Abstract
Double strand breaks (DSBs) are the most deleterious of the DNA lesions that initiate genomic instability and promote tumorigenesis. Cells have evolved a complex protein network to detect, signal, and repair DSBs. In mammalian cells, a key component in this network is H2AX, which becomes rapidly phosphorylated at Ser139 (γ-H2AX) at DSBs. Here we show that monoubiquitination of H2AX mediated by the RNF2-BMI1 complex is critical for the efficient formation of γ-H2AX and functions as a proximal regulator in DDR (DNA damage response). RNF2-BMI1 interacts with H2AX in a DNA damage-dependent manner and is required for monoubiquitination of H2AX at Lys119/Lys120. As a functional consequence, we show that the H2AX K120R mutant abolishes H2AX monoubiquitination, impairs the recruitment of p-ATM (Ser1981) to DSBs, and thereby reduces the formation of γ-H2AX and the recruitment of MDC1 to DNA damage sites. These data suggest that monoubiquitination of H2AX plays a critical role in initiating DNA damage signaling. Consistent with these observations, impairment of RNF2-BMI1 function by siRNA knockdown or overexpression of the ligase-dead RNF2 mutant all leads to significant defects both in accumulation of γ-H2AX, p-ATM, and MDC1 at DSBs and in activation of NBS1 and CHK2. Additionally, the regulatory effect of RNF2-BMI1 on γ-H2AX formation is dependent on ATM. Lacking their ability to properly activate the DNA damage signaling pathway, RNF2-BMI1 complex-depleted cells exhibit impaired DNA repair and increased sensitivity to ionizing radiation. Together, our findings demonstrate a distinct monoubiquitination-dependent mechanism that is required for H2AX phosphorylation and the initiation of DDR.
Keywords: Chromatin Histone Modification, DNA Damage, DNA Repair, E3 Ubiquitin Ligase, Ubiquitin
Introduction
Double strand break (DSB)3 formation in cells immediately triggers the recruitment of DNA damage signaling and repair proteins to the damaged loci, where these proteins form discrete nuclear foci (ionizing radiation (IR)-induced foci). The order and timing of the recruitment of DDR proteins is critical for detection, signaling, and repair of DSBs, which is necessary to maintain genomic stability (1, 2).
One of the initial events occurring at DNA damage loci is phosphorylation of H2AX, a histone H2A variant, at Ser139 of its carboxyl-terminal tail (γ-H2AX) by one or more members of the PI3K-like kinase group, including ATM, ATR, and DNA-PK (3–5). γ-H2AX decorates the chromatin flanking DSBs and recruits many early DDR proteins, such as MDC1 and BRIT1 (also known as MCPH1), to generate IR-induced foci (6–10).
In addition to phosphorylation of H2AX, ubiquitination of H2AX is an important epigenetic marker for DNA lesions in DDR (11–13). Recent studies have highlighted the function of RING-finger ubiquitin ligases RNF8 and RNF168 in promoting accumulation of repair proteins at DSBs in an MDC1-dependent manner (14–19). As expected, accumulating in vitro and in vivo studies have demonstrated that modifications of H2AX play a central role in regulating various cellular responses to DSBs, including DNA repair, cell cycle checkpoints, and tumor suppression (6, 20–23). How the function of H2AX is regulated remains a fundamental question that needs to be answered to elucidate the essential mechanisms controlling DDR.
To identify new and crucial factors regulating the function of H2AX in DDR, we conducted a proteomic analysis to systematically identify H2AX interacting proteins. To our surprise, we identified RNF2 and BMI1 as previously unknown binding partners of H2AX, RNF2 and BMI1 are known as RING-domain containing PcG proteins. They are core components in the polycomb repressive complex 1 and are required for H2A ubiquitination-associated transcriptional silencing (24). During the course of our study, several groups reported the involvement of BMI in DDR (25–27). However, the mechanisms by which the RNF2-BMI1 complex regulate DDR is still unclear. Here, our findings indicated that RNF2-BMI1 complex-mediated histone monoubiquitination is required for H2AX phosphorylation in the initiation of DDR.
