory products are dependent on the template sequence. For example, the AT-rich template contains more successive thymines than the minicircle used in most of the studies, which might lead to different consequences in impeding the polymerase activity. Another factor that could affect the extent of strand displacement synthesis of T7 DNA polymerase is its own exonuclease activity, because an exonuclease deficiency could lead to strand-displacement synthesis of 100–500 nt (42).
Acknowledgments
We thank Sharmistha Ghosh for helpful discussion and Steven Moskowitz (Advanced Medical Graphics) for illustrations.
This work was supported, in whole or in part, by National Institutes of Health Grant GM54397 (to C. C. R.).
- gp5
- T7 gene 5 DNA polymerase
- gp4
- gene 4 helicase-primase
- gp2.5
- gene 2.5 ssDNA-binding protein
- ssDNA
- single strand DNA
- dsDNA
- double strand DNA
- nt
- nucleotide(s)
- NTS
- non-template strand
- TS
- template strand
- trx
- thioredoxin.
REFERENCES
- 1. Hamdan S. M., Richardson C. C. (2009) Annu. Rev. Biochem. 78, 205–243 [DOI] [PubMed] [Google Scholar]
- 2. Kim S., Dallmann H. G., McHenry C. S., Marians K. J. (1996) Cell 84, 643–650 [DOI] [PubMed] [Google Scholar]
- 3. Mok M., Marians K. J. (1987) J. Biol. Chem. 262, 16644–16654 [PubMed] [Google Scholar]
- 4. Korhonen J. A., Pham X. H., Pellegrini M., Falkenberg M. (2004) EMBO J. 23, 2423–2429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Delagoutte E., von Hippel P. H. (2001) Biochemistry 40, 4459–4477 [DOI] [PubMed] [Google Scholar]
- 6. Schrock R. D., Alberts B. (1996) J. Biol. Chem. 271, 16678–16682 [DOI] [PubMed] [Google Scholar]
- 7. Cha T. A., Alberts B. M. (1989) J. Biol. Chem. 264, 12220–12225 [PubMed] [Google Scholar]
- 8. Lee J. B., Hite R. K., Hamdan S. M., Xie X. S., Richardson C. C., van Oijen A. M. (2006) Nature 439, 621–624 [DOI] [PubMed] [Google Scholar]
- 9. Stano N. M., Jeong Y. J., Donmez I., Tummalapalli P., Levin M. K., Patel S. S. (2005) Nature 435, 370–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Satapathy A. K., Kochaniak A. B., Mukherjee S., Crampton D. J., van Oijen A., Richardson C. C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 6782–6787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Richardson C. C. (1983) Cell 33, 315–317 [DOI] [PubMed] [Google Scholar]
- 12. Tabor S., Huber H. E., Richardson C. C. (1987) J. Biol. Chem. 262, 16212–16223 [PubMed] [Google Scholar]
- 13. Wuite G. J., Smith S. B., Young M., Keller D., Bustamante C. (2000) Nature 404, 103–106 [DOI] [PubMed] [Google Scholar]
- 14. Matson S. W., Tabor S., Richardson C. C. (1983) J. Biol. Chem. 258, 14017–14024 [PubMed] [Google Scholar]
- 15. Patel S. S., Picha K. M. (2000) Annu. Rev. Biochem. 69, 651–697 [DOI] [PubMed] [Google Scholar]
- 16. Dunn J. J., Studier F. W. (1983) J. Mol. Biol. 166, 477–535 [DOI] [PubMed] [Google Scholar]
- 17. Frick D. N., Richardson C. C. (2001) Annu. Rev. Biochem. 70, 39–80 [DOI] [PubMed] [Google Scholar]
- 18. Zhu B., Lee S. J., Richardson C. C. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 9099–9104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo S., Tabor S., Richardson C. C. (1999) J. Biol. Chem. 274, 30303–30309 [DOI] [PubMed] [Google Scholar]
