Abstract
Cytoplasmic caspase recruiting domain (CARD)-containing molecules often function in the induction of potent antimicrobial responses in order to protect mammalian cells from invading pathogens. Retinoic acid-induced gene-I (RIG-I) and nucleotide binding oligomerization domain 2 (NOD2) serve as key factors in the detection of viral and bacterial pathogens, and in the subsequent initiation of innate immune signals to combat infection. RIG-I and NOD2 share striking similarities in their cellular localization, both localize to membrane ruffles in non-polarized epithelial cells and both exhibit a close association with the junctional complex of polarized epithelia. Here we show that RIG-I and NOD2 not only colocalize to cellular ruffles and cell-cell junctions, but that they also form a direct interaction that is mediated by the CARDs of RIG-I and multiple regions of NOD2. Moreover, we show that RIG-I negatively regulates ligand-induced nuclear factor-κB (NF-κB) signaling mediated by NOD2, and that NOD2 negatively regulates type I interferon induction by RIG-I. We also show that the three main Crohn disease-associated mutants of NOD2 (1007fs, R702W, G908R) form an interaction with RIG-I and negatively regulate its signaling to a greater extent than wild-type NOD2. Our results show that in addition to their role in innate immune recognition, RIG-I and NOD2 form a direct interaction at actin-enriched sites within cells and suggest that this interaction may impact RIG-I- and NOD2-dependent innate immune signaling.
Keywords: Innate Immunity, Interferon, NF-κB (NF-κB), Protein Domains, Protein-Protein Interactions
Introduction
Several families of cytoplasmic caspase-recruiting domain (CARD)2-containing molecules function to induce potent antimicrobial responses to protect mammalian cells from pathogens. Whereas cellular helicases such as retinoic acid-induced gene-I (RIG-I) and melanoma differentiation-associated gene (MDA5) function in the detection of viral pathogens, nucleotide binding oligomerization domain (NOD)-like receptors such as NOD2 detect bacterial peptidoglycans (although recent work suggests that NOD2 might also play a role in viral detection (1)). CARDs are protein-protein interaction modules that play key roles in apoptosis and inflammatory signaling. Associations between these domains can occur through both homotypic and heterotypic interactions (2). RIG-I and MDA5 utilize CARD-dependent interactions to interact with a common downstream effector molecule (mitochondrial antiviral signaling (MAVS), also known as VISA/IPS-1/Cardif)) to induce type I interferon (IFN) production (3–5). Likewise, NOD2 utilizes its CARDs to induce both apoptosis and nuclear factor-κB (NF-κB) activation and to interact with downstream effectors (6).
Mutations in NOD2 are linked with the pathogenesis of Crohn disease (CD), a type of inflammatory bowel disease in which patients display significant inflammation of the lining of the digestive tract, causing severe diarrhea and abdominal pain (7, 8). CD-associated NOD2 mutations include two missense mutations, R702W and G908R, as well as a frameshift mutation, 1007fs. Individuals carrying two alleles of these mutations display a 20–40-fold increased risk of developing CD (7–9). All of these mutations localize within the leucine-rich repeat (LRR) regions of NOD2, which are critical for ligand-binding and downstream signaling and suggest that patients with CD may be unable to mount effective innate immune responses to bacterial (and perhaps viral) pathogens (10).
In addition to their roles in innate immune signaling, CARD-containing molecules have been shown to associate with the actin cytoskeleton and, in some cases, to participate in the regulation of cellular motility and migration. We have shown that RIG-I localizes to actin-enriched membrane ruffles and positively regulates cell migration (11). Others have also reported that loss of RIG-I inhibits both actin polymerization and actin distribution in macrophages following LPS stimulation (12). Similarly, NOD2 associates with the actin cytoskeleton and regulates cellular signaling in response to actin cytokskeletal modulation (13). However, some CD-associated mutants of NOD2 (particularly 1007fs) display altered membrane and cytoskeletal associations, indicating that these mutations directly impact proper NOD2 cytoskeletal localization (14, 15). Taken together, these studies suggest that molecules involved in innate immune and inflammatory responses may closely associate with the actin cytoskeleton and that this association may serve an important role in cellular motility and possibly in inflammatory signaling.
Our previous work showed that RIG-I localized to lamellipodia and to sites of cell-cell contact (11). This localization was strikingly similar to the localization of NOD2 shown by others (13, 15). RIG-I and NOD2 both contain CARDs, which function as protein-protein interaction modules. However, it remains unknown if there is an association between these two molecules or whether this association might play any functional role in innate immune signaling or CD. In this report, we show that RIG-I and NOD-2 colocalize to cellular ruffles and cell-cell junctions and form a direct interaction that is mediated by the CARDs of RIG-I and multiple regions of NOD2. Moreover, we show that RIG-I negatively regulates ligand-induced NF-κB signaling mediated by NOD2 and that NOD2 negatively regulates type I IFN induction via RIG-I. We further show that although mutations in NOD2 associated with Crohn disease still form an interaction with RIG-I, the 1007fs mutant of NOD2 alters the association of RIG-I with membrane ruffles and negatively regulates RIG-I-mediated signaling to a greater extent than wild type NOD2, indicating that there may be a functional effect of this mutation on RIG-I/NOD2 signaling. Our results show that in addition to their role in pattern recognition, RIG-I and NOD2 form a direct interaction at actin-enriched sites within cells and suggest that this interaction may impact RIG-I- and NOD-2-dependent innate immune signaling.
EXPERIMENTAL PROCEDURES
Cells
HT29 cells were purchased from the ATCC and cultured in McCoy's 5A medium supplemented with 10% fetal bovine serum and penicillin/streptomycin. HEK293, human osteosarcoma U2OS, HeLa (CCL-2), and Caco-2 cells were cultured in DMEM-H supplemented with 10% fetal bovine serum and penicillin/streptomycin. For all studies, cells were plated in collagen-coated 8-well culture slides (Nunc) or collagen-coated culture dishes. All cells were grown for a minimum of 24- (HEK293, HeLa, U2OS) or 48- (HT-29, Caco-2) h prior to study. Cells were screened for mycoplasma using a PCR-based mycoplasma test (Takara Bio USA, Madison, WI) to prevent abnormalities in cellular morphology, transfection, and growth.
Plasmids, siRNAs, and Transfections
EGFP- and DsRed-RIG-I constructs have been described (11). HA- and EGFP-NOD2 constructs were kindly provided by Sylvia Legrand-Poels and Vincent Ollendorff, respectively. Mutations of NOD2 were performed using Stratagene Quickchange mutagenesis per the manufacturer's instruction. pGEX-4T-RIG-I was constructed by inserting the CARDs of RIG-I into the SalI and NotI sites of pGEX-4T. pET-SUMO-NOD2 constructs were constructed using the ChampionTM pET SUMO Expression System (Invitrogen) following PCR amplification. RIG-I On-Target PLUS pooled siRNAs were purchased from Dharmacon.
Plasmid transfections were performed using FuGENE 6 according to the manufacturer's protocol (Roche Applied Science). Following transfection, cells were plated as described above and used 24–72 h later. HT-29 and HeLa cells were transfected by Amaxa Nuclefection (HT-29: solution R and program W-17; HeLa: solution R and program I-13).