EXPERIMENTAL PROCEDURES
Cells and Cell Lines
U2OS cells were maintained in McCoy's 5A medium supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin. 293T and mouse embryonic fibroblast (MEF) cell lines were grown in Dulbecco's modified Eagle's medium supplemented with 10% FBS, penicillin, and streptomycin. Anti-RNF2, anti-H2AX, and anti-BMI1 were purchased from Abcam; anti-p-ATM and anti-γ-H2AX were purchased from Upstate Biotechnology; and anti-MDC1 and anti-Bmi1 were purchased from Sigma. Anti-FK2 was purchased from Enzo Life Sciences (incorporating BIOMOL and Alexis). H2AX-null MEFs and AT cells were obtained from Dr. Stephen J. Elledge (Harvard University). Reconstituted stable H2AX−/− MEFs expressing H2AX WT and H2AX K119R/K120R were generated by infection with retroviral vector containing hygromycin-resistance markers. Forty-eight hours after infection, cells were cultured in medium containing 100 μg/ml of hygromycin B. After 2 weeks of selection, the hygromycin-resistance colonies were determined by immunoblotting to assess the expression of wild-type or mutant H2AX.
Plasmids, siRNAs, and Transfection
Myc-RNF2 was provided by Dr. Aaron Ciechanover (Technion-Israel Institute of Technology, Israel). HA-RNF2 was provided by Dr. Seongman Kang (Korea University, Korea). GST-RNF2 was provided by Dr. Titia K. Sixma (Netherlands Cancer Institute, Netherlands). FLAG-H2AX was provided by Dr. Claudia Lukas (Institute of Cancer Biology and Centre for Genotoxic Research, Denmark). GST-H2AX and GST-BMI1 were generated by polymerase chain reaction (PCR) from MCF-10A cells, by using primers with restriction enzyme sites, and subcloned into pGEX5X-2 plasmids (GE Healthcare) in-frame. All mutations described were generated by using the QuikChange Site-directed Mutagenesis Kit (Stratagene). Plasmids were verified by DNA sequencing. The siRNA duplexes were 19 base pairs with a 2-base deoxynucleotide overhang. ON-TARGET SMARTpool siRNAs against RNF2, H2AX, and BMI1 were purchased from Dharmacon Research. The sequences of RNF2 siRNA2 and siRNA3 oligonucleotides were GAGAAAUACUGGAAAGUGA and GCACAGACGAGAUACAUAA, respectively. Control siRNAs were purchased from Dharmacon. All plasmid transfections were performed by using FuGENE 6 (Roche Applied Science). Cells were transfected with siRNA duplexes using Oligofectamine (Invitrogen), following the manufacturer's instructions. Transfection in MEFs was performed as standard virus infection.
Immunoblot, Immunoprecipitation, and Immunofluorescent Analysis
For immunoblotting, cells were sonicated in urea buffer (8 m urea, 150 mm β-mercaptoethanol, 50 mm Tris-HCl, pH 7.5), and cellular debris was removed by centrifugation. Protein concentration was determined by using the Bio-Rad Protein determination reagent. Proteins were loaded on an SDS-PAGE gel and transferred to nitrocellulose, and immunoblots were performed using the appropriate antibodies. For immunoprecipitation, cells were resuspended in low-salt permeabilization buffer (10 mm Hepes, pH 7.4, 10 mm KCl, 0.05% Nonidet P-40 and protease inhibitors (Roche Applied Science)) on ice for 10 min. Then, nuclei were recovered and resuspended in 0.2 m HCl overnight. The soluble fraction was neutralized with 1 m Tris-HCl, pH 8.0. Protein concentration was determined by using the Bio-Rad Protein determination reagent. The clarified extract was incubated with the appropriate HA or Myc fusion protein-agarose overnight at 4 °C. The immunocomplexes were washed three times in RIPA buffer, boiled in SDS sample buffer, and analyzed by immunoblotting as described above.