- 20. Lee S. J., Richardson C. C. (2004) J. Biol. Chem. 279, 23384–23393 [DOI] [PubMed] [Google Scholar]
- 21. Zhu B., Lee S. J., Richardson C. C. (2009) J. Biol. Chem. 284, 23842–23851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Donmez I., Patel S. S. (2006) Nucleic Acids Res. 34, 4216–4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bernstein J. A., Richardson C. C. (1988) Proc. Natl. Acad. Sci. U.S.A. 85, 396–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bernstein J. A., Richardson C. C. (1988) J. Biol. Chem. 263, 14891–14899 [PubMed] [Google Scholar]
- 25. Bernstein J. A., Richardson C. C. (1989) J. Biol. Chem. 264, 13066–13073 [PubMed] [Google Scholar]
- 26. Zhang H., Lee S. J., Zhu B., Tran N. Q., Tabor S., Richardson C. C. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 9372–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hamdan S. M., Marintcheva B., Cook T., Lee S. J., Tabor S., Richardson C. C. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 5096–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hamdan S. M., Johnson D. E., Tanner N. A., Lee J. B., Qimron U., Tabor S., van Oijen A. M., Richardson C. C. (2007) Mol. Cell 27, 539–549 [DOI] [PubMed] [Google Scholar]
- 29. Cox M. M., Goodman M. F., Kreuzer K. N., Sherratt D. J., Sandler S. J., Marians K. J. (2000) Nature 404, 37–41 [DOI] [PubMed] [Google Scholar]
- 30. Heller R. C., Marians K. J. (2006) Nat. Rev. Mol. Cell Biol. 7, 932–943 [DOI] [PubMed] [Google Scholar]
- 31. McInerney P., O'Donnell M. (2007) J. Biol. Chem. 282, 25903–25916 [DOI] [PubMed] [Google Scholar]
- 32. Kuzminov A. (1995) Mol. Microbiol. 16, 373–384 [DOI] [PubMed] [Google Scholar]
- 33. Lee J., Chastain P. D., 2nd, Kusakabe T., Griffith J. D., Richardson C. C. (1998) Mol. Cell 1, 1001–1010 [DOI] [PubMed] [Google Scholar]
- 34. Rezende L. F., Hollis T., Ellenberger T., Richardson C. C. (2002) J. Biol. Chem. 277, 50643–50653 [DOI] [PubMed] [Google Scholar]
- 35. Ahnert P., Patel S. S. (1997) J. Biol. Chem. 272, 32267–32273 [DOI] [PubMed] [Google Scholar]
- 36. Ghosh S., Marintcheva B., Takahashi M., Richardson C. C. (2009) J. Biol. Chem. 284, 30339–30349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matson S. W., Richardson C. C. (1983) J. Biol. Chem. 258, 14009–14016 [PubMed] [Google Scholar]
- 38. Rezende L. F., Willcox S., Griffith J. D., Richardson C. C. (2003) J. Biol. Chem. 278, 29098–29105 [DOI] [PubMed] [Google Scholar]
- 39. Kaplan D. L. (2000) J. Mol. Biol. 301, 285–299 [DOI] [PubMed] [Google Scholar]
- 40. Nelson S. W., Benkovic S. J. (2010) J. Mol. Biol. 401, 743–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Johnson D. S., Bai L., Smith B. Y., Patel S. S., Wang M. D. (2007) Cell 129, 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lechner R. L., Engler M. J., Richardson C. C. (1983) J. Biol. Chem. 258, 11174–11184 [PubMed] [Google Scholar]
- 43. Masamune Y., Richardson C. C. (1971) J. Biol. Chem. 246, 2692–2701 [PubMed] [Google Scholar]
- 44. Soengas M. S., Esteban J. A., Lázaro J. M., Bernad A., Blasco M. A., Salas M., Blanco L. (1992) EMBO J. 11, 4227–4237 [DOI] [PMC free article] [PubMed] [Google Scholar]