Generation of Stable RIG-I Knockdown Cells
HeLa (CCL-2) cells stably expressing control and RIG-I shRNA vectors were constructed as follows: cells were transfected with control and human RIG-I shRNA vectors (psiRNA-h7SKGFPzeo plasmids with GFP::Zeo fusion genes (Invivogen)) as described above. Forty-eight hours following transfection, cells were placed in 100 μg/ml zeocin and after 2 weeks, individual zeocin-resistant clones were selected. Clones were expanded and screened for GFP expression and level of RIG-I knockdown. The clone with the highest level of knockdown was used in all experiments. To stably maintain cells, zeocin was maintained throughout.
Purification of Recombinant Proteins
The pET-SUMO-NOD2 or pGEX4T-RIG-I CARDs expression vectors were introduced into competent Escherichia coli BL21 (DE3) cells (Invitrogen). Overnight cultures grown in LB were diluted 1:5 into a final culture volume of 250 ml. At an A600 of 0.6 cultures were exposed to 1 mm isopropyl-1-thio-β-d-galactopyranoside (Fisher), and bacteria collected after 2 h at 37 °C. Bacterial cell pellets were resuspended in phosphate-buffered saline containing 10 mm 2-mercaptoethanol and 1% Triton X-100. The cells were lysed by sonication, and the cell debris was removed by centrifugation at 4,000 rpm for 30 min. For purification of SUMO-NOD2 constructs, the supernatant was incubated with a 20% slurry of Ni-NTA-agarose beads for 2 h at 4 °C. The beads were washed four times with cold phosphate-buffered saline, and the protein was eluted from the beads with 500 mm imidazole. For purification of GST-RIG-I-CARDs, the supernatant was incubated with glutathione-Sepharose 4B beads for 2 h at 4 °C, and beads washed with phosphate-buffered saline. Purified protein was eluted with glutathione (10 mm).
Antibodies
Goat polyclonal antibody specific for RIG-I (L-15) and blocking peptide (L-15P) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Rabbit polyclonal and mouse monoclonal antibodies directed against GFP (FL, B-2), HA (Y-11, F-7), GAPDH (sc-25778), and DsRed (F9) were purchased from Santa Cruz Biotechnology. Mouse monoclonal anti-FLAG (M2) was purchased from Sigma. Mouse monoclonal NOD2 (sc-56168) was purchased from Santa Cruz Biotechnology. Rabbit anti-RIG-I polyclonal antibody was purchased from Cell Signaling Technologies. Alexa Fluor-conjugated secondary antibodies and phalloidin were purchased from Invitrogen. Anti-GST mouse monoclonal was purchased from Millipore. Anti-His6 chicken polyclonal antibody was purchased from Thermo Scientific.
Immunofluorescence Microscopy
Cultures were washed and fixed with either 4% paraformaldehyde or with ice-cold methanol/acetone (3:1). Cells were then permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS) and incubated with the indicated primary antibodies for 1 h at room temperature (RT). Following washing, cells were incubated with secondary antibodies for 30 min at room temperature, washed, and mounted with Vectashield (Vector Laboratories, Burlingame, CA) containing 4′,6-diamidino-2-phenylindole (DAPI). Images were captured using an Olympus IX81 inverted microscope equipped with a motorized Z-axis drive. Images were generated by multiple-section stacking (0.2 μm stacks) and deconvolved using a calculated point-spread function (Slidebook 5.0). Alternatively, images were acquired on a confocal laser-scanning microscope (FV1000, Olympus). Images were processed using Adobe Photoshop CS3 (Adobe, San Jose, CA).
Reporter Gene Assays
Activation of IFNβ or NF-κB promoters were measured by reporter assay. HEK293 or HeLa psiRNA cells were transfected in 24-well plates (as described above) with p-125 luc (IFNβ) or NF-κB reporter plasmid together with the indicated plasmids. Luciferase activity was measured by a luciferase assay system (Promega). All experiments were performed in triplicate and conducted a minimum of three times.
Inhibition of RIG-I-mediated Gene Induction by NOD2
HEK293 cells (3 × 105 cells/well in 24-well plates) were co-transfected with the indicated NOD2 constructs and ISG56-Luciferase reporter (0.4 μg) and β-actin Renilla luciferase reporter (0.012 μg) as described previously (16). Twenty-four hours post-transfection, cells were collected by trypsin digestion and seeded onto a 96-well plate (the remaining cells were collected for immunoblotting). Twenty-four hours later, cells were stimulated with Sendai virus (multiplicity of infection (MOI) = 10) for 16 h and luciferase activities were measured using the Dual-Glo luciferase assay system (Promega, Madison, WI). Results are expressed as fold induction of ISG56-luciferase relative to that of mock infected cells after normalizing to Renilla luciferase.
Immunoblot Analysis
Cell lysates were prepared with RIPA buffer (50 mm Tris-HCl, pH 7.4; 1% Nonidet P-40; 0.25% sodium deoxycholate; 150 mm NaCl; 1 mm EDTA; 1 mm phenylmethanesulfonyl fluoride; 1 mg/ml aprotinin, leupeptin, and pepstatin; 1 mm sodium orthovanadate), and insoluble material was cleared by centrifugation for 5 min at 4 °C. Lysates (30–50 mg) were loaded onto 4–20% Tris-HCl gels (Bio-Rad) and transferred to polyvinylidene difluoride membranes. Membranes were blocked overnight in 5% nonfat dry milk or 3% bovine serum albumin, probed with the indicated antibodies, and developed with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechology), and SuperSignal West Pico or sWest Dura chemiluminescent substrates (Pierce Biotechnology).
Immunoblots of endogenous RIG-I and NOD2 were performed using an Odyssey Infrared Imaging System (LI-COR Biosciences). Whole-cell lysates from the indicated cells (50 μg) were loaded onto 4–20% Tris-HCl gels, separated electrophoretically, and transferred to nitrocellulose membranes. Membranes were blocked in Odyssey Blocking buffer and then incubated with the appropriate antibodies overnight at 4 °C in Odyssey Blocking buffer. Following washing, membranes were incubated with anti-rabbit or anti-mouse antibodies conjugated to IRDye 680 or 800 CW and visualized with the Odyssey Infrared Imaging System according to the manufacturer's instructions.
Immunoprecipitations
For immunoprecipitations, HEK293 cells transiently transfected with the indicated plasmids were lysed with EBC buffer (50 mm Tris, pH 8.0, 120 mm NaCl, 0.5% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, and 0.5 μg/ml pepstatin). Insoluble material was cleared by centrifugation. Lysates were incubated with the indicated antibodies in EBC buffer for 1 h at 4 °C followed by the addition of Sepharose G beads for an additional 1 h at 4 °C. After centrifugation, the beads were washed in NETN buffer (150 mm NaCl, 1 mm EDTA, 50 mm Tris-HCl (pH 7.8), 1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, 0.5 μg/ml leupeptin, and 0.5 μg/ml pepstatin), then heated at 95 °C for 10 min in Laemmli sample buffer. Following a brief centrifugation, the supernatant was immunblotted with the indicated antibodies as described above.