In Vitro Ubiquitination Assay
GST-RNF2, GST-BMI1, GST-H2AX, GST-RNF2 H69Y, GST-H2AX, GST-H2AX K96R, K119R, and K120R were purified on glutathione-Sepharose (Sigma). The expression levels of protein were further confirmed by immunoblotting. Reactions were performed in 30 μl of ubiquitylation buffer (50 mm Tris, pH 7.5, 2.5 mm MgCl2, 0.5 mm DTT) containing UB-activating enzyme E1, UBCH5c, biotinylated UB, 0.2 mm ATP, and 1 μg of indicated GST fusion protein. The reactions were thoroughly mixed and incubated at 37 °C for 60 min. The reaction was then stopped by the addition of Laemmli sample buffer, and proteins were resolved by SDS-PAGE.
Colony Formation Assay
U2OS cells transfected with siRNA were seeded at 500 cells per 6-cm dish and irradiated with various doses of IR. Cells were incubated for 14 days at 37 °C to allow colonies to form. Colonies were stained with 2% crystal violet and counted. Colonies were defined as groups of 50 or more cells.
Comet Assay
DSB repair was analyzed by neutral comet assay using the Trevigen's Comet Assay kit (4250–050-K) according to the manufacturer's instruction.
Statistical Analysis
All statistical analysis was done by one-tailed Student's t test.
RESULTS
RNF2-BMI1 Interacts with H2AX in a DNA Damage-induced Manner
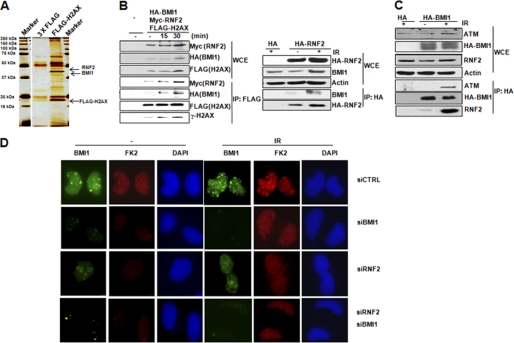
We conducted FLAG-H2AX affinity purification to identify H2AX interacting proteins. From the results of proteomic analysis, we identified RNF2 and BMI1 binding partners of H2AX (Fig. 1A). The specificity of this interaction was confirmed by co-immunoprecipitation of endogenous γ-H2AX and RNF2 (supplemental Fig. S1). More importantly, the interactions among BMI1, RNF2, and H2AX were all remarkably enhanced after IR-induced DNA damage (Fig. 1B). In addition, the interaction between BMI1 and ATM was also enhanced in response to IR (Fig. 1C). These data suggest that the interactions among RNF2-BMI1, H2AX, and ATM have functional significance in DNA damage response. To further support the role of RNF2-BMI1 in signaling DDR, we showed BMI1 to form radiation-induced nuclear foci, which colocalize with p-ATM, γ-H2AX, and MDC1 (supplemental Fig. S2). By using anti-mono- and polyubiquitinylated conjugates FK2 mouse antibody, we showed that BMI1 foci colocalize with radiation-induced nuclear foci formation of DNA damage-induced ubiquitin conjugates (supplemental Fig. S2). More importantly, knockdown RNF2, BMI1, or both significantly reduced FK2 foci formation, suggesting impaired accumulation of ubiquitin conjugates on DNA damage loci (Fig. 1D).
FIGURE 1.
RNF2-BMI1 interacts with H2AX in response to DNA damage. A, silver staining of the H2AX complex separated by SDS-PAGE. 293T cells were transfected with the indicated plasmids, and FLAG-H2AX complexes were purified from whole cell extracts and analyzed by mass spectrometry. B, left, co-immunoprecipitation of RNF2 and BMI1 with H2AX from 293T cells transfected with Myc-RNF2, HA-BMI1, and FLAG-H2AX was analyzed by Western blot analysis at the indicated time points after IR (4 gray). Right, co-immunoprecipitation of BMI1 with RNF2 from 293T cells transfected with HA-RNF2 15 min after IR (4 gray). C, co-immunoprecipitation of ATM and RNF2 with BMI1 from 293T cells transfected with HA-BMI1 15 min after IR (4 gray). D, U2OS cells were transfected with the indicated siRNAs. Seventy-two h later, cells were exposed to IR (4 gray) and analyzed by immunofluorescence assay with FK2 and BMI1 antibodies.
RNF2-BMI1 Complex Regulates Monoubiquitination of H2AX at Lys120
As a component of polycomb repressive complex 1, RNF2 has been implicated in constitutive monoubiquitination of H2A Lys119 at repressed chromatin in gene silencing (24), where BMI1 functions as an enhancer of RNF2 E3 ligase activity to promote the H2A ubiquitination (28). More recently, RNF2 has also been found to mediate nucleotide excision repair-dependent monoubiquitination of H2A at Lys119 in a γ-H2AX-independent manner (29). Based on the known E3 ligase activity of RNF2-BMI1 on chromatin, our results raised the interesting possibility that as binding partners of H2AX, RNF2-BMI1 may mediate a unique monoubiquitination mechanism targeting H2AX and regulating DNA damage signaling. This function of RNF2-BMI1 in H2AX ubiquitination could potentially be distinct from the role of RNF8-mediated H2AX ubiquitination, which primarily regulates diubiquitination and polyubiquitination and has little effects on IR-induced monoubiquitination of γ-H2AX (17).
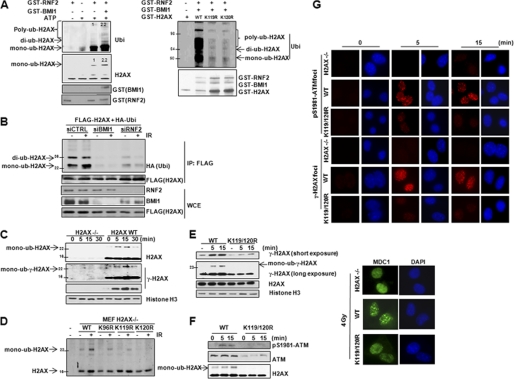
To test this hypothesis, we first determined whether H2AX is a direct substrate for E3 ligase activity of RNF2-BMI1. We performed an in vitro ubiquitination assay to confirm that RNF2 is an E3 ligase for monoubiquitination of H2AX (Fig. 2A and supplemental Fig. S3). We found that H2AX ubiquitination in vitro was induced by wild-type but not by ligase-dead (H69Y) RNF2. Notably, the presence of BMI1 functions as a co-activator of RNF2-mediated H2AX ubiquitination (Fig. 2A, left panel). This result is consistent with an established role of BMI1 as an enhancer of RNF2 E3 ligase activity in gene silencing (28). Next we conducted mutation analysis to identify specific ubiquitination site(s) required for RNF2-BMI1-mediated H2AX monoubiquitination. Protein sequence analysis indicates that there are three potential lysine (Lys) sites for H2AX ubiquitination. Thereby, we generated individual H2AX mutants by replacing Lys96, Lys119, or Lys120 with arginine (Arg). We found that H2AX K119R and K120R abolished RNF2-mediated ubiquitination of H2AX (Fig. 2A, right panel) (28). Then, we tested whether RNF2-BMI1 mediates monoubiquitination of H2AX in vivo. We performed immunoprecipitation of ubiquitinated H2AX in RNF2 or BMI1-depleted cells. As shown in Fig. 2B, when RNF2 or BMI1 were depleted, monoubiquitination of H2AX was reduced in both the presence and absence of IR. To further confirm that H2AX is a substrate for RNF2-BMI1 E3 ligase, we performed an in vivo ubiquitination assay. Overexpression of wild-type RNF2 but not ligase-dead RNF2 promotes monoubiquitination of H2AX (supplemental Fig. S4). These data indicated that RNF2-BMI1 is an E3 ligase for H2AX and is required for H2AX monoubiquitination at Lys119 and Lys120.
FIGURE 2.