In Vitro Binding Assays
SUMO- or SUMO-NOD2-purified proteins conjugated to Ni-NTA beads were added to EBC buffer and incubated with purified GST or GST-RIG-I CARDs beads for 2 h at 4 °C. Beads were sedimented by centrifugation, washed in NETN buffer, and heated at 95 °C for 10 min in Laemmli sample buffer. Following a brief centrifugation, the supernatant was run on a 4–20% Tris-HCl gel and transferred to a polyvinylidene difluoride membrane. The membranes were blocked overnight in 3% bovine serum albumin at 4 °C and incubated with anti-GST monoclonal antibody. Alternatively, samples were run on 4–20% gels and stained with Coomassie Blue to control for loading of GST and GST fusion proteins.
Statistical Analysis
Data are presented as mean ± standard deviation. A one-way analysis of variance (ANOVA) and Bonferroni's correction for multiple comparisons were used to determine statistical significance (p < 0.05). Data are representative of experiments performed at least three times.
RESULTS
RIG-I and NOD2 Colocalize to Actin-enriched Membrane Ruffles and Sites of Cell Contact
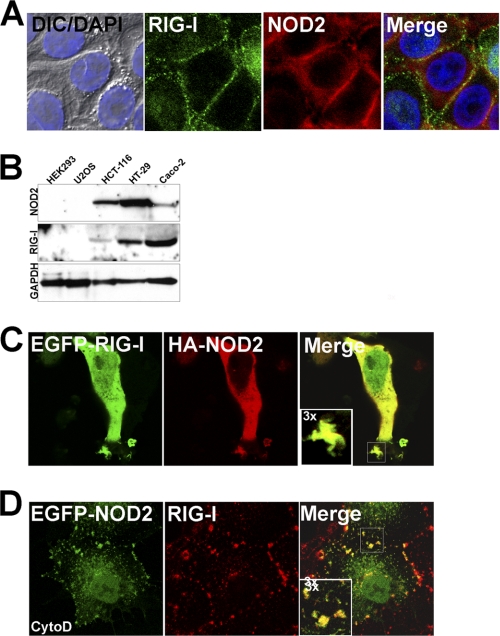
To determine whether RIG-I and NOD2 colocalize to similar cellular domains, we performed immunofluorescence microscopy in human intestinal epithelial HT-29 cells (which endogenously express high levels of both RIG-I and NOD2 (Fig. 1B)) and in human embryonic kidney HEK293 overexpressing NOD2. We found that endogenous RIG-I and NOD2 colocalized to cellular junctions in confluent HT29 cells (Fig. 1A) and to actin-enriched membrane ruffles when HT29 cells were grown under subconfluent conditions (not shown). This staining was specific for RIG-I as incubation with a blocking peptide abolished RIG-I signal (supplemental Fig. S1). NOD2 localizes predominantly to membrane ruffles and sites of cell contact when overexpressed in HEK293 cells (7). (HEK293 cells do not express endogenous NOD2.) We found that overexpressed HA-tagged NOD2 colocalized with overexpressed RIG-I (Fig. 1C). These results suggest that RIG-I and NOD2 localize to similar cellular domains and that both molecules associate with the actin cytoskeleton.
FIGURE 1.
RIG-I and NOD2 colocalize at cell-cell junctions and membrane ruffles. A, HT-29 cells grown under confluent conditions were fixed and stained with anti-RIG-I (green) and anti-NOD2 (red) antibodies. At left are shown differential interference contrast (DIC) and DAPI (nuclei, blue) merged images. B, lysates (50 μg) from the indicated cell types were immunoblotted for RIG-I, NOD2, and GAPDH as a loading control using a LI-COR Odyssey infrared imaging system. C, HEK293 cells were transfected with EGFP-RIG-I and HA-NOD2 and fixed and stained for HA (red). D, Caco-2 cells were transfected with EGFP-NOD2 and 48 h post-transfection, exposed to 10 μm cytochalasin D (cytoD) for 60 min. Cells were then fixed and stained for endogenous RIG-I (red). Data are representative of experiments performed at least three times.
Disruption of the actin cytoskeleton has been linked to the activation or NF-κB- and interferon regulatory factor (IRF)-3-dependent signaling (11, 13, 17, 18). In addition, the localization of NOD2 and RIG-I have both been shown to be altered by treatment of cells with cytochalasin D (cytoD), an actin depolymerizing agent (11, 13). We found that NOD2 and endogenous RIG-I remained colocalized when polarized intestinal epithelial Caco-2 cells expressing EGFP-fused NOD2 were exposed to cytoD, confirming that the site of NOD2 and RIG-I localization is sensitive to perturbations of the actin cytoskeleton (Fig. 1D). Moreover, these data suggest that NOD2 and RIG-I may form a direct association that remains intact even when the actin cytoskeleton is disrupted.
RIG-I Interacts with NOD2
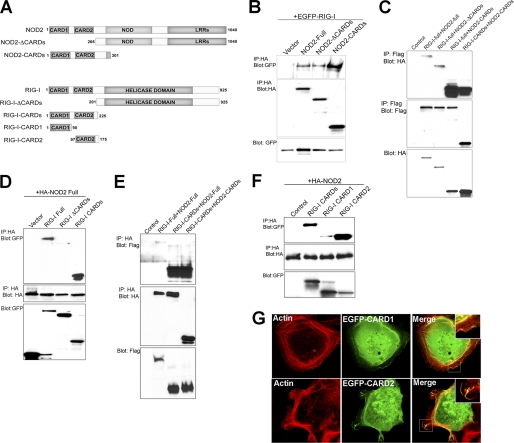
We found that overexpressed NOD2 and RIG-I colocalized within membrane ruffles in HEK293 cells (Fig. 1C), and to at sites of cell contact (Fig. 1A). Because of their colocalization, we determined whether RIG-I and NOD2 interact by performing co-immunoprecipitation assays. We examined the interaction between Flag-tagged RIG-I (Flag-RIG-I) and HA-tagged NOD2 (HA-NOD2) from lysates of transiently transfected HEK293 cells. We found that Flag-RIG-I co-immunoprecipitated with HA-NOD2 (Fig. 2C) and vice versa (Fig. 2E). To confirm these findings, we also performed co-immunoprecipitation studies in HEK293 cells transiently transfected with EGFP-RIG-I and HA-NOD2. Similar to our findings with Flag-RIG-I, we found that EGFP-RIG-I interacted with HA-NOD2, indicating that the interaction is not tag-dependent (Fig. 2B).
FIGURE 2.