Monoubiquitination of H2AX mediated by RNF2-BMI1 regulates DNA damage signaling. A, wild-type (left) and mutant GST-H2AX (right) proteins were incubated with adenosine triphosphate, E1, E2 along with GST, or GST-RNF2 or GST-BMI1 proteins for in vitro ubiquitination of H2AX. B, FLAG-H2AX was overexpressed in U2OS cells transfected with control siRNA, RNF2 siRNA, or BMI1 siRNA. Fifteen min after IR (4 gray), in vivo ubiquitination of H2AX was detected by the indicated antibodies after immunoprecipitation. C, H2AX-deficient MEFs were stably reconstituted with wild-type H2AX and harvested at the indicated time points after IR (4 gray). D, H2AX-deficient MEFs were transiently reconstituted with the indicated H2AX constructs and monoubiquitination of H2AX was detected 15 min after IR (4 gray). E, H2AX-deficient MEFs were stably reconstituted with wild-type H2AX or the H2AX K119R/K120R mutant. The γ-H2AX expression level was detected at the indicated time points after IR (4 gray). F, H2AX-deficient MEFs were stably reconstituted with wild-type H2AX or the H2AX K119R/K120R mutant. Chromatin fractionation was harvested at the indicated time points (IR 4 gray) to detect chromatin-bound ATM. G, radiation-induced nuclear foci formation was analyzed by using antibodies against p-ATM and γ-H2AX after IR (4 gray, 15 min) in H2AX-deficient MEFs stably reconstituted with wild-type H2AX or H2AX K119R/K120R mutant.
As we observed that RNF2-BMI1 interacts with H2AX in a DNA damage-dependent manner and BMI1, a co-activator of RNF2 E3 ligase activity is recruited to DSBs after DNA damage, we speculated that enhanced interaction between the BMI1-RNF2-H2AX complex might promote RNF2-mediated H2AX ubiquitination upon DNA damage. Thus, in the next experiment, we tested whether H2AX monoubiquitination could be induced during DNA damage. We reconstituted wild-type H2AX in H2AX-deficient MEFs and observed that monoubiquitination of H2AX was induced quickly 5 min after IR. The level of H2AX monoubiquitination returned to the undamaged basal level 30 min after IR (Fig. 2C). These observations indicate that monoubiquitination of H2AX is one of the early modifications involved in the dynamic cellular responses to DSBs. Furthermore, by reconstitution of individual H2AX mutants (K96R, K119R, and K120R) into H2AX-deficient MEFs, we found that the H2AX mutant K120R had significantly impaired ubiquitination in response to IR (Fig. 2D), suggesting that H2AX Lys120 is the major ubiquitination site required for DNA damage-induced monoubiquitination of H2AX in vivo.
Monoubiquitination of H2AX Is Required for the Recruitment of DNA Damage Responsive Proteins
Having determined that H2AX monoubiquitination occurs in a DNA damage-dependent manner, we next investigated whether the RNF2-mediated H2AX monoubiquitination has a functional significance in regulating DNA damage signaling. First, we disrupted ubiquitination events by depleting ubiquitin. Interestingly, we found that knockdown of ubiquitin reduced the presence of ubiquitinated γ-H2AX and concurrently decreased H2AX phosphorylation in response to DSBs at an early stage (supplemental Fig. S5). These findings suggested that a novel ubiquitination event, which is distinct from the ubiquitination process mediated by RNF8 and RNF168, is indeed required for the formation of γ-H2AX.
We next tested the possibility that RNF2-mediated H2AX monoubiquitination might orchestrate H2AX phosphorylation in response to DNA damage. We stably reconstituted H2AX-deficient MEFs with wild-type H2AX or H2AX K119R/K120R mutant lacking both RNF2-mediated monoubiquitination sites. It has been reported that alteration of the H2A/H2AX ratio might affect DSB repair (30, 31). As shown under supplemental Fig. S6A, the expression levels of exogenous H2AX (H2AXwt and H2AXK119R/K120R) in H2AX−/− MEFs were similar to the expression level of endogenous H2AX in H2AX+/+ MEFs. These data indicated that our stably reconstituted expression system did not change the ratio of H2A/H2AX. As shown in Fig. 2E, the H2AX mutant significantly reduced H2AX phosphorylation compared with wild-type H2AX. Based on our observation that wild-type and mutant H2AX proteins have the same levels of chromatin binding, we could rule out the possibility that impaired H2AX phosphorylation was due to incorrect incorporation of the mutant H2AX in chromatin. To further confirm that ubiquitination of H2AX is critical for regulating H2AX phosphorylation, we introduced the mutant H2AX K120R into U2OS cells. Interestingly, overexpression of the H2AX K120R mutant significantly impaired endogenous γ-H2AX formation (supplemental Fig. S6B). This result indicated that H2AX K120R has a dominant-negative effect on H2AX phosphorylation. It is worth noting that although our findings suggested that RNF2 can monoubiquitinate both Lys119 and Lys120 of H2AX, ubiquitination of H2AX Lys120 is more important for regulating DDR signaling, which is consistent with observed DNA damage-induced monoubiquitination at this site.
To gain mechanistic insights into how monoubiquination of H2AX (Lys120) might regulate H2AX phosphorylation, we tested whether the K120R H2AX mutant could affect the recruitment of ATM, which is an upstream kinase initiating γ-H2AX formation at an early stage of DDR and plays a central role in signaling transduction induced by DSBs. Compared with wild-type H2AX stably reconstituted MEFs, the recruitment of p-ATM (Ser1981, the activated form of ATM) and total ATM to chromatin was remarkably impaired in mutant H2AX (Lys119/Lys120) MEF cells (Fig. 2F). However, the level of p-ATM in total cell lysates was not changed (supplemental Fig. S7A), suggesting the defective recruitment of ATM to DSBs rather than the defective ATM activation was caused by the H2AX K120R mutation. By using radiation-induced nuclear foci assay with cytoskeleton stripping buffer, we also observed that the H2AX (K119R/K120R) mutant significantly reduced p-ATM foci formation (Fig. 2G), supporting the functional role of monoubiquitination of H2AX Lys120 in recruiting activated ATM to DSBs. Consistent with the lack of p-ATM recruitment, γ-H2AX foci formation was markedly reduced in H2AX Lys119/Lys120 mutant MEFs compared with wild-type H2AX MEFs (Fig. 2G). These observations were further confirmed by transient reconstitution of H2AX-deficient MEFs with wild-type H2AX or the H2AX K120R mutant. Mutant H2AX showed impaired foci formation of p-ATM and γ-H2AX (supplemental Fig. S8). Recruitment of MDC1 by γ-H2AX amplifies H2AX phosphorylation (32), possibly by tethering ATM at DSBs (4). As a result from reduced γ-H2AX formation, we also found that H2AX K120R impaired MDC1 foci formation (Fig. 2G and supplemental Fig. S8). To further demonstrate the involvement of monoubiquitination of H2AX in recruiting ATM in response to DSBs, we conducted biochemistry analysis and found that the H2AX Lys120 mutant had reduced binding affinity for ATM compared with wild-type H2AX (supplemental Fig. S7B). These data collectively showed that monoubiquination of H2AX at Lys120 potentially provides a unique histone marker for the recruitment of ATM and thereby facilitates γ-H2AX formation at an early stage of DDR signaling.