RIG-I and NOD2 interact. A, schematic of wild type and truncated RIG-I and NOD2 proteins used in co-immunoprecipitation studies. Numbers represent amino acid residues. B, HEK293 cells were transfected with EGFP-RIG-I and vector control or HA-tagged NOD2 full, NOD2 ΔCARDs, or NOD2 CARDs. Following transfection (∼48 h), cells were lysed and immunoprecipitations performed with anti-HA antibody and immunoblots performed with anti-GFP (top) or anti-HA (middle) antibodies. In parallel, immunoblots were performed for GFP in lysates of transfected cells (bottom). C, HEK293 cells were transfected with vector control or Flag-tagged RIG-I or RIG-I CARDs constructs and HA-tagged NOD2, NOD2 ΔCARDs, or NOD2 CARDs. Following transfection (∼48 h), cells were lysed and immunoprecipitations performed with anti-Flag antibody and immunoblots performed with anti-HA (top) or anti-Flag (middle) antibodies. In parallel, immunoblots were performed for HA in lysates of transfected cells (bottom). D, HEK293 cells were transfected with vector control or EGFP-tagged RIG-I, RIG-I ΔCARDs, or RIG-I CARDs constructs and HA-tagged NOD2. Following transfection (∼48 h), cells were lysed and immunoprecipitations performed with anti-HA antibody and immunoblots performed with anti-GFP (top) or anti-HA (middle) antibodies. In parallel, immunoblots were performed for GFP in lysates of transfected cells (bottom). E, HEK293 cells were transfected with vector control or Flag-tagged RIG-I or RIG-I CARDs constructs and HA-tagged NOD2 or NOD2 CARDs. Following transfection (∼48 h), cells were lysed and immunoprecipitations performed with anti-HA antibody and immunoblots performed with anti-Flag (top) or anti-HA (middle) antibodies. In parallel, immunoblots were performed for Flag in lysates of transfected cells (bottom). F, top, HEK293 cells were transfected with vector control or EGFP-tagged RIG-I CARDs, CARD1, or CARD2 and HA-tagged NOD2. Following transfection (∼48 h), cells were lysed and immunoprecipitations performed with anti-HA antibody and immunoblots performed with anti-GFP (top) or anti-HA (middle) antibodies. In parallel, immunoblots were performed for GFP in lysates of transfected cells (bottom). G, confocal micrographs of U2OS cells transfected with EGFP-RIG-I CARD1 or CARD2 and costained for actin (red) 48 h following transfection.
The CARDs of RIG-I Are Required for NOD2 Interactions
Many pro-apoptotic and pro-inflammatory molecules contain CARDs, which serve to mediate specific interactions with downstream CARD-containing molecules and thus promote signaling. For example, CARD-CARD associations between RIG-I and its downstream effector MAVS/VISA/IPS-1/Cardif ultimately leading to the induction of type I IFNs (3, 5, 19). Because RIG-I and NOD2 both contain CARDs, we determined whether these domains were necessary for their interaction. To do this, we performed co-immunoprecipitation studies with constructs of RIG-I and NOD2 lacking both CARDs or containing the tandem CARDs alone (Fig. 2A). We found that although deletion of the CARDs of NOD2 (ΔCARDs) did not alter its interaction with full-length RIG-I, the CARDs of NOD2 expressed alone exhibited an enhanced ability to interact with RIG-I (Fig. 2, B, C, E). Likewise, we found that expression of the CARDs of NOD2 and RIG-I alone exhibited an increased capacity to associate with one another compared with the level of association between full-length or ΔCARDs NOD2 and RIG-I (Fig. 2C, lanes 4 and 5). Although the CARDs of NOD2 were dispensable for its interaction with RIG-I, we found that deletion of the CARDs of RIG-I diminished its interaction with NOD2 markedly, as evidenced by a lack of significant co-immunoprecipitation of HA-NOD2 and either or EGFP-RIG-I ΔCARDs (Fig. 2D) or Flag-RIG-I ΔCARDs (not shown). Furthermore, we found that expression of either the Flag- or EGFP-tagged CARDs of RIG-I alone were sufficient to co-immunoprecipitate both full-length HA-NOD2 and HA-NOD2 CARDs (Fig. 2, C, D, and E). Similar to our findings with NOD2, we found that expression of the CARDs of RIG-I alone exhibited significantly more association with NOD2 than full-length RIG-I (Fig. 2E, lanes 3 and 4). These data show that the CARDs of RIG-I are necessary and sufficient to mediate NOD2 interactions, but that the CARDs of NOD2 are dispensable.
The Second CARD of RIG-I Primarily Mediates NOD2 Interactions
We found that expression of the CARDs of RIG-I were both necessary and sufficient to mediate its interaction with NOD2 (Fig. 2, C–E). As RIG-I contains two tandem CARDs, we next investigated which of these domains was responsible for mediating its interaction with NOD2. To do this, we expressed either the first (CARD1) or second (CARD2) CARD of RIG-I and assessed the ability of these domains to co-immunoprecipitate full-length NOD2. We found that the extent of co-immunoprecipitation between RIG-I CARD2 was significantly more than that of CARD1, but that both domains were capable of immunoprecipitating NOD2 (Fig. 2F). Furthermore, we found that although both RIG-I CARD1 and CARD2 were associated with actin-enriched membrane ruffles, the extent of association with CARD2 was enhanced (Fig. 2G), supporting a role for CARD2 in mediating the localization of RIG-I to the actin cytoskeleton and its association with NOD2.
RIG-I and NOD2 Interact in Vitro
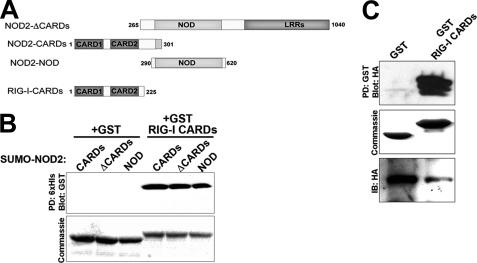
Our co-immunoprecipitation studies indicated that RIG-I and NOD2 formed an interaction when overexpressed in HEK293 cells. To determine if this interaction was direct, we tested whether a GST fusion protein containing both CARDs of RIG-I (GST-RIG-I CARDs) could precipitate the CARDs of NOD2 (CARDs), NOD2 lacking its CARDs (CARDs), or the nucleotide-binding oligomerization domain of NOD2 alone (NOD) prepared as a His6-tagged SUMO fusion proteins (SUMO-CARDs, SUMO-ΔCARDs, or SUMO-NOD). SUMO-NOD2 fusion protein were immobilized on Ni-NTA agarose beads and incubated with purified GST-RIG-I-CARDs. Beads were washed, and precipitated proteins were blotted for GST. All three SUMO-NOD2 proteins precipitated GST-RIG-I-CARDs, but not GST alone (Fig. 3B).
FIGURE 3.
RIG-I and NOD2 interact in vitro. A, schematic of the truncated RIG-I and NOD2 proteins used in in vitro pulldown studies. Numbers represent amino acid residues. B, purified His6-tagged SUMO-NOD2 CARDs, ΔCARDs, or NOD conjugated to Ni-NTA beads was added to pull-down buffer containing GST or GST-RIG-I CARDs. Precipitated proteins were subjected to Western blot analysis with a monoclonal anti-GST antibody (top). Alternatively, samples were run on an SDS-polyacrylamide gel and stained with Coomassie blue to control for loading of GST and GST-RIG-I CARDs (bottom). C, HEK293 cells transfected with HA-NOD2 were lysed 48 h following transfection and incubated with GST or GST-RIG-I CARDs conjugated to Sepharose beads. Precipitated proteins were subjected to Western blot analysis with anti-HA antibody (top). Alternatively, supernatant (representative of un-precipitated proteins) were subjected to immunoblot for HA (bottom). In parallel, samples were run on an SDS-polyacrylamide gel and stained with Coomassie Blue to control for loading of GST and GST-RIG-I CARDs (middle).
To further confirm a direct interaction between RIG-I and NOD2, we overexpressed full-length HA-tagged NOD2 in HEK293 cells and performed in vitro binding experiments with GST- RIG-I-CARDs (or GST alone) immobilized on glutathione-Sepharose beads in lysates of transfected cells. We found that GST-RIG-I-CARDs, but not GST alone, was capable of precipitating full-length overxpressed NOD2 (Fig. 3C). Taken together, these results confirm that the CARDs of RIG-I interact directly with multiple domains of NOD2.
NOD2 Expression Negatively Regulates RIG-I-mediated IFNβ Signaling
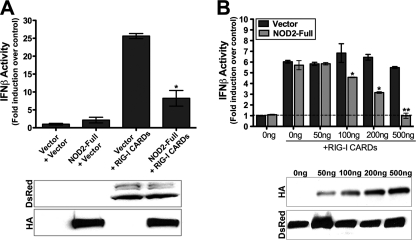
The C-terminal domain of RIG-I serves as a regulatory repressor domain that masks the exposure of the CARDs to prevent downstream signaling in the absence of stimulus. The CARDs of RIG-I thus function as signal activators and when overexpressed alone, lead to constitutive type I IFN induction (20). Because we observed an interaction between RIG-I and NOD2, we assessed the effects of NOD2 overexpression on type I IFN signaling mediated by the CARDs of RIG-I by using an IFNβ luciferase reporter assay. HEK293 cells do not express detectable levels of endogenous NOD2 and thus serve as an ideal model for these studies. To determine if NOD2 inhibited signaling by RIG-I CARDs, we expressed the CARDs alone of RIG-I fused to DsRed at the N terminus (DsRed-RIG-I CARDs) in the absence or presence of NOD2. As expected, when overexpressed alone, DsRed-RIG-I CARDs led to pronounced IFNβ promoter activity (Fig. 4A). However, when HA-tagged full-length NOD2 was co-expressed with DsRed-RIG-I CARDs, there was a significant decrease in the level of IFNβ promoter activity, despite similar levels of DsRed-RIG-I CARDs expression (Fig. 4A). Furthermore, we found an increasing dose-dependent inhibition of RIG-I CARDs-mediated IFNβ induction with increasing amounts of NOD2 transfection (Fig. 4B). These results suggest that NOD2 acts as a negative regulator of RIG-I signaling, presumably by sequestering the CARDs of RIG-I at a subcellular localization distinct from the site of MAVS distribution (mitochondria/peroxisomes (4, 21)).
FIGURE 4.
NOD2 negatively regulates RIG-I-CARDs-mediated IFN signaling. A, luciferase assay (expressed as fold IFNβ induction versus vector controls) from HEK293 cells transfected with vector, EGFP-NOD2 (300 ng) or DsRed-RIG-I CARDs (300 ng) alone or in combination and an IFNβ promoter luciferase construct (100 ng). B, luciferase assay (expressed as fold IFNβ induction versus vector controls) from HEK293 cells transfected with vector or DsRed-RIG-I CARDs (200 ng) and the indicated concentrations of EGFP-NOD2 and an IFNβ promoter luciferase construct (100 ng). Data are shown as mean ± standard deviation. Asterisks indicate p values < 0.05 (*) or <0.001 (**). Immunoblots from lysates are shown at the bottom of A and B.
RIG-I CARD2 Expression Negatively Regulates MDP-stimulated NOD2 Signaling
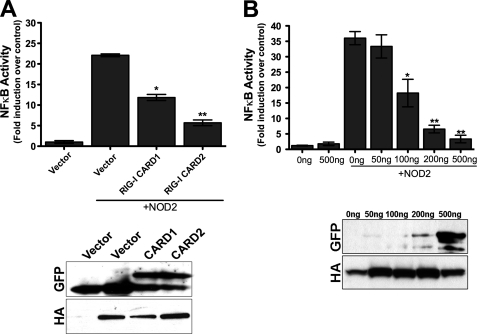
Muramyl dipeptide (MDP), a peptidoglycan motif common to all bacteria, is the ligand for NOD2 (22). Activation of NOD2 by MDP results in the activation of the NF-κB signaling pathway to stimulate antibacterial innate immune signaling (6, 8). Because we found that expression of NOD2 partially abrogated RIG-I-mediated signal transduction, we determined whether RIG-I might function as a negative regulator of MDP-generated NOD2 signaling. We found that expression of NOD2 alone led to a significant increase in NF-κB signal transduction that was significantly inhibited by RIG-I CARD2, and to a lesser extent, RIG-I CARD1 (Fig. 5A). These results are consistent with our co-immunoprecipitation studies indicating that the second CARD of RIG-I interacted with NOD2 with better efficiency that CARD1 (Fig. 2F). Moreover, we found that expression of increasing concentrations of RIG-I CARD2 (in the presence of overexpressed NOD2) abolished MDP-mediated activation of NF-κB signaling through NOD2 (Fig. 5B), indicating that RIG-I negatively regulates NF-κB signaling via NOD2.
FIGURE 5.
RIG-I CARD2 negatively regulates MDP-induced NOD2 inflammatory signaling. A, luciferase assay (expressed as fold NF-κB induction versus vector controls) from HEK293 cells transfected with HA-NOD2 (300 ng) and either vector or EGFP-RIG-I CARD1 or CARD2 (300 ng) and an NF-κB promoted luciferase construct (100 ng). B, luciferase assay (expressed as fold NF-κB induction versus vector controls) from HEK293 cells transfected with vector or EGFP-RIG-I CARD2 (200 ng) at the indicated concentrations and HA-NOD2 and an NF-κB promoter luciferase construct (100 ng). Transfected cells were exposed to 10 μg/ml MDP for 18 h and luciferase measured. Data are shown as mean ± S.D. Asterisks indicate p values < 0.05 (*) or <0.001 (**). Immunoblots from lysates are shown at the bottom of A and B.