RNF2-BMI1 Complex Regulates DNA Damage Response in an ATM-dependent Manner
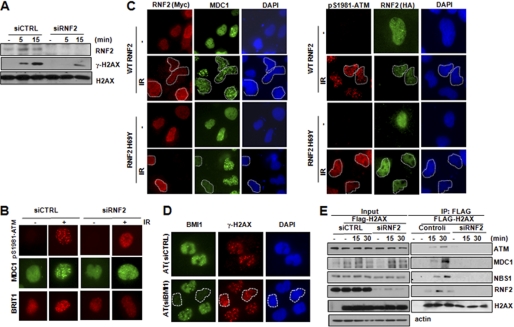
RNF2-BMI1 possesses the E3 ligase activity for H2AX monoubiquitination. Having determined the role of H2AX monoubiquitination at Lys120 in regulating DDR signaling, we then sought to dissect the functional involvement of RNF2-BMI1 in DDR. We first tested whether RNF2-BMI1 could regulate H2AX phosphorylation in response to DNA damage. Interestingly, by using Western blot analysis, we found that in cells with siRNA depletion of RNF2, the total level of γ-H2AX (Fig. 3A) and the monoubiquitination of H2AX (supplemental Fig. S9A) were significantly reduced 15 min after exposure to IR. Consistent with this observation, at an early stage of DDR, BMI1 or RNF2 deficiency significantly impaired p-ATM and γ-H2AX foci formation and consequently the recruitment of various downstream DNA damage responsive proteins to DSBs including MDC1, BRIT1, BRCA1, and 53BP1 as indicated by reduced foci formation 15 min after IR (Fig. 3B and supplemental Fig. S10). Although we found that foci formation of p-ATM (p-S1981) was markedly repressed in RNF2 knockdown cells, the total level of p-ATM was not changed by RNF2 depletion (supplemental Fig. S9B). These findings indicated that RNF2 deficiency leads to a specific defect in the recruitment of p-ATM to DSBs, which is consistent with the impaired function of the H2AX K120R mutant in p-ATM recruitment.
FIGURE 3.
RNF2-BMI1 is involved in regulating DNA damage response. A, U2OS cells were transfected with the indicated siRNAs. Analysis of H2AX phosphorylation was performed by immunoblotting at the indicated time points after exposure to IR (4 gray). B, U2OS cells were transfected with control siRNA and RNF2 siRNA. Seventy-two hours later, cells were exposed to IR (4 gray) and analyzed by immunofluorescence assay with the indicated antibodies 15 min after IR. C, U2OS cells were transfected with wild-type or ligase-dead mutant RNF2 tagged by hemagglutinin (HA). Forty-eight hours later, cells were exposed to IR (4 gray), and 15 min after that, p-ATM (right) or MDC1 (left) foci formation were analyzed with immunofluorescence assay. Expression of ectopic HA-tagged protein in cells was identified by anti-HA antibody. D, A-T human fibroblast cells were transfected with control siRNA or BMI1 siRNA. Seventy-two hours later, cells were analyzed by immunofluorescence assay with γ-H2AX antibody 15 min after IR (4 gray). E, co-immunoprecipitation of H2AX, MDC1, MDC1, and ATM from U2OS cells transfected with control siRNA or RNF2 siRNA with FLAG-H2AX at the indicated the time points after IR (4 gray).
To further confirm the function of RNF2 in regulating the early DDR, we conducted a series of experiments to disrupt RNF2 function. First, we transfected cells with a vector that encodes GFP-tagged shRNA with specificity against RNF2. This vector allowed us to identify GFP-positive cells in which RNF2 expression was successfully depleted. GFP-RNF2 shRNA-expressing cells displayed reduced levels of both γ-H2AX and BRIT1 foci formation (supplemental Fig. S11). Moreover, we investigated whether the E3 ligase activity of RNF2 is required for its function in DDR. We transfected wild-type or ligase-dead RNF2 into cells. Abrogation of RNF2 E3 ligase activity reduced p-ATM (p-S1981) and MDC1 foci formation via a dominant-negative effect (Fig. 3C).