Crohn Disease-associated Mutants of NOD2 Interact with RIG-I
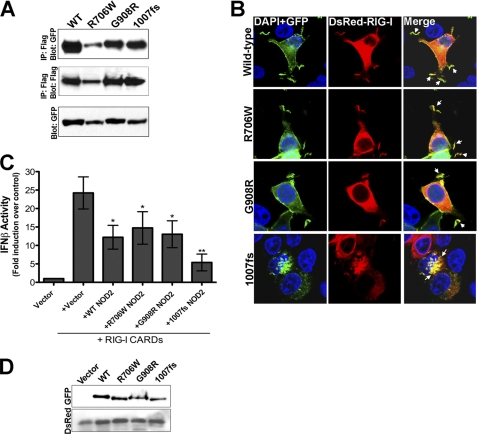
Mutations in the NOD2 gene are associated with an increased risk of CD (7, 8). Three mutations within the leucine-rich repeat region (LRR) of NOD2 have been associated with the development of CD: R702W, G908R, and a C-insertion mutation at nucleotide 3020 that results in a frameshift at the second nucleotide of codon 1007 and a truncated gene product (1007fs). Because CD-associated NOD2 mutations may result in altered associations with binding partners, we investigated the ability of RIG-I to interact with these mutants (R702W, G908R, and 1007fs). To do this, we constructed EGFP-fused NOD2 containing CD-associated mutations and assessed the ability of these mutants to co-immunoprecipitate RIG-I in transiently transfected HEK293 cells. We found that there were no significant differences between the level of RIG-I associated with CD-associated NOD2 mutants in comparison to wild-type, indicating that CD-associated mutations in NOD2 retain their ability to bind RIG-I (Fig. 6A).
FIGURE 6.
Crohn disease-associated mutants of NOD2 associate with RIG-I. A, HEK293 cells were transfected with Flag-RIG-I (250 ng) and EGFP-tagged wild-type (WT), R706W, G908R, or 1007fs NOD2 (250 ng). Following transfection (∼48 h), cells were lysed, and immunoprecipitations performed with anti-Flag antibody and immunoblots performed with anti-GFP (top) or anti-Flag (middle) antibodies. In parallel, immunoblots were performed for GFP in lysates of transfected cells (bottom). B, confocal micrographs of HEK293 cells transfected with EGFP-NOD2 WT, R706W, G908R, or 1007fs NOD2 (250 ng) and DsRed-RIG-I (250 ng) 48 h following transfection. Arrows denote sites of colocalization. C, luciferase assay (expressed as fold IFNβ induction versus vector controls) from HEK293 cells transfected with vector or EGFP-WT, R706W, G908R, or 1007fs NOD2 (250 ng) and DsRed-RIG-I CARDs (200 ng) and a IFNβ-promoted luciferase construct (100 ng). Data are shown as mean ± S.D. Asterisks indicate p values < 0.05 (*) or <0.001 (**). D, immunoblots for GFP and DsRed from lysates in C.
As we found that all three CD-associated NOD2 mutants retained their ability to associate with RIG-I, we next determined whether they exhibited similar patterns of cellular colocalization with RIG-I. Previous studies have shown that although both R702W and G908R NOD2 mutants retain their localization to membrane ruffles, 1007fs NOD2 relocalizes from the plasma membrane to intracellular vesicles (15). We found that similar to wild-type NOD2, R702W, and G908R NOD2 colocalized with RIG-I to actin-enriched membrane ruffles (Fig. 6B). Similar to previous reports (15), we found that 1007fs NOD2 no longer localized to membrane ruffles and was instead heavily associated with large intracellular vesicles (Fig. 6B). Interestingly, we found that RIG-I was relocalized to these vesicles in the presence of 1007fs NOD2 (Fig. 6B).
Crohn Disease-associated Mutants Negatively Regulate RIG-I-mediated IFNβ Induction
We found that coexpression of NOD2 negatively regulated IFNβ induction via the CARDs of RIG-I (Fig. 4, A and B). Because we also observed interactions between CD-associated mutants of NOD2 and RIG-I, we next determined whether these mutants would display differences in their capacities to inhibit RIG-I-mediated type I IFN induction. We found that similar to WT NOD2, all three CD-associated NOD2 mutants exhibit negative regulation of IFNβ signaling mediated by the CARDs of RIG-I (Fig. 6, C and D). However, 1007fs NOD2 exhibited the most pronounced inhibition of RIG-I signaling (Fig. 6C).
NOD2 and CD-associated NOD2 Mutants Inhibit Endogenous RIG-I-mediated Antiviral Signaling
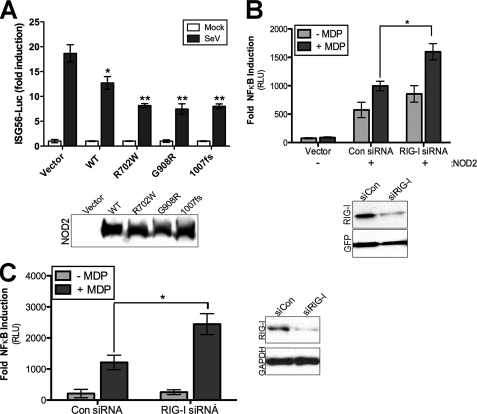
RIG-I is a key component in the initiation of type I IFN induction in response to virus infections. Because we observed an inhibition of type I IFN induction via overexpression of RIG-I with NOD2, we next determined whether NOD2 overexpression could attenuate endogenous RIG-I-mediated antiviral signaling. To that end, we transfected both wild-type and CD-associated NOD2 mutant constructs with an interferon stimulated gene (ISG) 56-Luciferase reporter and then infected cells with Sendai virus (SeV), which is detected by RIG-I (23). We found that both wild-type NOD2, and to a greater extent CD-associated NOD2 mutants, inhibited ISG56 luciferase induction in response to SeV infection (Fig. 7A), indicating that NOD2 acts as a negative regulator of RIG-I antiviral signaling.
FIGURE 7.
RIG-I suppresses NOD2 in intestinal cells. A, luciferase assay from HEK293 cells transfected with the indicated HA-NOD2 constructs and ISG56-Luciferase reporter. Forty-eight hours post-transfection, cells were stimulated with Sendai virus (100 HAU/ml) for 16 h and luciferase activities measured. Results are expressed as fold induction of ISG56-luciferase relative to that of mock-infected cells after normalizing to Renilla luciferase. Immunoblots of HA-NOD2 expression are shown in the bottom panel. B, top, luciferase assay (expressed as fold NF-κB induction versus vector controls) from HeLa cells stably expressing control or RIG-I shRNA and transfected with HA-NOD2 (250 ng) plus NF-κB promoter luciferase (100 ng) and MDP (10 μg/ml) for 24 h. Bottom, immunoblots from lysates shown at top. C, left, luciferase assay (expressed as fold NF-κB induction versus untransfected controls) from HT29 cells transfected with control (Con) or RIG-I siRNAs plus NF-κB promoter luciferase (250 ng). Forty-eight hours following transfection, cells were exposed to MDP (20 μg/ml) for 14 h and luciferase activity measured. Right, immunoblots from lysates at left. Data are shown as mean ± S.D. Asterisks indicate p values < 0.05 (*) or <0.001 (**).
Endogenous RIG-I Knockdown Enhances NOD2-mediated NF-κB Signaling
Because we observed an inhibition of RIG-I mediated antiviral signaling by NOD2 overexpression, we next determined the impact of down-regulation of endogenous RIG-I on MDP-mediated NOD2 responses. We first generated a stable HeLa cell line expressing a RIG-I shRNA and assessed the impact of RIG-I down-regulation on NOD2-mediated MDP signaling. (As HeLa cells do not express endogenous NOD2, we performed these studies using overexpressed NOD2.) We found that there was an enhancement of NOD2-mediated NF-κB signaling in response to MDP treatment in HeLa cells in which RIG-I expression was down-regulated (Fig. 7B).
As a correlate to these studies, we performed similar experiments using HT-29 cells, which express both endogenous RIG-I and NOD2. Similar to our findings with overexpressed NOD2, we found that down-regulation of RIG-I expression via transient siRNA transfection enhanced NF-κB signaling in response to MDP treatment (Fig. 7C). These data show that RIG-I acts as a negative regulator of NOD2 signaling in human intestinal epithelial cells.
DISCUSSION
RIG-I and NOD2 are central components in the innate immune response against viral and bacterial pathogens. Here we show that RIG-I and NOD2 form a direct interaction at membrane ruffles and that this interaction may serve to sequester and inhibit RIG-I and NOD2 from downstream inflammatory signaling. We also show that mutations in NOD2 associated with CD still interact with RIG-I and also serve as negative regulators of RIG-I-dependent type I IFN induction. Although two CD-associated mutations, R706W and G908R, localized with RIG-I to membrane ruffles, the 1007fs mutation (the most common NOD2 mutation associated with CD) localized with RIG-I to large intracellular vesicles. These data highlight a role for the cooperative signaling that may exist between RIG-I and NOD2 and suggest that this cooperation may have implications for antimicrobial signaling in CD.
Although a role for NOD2 in gastrointestinal pathology is well established, less is known regarding whether RIG-I plays any role in maintaining inflammatory signaling in the GI tract. Interestingly, mice lacking RIG-I expression exhibit a colitis-like phenotype (24), although it is unclear whether the association between NOD2 and RIG-I plays any role in this process. Our data show that RIG-I and NOD2 colocalize to sites of cellular junctions and actin-enriched membrane ruffles of intestinal epithelial cells (IECs) (Fig. 1A and not shown). IECs represent a unique model for the study of innate immunity as the intestinal mucosa interacts with a variety of foreign invading viruses and bacteria, but is also continually in contact with a vast array of commensal bacteria. Patients with CD exhibit excessive inflammation in response to commensal flora. Although the precise molecular mechanisms for this enhanced inflammation have remained elusive, recent work suggests that defects in NOD2-mediated autophagy are a component of CD etiology. The interaction between NOD2 and the autophagy-regulating factor ATG16L1 is involved in this process and cells expressing 1007fs NOD2 exhibited impaired autophagosome formation, indicating localization of NOD2 to the membrane is a central feature of this process (25). Our data show that the interaction between NOD2 and RIG-I functions to negatively regulate innate immune signaling via both molecules. Although 1007fs NOD2 and RIG-I still interact, they both relocalize from the membrane to intracellular vesicles. Furthermore, 1007fs NOD2 negatively regulates signaling via the CARDs of RIG-I to a greater extent than either wild type NOD2 of other CD-associated mutants of NOD2 (Fig. 6C). These data suggest that the membrane association of NOD2 and RIG-I is an important regulator of their innate immune signaling and that the relocalization of 1007fs NOD2 and RIG-I from the membrane to intracellular vesicles may alter innate immune signaling against both viral and bacterial pathogens.
Our results demonstrating that NOD2 expression inhibits IFNβ induction by the CARDs of RIG-I would suggest that NOD2 acts as a negative regulator of RIG-I signaling. Likewise, we found that the primary CARD of RIG-I responsible for mediating NOD2 interactions, CARD2, inhibited NOD2-mediated NF-κB induction in response to MDP. A primary feature of CARD-dependent RIG-I signaling is the association between the CARDs or RIG-I and MAVS (3, 5, 19). Similarly, the CARDs of NOD2 are required for its association with receptor-interacting serine-threonine kinase 2 (RIPK2), which is involved in the initiation of NF-κB signaling required for the induction of proinflammatory cytokines (8, 26, 27). It thus seems likely that the negative regulation of RIG-I signaling induced by NOD2 expression, and vice versa, is a consequence of the sequestering of the CARDs of RIG-I and NOD2 away from MAVS or RIPK2, respectively. The sequestration is likely to occur at membrane ruffles and/or sites of cell contact, as these are the primary cellular domains where we observed NOD2 and RIG-I to be localized. Given that endogenous NOD2 expression is generally low in the intestine, but can be induced by inflammatory mediators (28–30), the association between NOD2 and RIG-I may serve to alter a primary feature of innate immune signaling, particularly under conditions associated with high levels of inflammation.
Although NOD2 has been linked to susceptibility to CD, it is clear that there are several factors at play that might contribute to the inflammatory state present in the intestinal tracts of patients with CD. These can include genetic factors (such as NOD2 or ATG16L1), the composition of commensal bacterial flora, and even infection with viral pathogens. Recent work has shown that norovirus infections can trigger the onset of inflammation in an ATG16L1 mouse model of CD (31). Thus, there is likely to exist a complex interplay between viral pathogens and susceptibility genes in CD. Our data indicate that the interaction between RIG-I and NOD2 may serve a functional role in regulating type I IFN signaling. We found that 1007fs NOD2 led to the relocalization of RIG-I from membrane ruffles to cytoplasmic vesicles, thus sequestering RIG-I from its normal cellular location. The sequestration of RIG-I could have a pronounced impact on its ability to signal in an antiviral capacity and may enhance the susceptibility of 1007fs NOD2-expressing cells to some viral pathogens, although further studies are required to provide a functional link for these findings.
RIG-I and NOD2 function as key factors in the detection of viral and bacterial pathogens and in the subsequent initiation of innate immune signals to combat these infections. Here we show that RIG-I and NOD2 form an interaction and that this interaction may serve an important function in the regulation of innate immune signaling. Our data support a role for RIG-I and NOD2 associations in the regulation of innate immune signaling and suggest that this regulation may be altered by the main CD-associated mutation of NOD2.
Supplementary Material
Acknowledgment
We thank Tianyi Wang for sharing reagents and providing helpful suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants R01AI081759 (to C. B. C.) and U24AI082673 (to S. N. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- CARD
- caspase activation and recruitment domain
- CD
- Crohn disease
- cytoD
- cytochalasin D
- IFN
- interferon
- IRF
- interferon regulatory factor
- IEC
- intestinal epithelial cell
- LRR
- leucine-rich repeat
- MDA5
- melanoma differentiation-associated gene
- NOD2
- nucleotide oligimerization domain-2
- RIG-I
- retinoic acid-induced gene-I.
REFERENCES
- 1. Sabbah A., Chang T. H., Harnack R., Frohlich V., Tominaga K., Dube P. H., Xiang Y., Bose S. (2009) Nat. Immunol. 10, 1073–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hong G. S., Jung Y. K. (2002) J. Biochem. Mol. Biol. 35, 19–23 [DOI] [PubMed] [Google Scholar]
- 3. Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., Tschopp J. (2005) Nature 437, 1167–1172 [DOI] [PubMed] [Google Scholar]
- 4. Seth R. B., Sun L., Ea C. K., Chen Z. J. (2005) Cell 122, 669–682 [DOI] [PubMed] [Google Scholar]
- 5. Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K. J., Takeuchi O., Akira S. (2005) Nat. Immunol. 6, 981–988 [DOI] [PubMed] [Google Scholar]
- 6. Ogura Y., Inohara N., Benito A., Chen F. F., Yamaoka S., Nunez G. (2001) J. Biol. Chem. 276, 4812–4818 [DOI] [PubMed] [Google Scholar]
- 7. Hugot J. P., Chamaillard M., Zouali H., Lesage S., Cézard J. P., Belaiche J., Almer S., Tysk C., O'Morain C. A., Gassull M., Binder V., Finkel Y., Cortot A., Modigliani R., Laurent-Puig P., Gower-Rousseau C., Macry J., Colombel J. F., Sahbatou M., Thomas G. (2001) Nature 411, 599–603 [DOI] [PubMed] [Google Scholar]
- 8. Ogura Y., Bonen D. K., Inohara N., Nicolae D. L., Chen F. F., Ramos R., Britton H., Moran T., Karaliuskas R., Duerr R. H., Achkar J. P., Brant S. R., Bayless T. M., Kirschner B. S., Hanauer S. B., Nuñez G., Cho J. H. (2001) Nature 411, 603–606 [DOI] [PubMed] [Google Scholar]
- 9. Hampe J., Frenzel H., Mirza M. M., Croucher P. J., Cuthbert A., Mascheretti S., Huse K., Platzer M., Bridger S., Meyer B., Nürnberg P., Stokkers P., Krawczak M., Mathew C. G., Curran M., Schreiber S. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 321–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marks D. J., Harbord M. W., MacAllister R., Rahman F. Z., Young J., Al-Lazikani B., Lees W., Novelli M., Bloom S., Segal A. W. (2006) Lancet 367, 668–678 [DOI] [PubMed] [Google Scholar]
- 11. Mukherjee A., Morosky S. A., Shen L., Weber C. R., Turner J. R., Kim K. S., Wang T., Coyne C. B. (2009) J. Biol. Chem. 284, 6486–6494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kong L., Sun L., Zhang H., Liu Q., Liu Y., Qin L., Shi G., Hu J. H., Xu A., Sun Y. P., Li D., Shi Y. F., Zang J. W., Zhu J., Chen Z., Wang Z. G., Ge B. X. (2009) Cell Host Microbe 6, 150–161 [DOI] [PubMed] [Google Scholar]
- 13. Legrand-Poels S., Kustermans G., Bex F., Kremmer E., Kufer T. A., Piette J. (2007) J. Cell Sci. 120, 1299–1310 [DOI] [PubMed] [Google Scholar]
- 14. McDonald C., Chen F. F., Ollendorff V., Ogura Y., Marchetto S., Lécine P., Borg J. P., Nuñez G. (2005) J. Biol. Chem. 280, 40301–40309 [DOI] [PubMed] [Google Scholar]
- 15. Barnich N., Aguirre J. E., Reinecker H. C., Xavier R., Podolsky D. K. (2005) J. Cell Biol. 170, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu J., Smith K., Hsieh P. N., Mburu Y. K., Chattopadhyay S., Sen G. C., Sarkar S. N. (2010) J. Immunol. 184, 5768–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kustermans G., El Benna J., Piette J., Legrand-Poels S. (2005) Biochem. J. 387, 531–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Németh Z. H., Deitch E. A., Davidson M. T., Szabó C., Vizi E. S., Haskó G. (2004) J. Cell Physiol. 200, 71–81 [DOI] [PubMed] [Google Scholar]
- 19. Xu L. G., Wang Y. Y., Han K. J., Li L. Y., Zhai Z., Shu H. B. (2005) Mol. Cell 19, 727–740 [DOI] [PubMed] [Google Scholar]
- 20. Yoneyama M., Kikuchi M., Matsumoto K., Imaizumi T., Miyagishi M., Taira K., Foy E., Loo Y. M., Gale M., Jr., Akira S., Yonehara S., Kato A., Fujita T. (2005) J. Immunol. 175, 2851–2858 [DOI] [PubMed] [Google Scholar]
- 21. Dixit E., Boulant S., Zhang Y., Lee A. S., Odendall C., Shum B., Hacohen N., Chen Z. J., Whelan S. P., Fransen M., Nibert M. L., Superti-Furga G., Kagan J. C. (2010) Cell 141, 668–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Girardin S. E., Boneca I. G., Viala J., Chamaillard M., Labigne A., Thomas G., Philpott D. J., Sansonetti P. J. (2003) J. Biol. Chem. 278, 8869–8872 [DOI] [PubMed] [Google Scholar]
- 23. Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K. J., Yamaguchi O., Otsu K., Tsujimura T., Koh C. S., Reis e Sousa C., Matsuura Y., Fujita T., Akira S. (2006) Nature 441, 101–105 [DOI] [PubMed] [Google Scholar]
- 24. Wang Y., Zhang H. X., Sun Y. P., Liu Z. X., Liu X. S., Wang L., Lu S. Y., Kong H., Liu Q. L., Li X. H., Lu Z. Y., Chen S. J., Chen Z., Bao S. S., Dai W., Wang Z. G. (2007) Cell Res. 17, 858–868 [DOI] [PubMed] [Google Scholar]
- 25. Travassos L. H., Carneiro L. A., Ramjeet M., Hussey S., Kim Y. G., Magalhães J. G., Yuan L., Soares F., Chea E., Le Bourhis L., Boneca I. G., Allaoui A., Jones N. L., Nuñez G., Girardin S. E., Philpott D. J. (2010) Nat. Immunol. 11, 55–62 [DOI] [PubMed] [Google Scholar]
- 26. Inohara N., Ogura Y., Chen F. F., Muto A., Nuñez G. (2001) J. Biol. Chem. 276, 2551–2554 [DOI] [PubMed] [Google Scholar]
- 27. Abbott D. W., Wilkins A., Asara J. M., Cantley L. C. (2004) Curr. Biol. 14, 2217–2227 [DOI] [PubMed] [Google Scholar]
- 28. Gutierrez O., Pipaon C., Inohara N., Fontalba A., Ogura Y., Prosper F., Nunez G., Fernandez-Luna J. L. (2002) J. Biol. Chem. 277, 41701–41705 [DOI] [PubMed] [Google Scholar]
- 29. Hisamatsu T., Suzuki M., Reinecker H. C., Nadeau W. J., McCormick B. A., Podolsky D. K. (2003) Gastroenterology 124, 993–1000 [DOI] [PubMed] [Google Scholar]
- 30. Begue B., Dumant C., Bambou J. C., Beaulieu J. F., Chamaillard M., Hugot J. P., Goulet O., Schmitz J., Philpott D. J., Cerf-Bensussan N., Ruemmele F. M. (2006) J. Cell. Physiol. 209, 241–252 [DOI] [PubMed] [Google Scholar]
- 31. Cadwell K., Patel K. K., Maloney N. S., Liu T. C., Ng A. C., Storer C. E., Head R. D., Xavier R., Stappenbeck T. S., Virgin H. W. (2010) Cell 141, 1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.