Together, these findings strongly suggested that RNF2-BMI1 coordinates early DDR events potentially via mediating H2AX monoubiquitination. As H2AX monoubiquitination at Lys120 provides a unique histone marker to facilitate the recruitment of ATM in DDR, next we determined whether the regulatory effect of RNF2-BMI1 on γ-H2AX formation is dependent on ATM signaling. First of all, we examined the effects of BMI1 knockdown on γ-H2AX foci formation in a primary human fibroblast A-T cell line lacking functional ATM. It has been shown that although A-T cells have a slower kinetics of γ-H2AX formation, by 30 min after IR, A-T cells have a similar number of γ-H2AX foci compared with the control cell line, which is largely due to the compensatory effect from DNA-dependent protein kinase (DNA-PK) (33). As indicated in Fig. 3D, BMI1 knockdown did not affect γ-H2AX foci formation in A-T cells. Second, we observed that RNF2 knockdown had a similar effect on the kinetics of γ-H2AX formation as the ATM inhibitor (supplemental Fig. S12). Consistent with a potential compensatory effect from DNA-PK, we found that phosphorylation of H2AX in RNF2 knockdown cells was comparable with its level in control cells 60 min after IR. This result suggested that DNA-PK might compensate for impaired ATM function due to RNF2 deficiency at the late stage of DNA damage response. More importantly, this increased phosphorylation of H2AX 60 min after IR was completely abolished by the DNA-PK inhibitor but not by the ATM/ATR inhibitor (supplemental Fig. S12). This finding suggested that the effect of RNF-2-BMI1 on H2AX phosphorylation at a very early stage of DNA damage response is dependent on ATM and potentially through recruiting ATM via H2AX monoubiquitination. To further address the function of RNF2 in regulating the recruitment of ATM to damaged site, next we tested whether RNF2 deficiency leads to impairment of the interaction among H2AX, MDC1, and NBS1, because it has been well known that these early DNA damage-responsive proteins are involved in the recruitment of ATM to the damaged site (32, 34). As shown in Fig. 3E, depletion of RNF2 in cells significantly impaired the interactions among H2AX, MDC1, ATM, and NBS1 after exposure to IR.
RNF2-BMI1 Depletion Causes Hypersensitivity to IR and Impaired DSB Repair
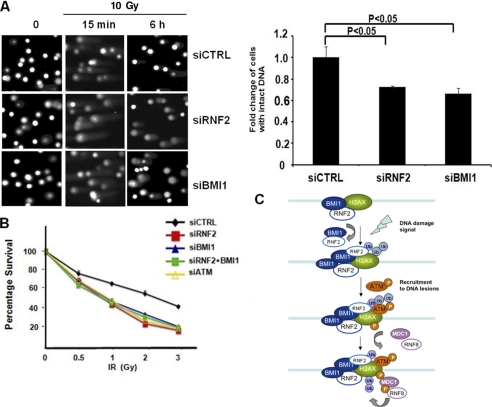
Finally, we examined the biological consequences of defective DDR in RNF2-depleted cells. We first used a neutral comet assay to test whether RNF2-depleted cells resulted in accumulation of DSBs due to impaired DNA repair. Compared with control cells, RNF2-depleted cells showed significantly longer comet tails 6 h after exposure to IR (Fig. 4A), suggesting impaired DNA repair of IR-induced DSBs in RNF2- and BMI1-deficient cells. In addition, we also found that RNF2-depleted cells showed longer comet tails even without irradiation. This result indicated that RNF2 may prevent the accumulation of endogenous DNA damage. Consistent with this notion, cells with deficiency in the RNF2-BMI1 complex exhibited longer comet tails in the absence of radiation. These data indicated that the RNF2-BMI1 complex was involved in controlling endogenous and exogenous DNA damage lesions to maintain genomic stability. Next, we tested whether RNF2-BMI1 complex deficiency leads to increased sensitivity to IR due to the lack of DSB repair. Knockdown cells were plated at low density, irradiated with IR, and tested for their ability to form colonies. We found that RNF2-BMI1 complex-depleted cells were hypersensitive to IR compared with control cells, which was consistent with the reduced cell survival in ATM-knockdown cells (Fig. 4B). Our data therefore highlight a vital role of RNF2 in maintaining genome stability and cell viability. Indeed, impaired RNF2 function in DDR and genome maintenance may contribute to the dramatic developmental defects observed in RNF2 knock-out mice (35).
FIGURE 4.
RNF2 depletion causes impaired DSB repair and hypersensitivity to IR. A, neutral comet assay was performed in U2OS cells transfected with control, BMI1 siRNA, or RNF2 siRNA at the indicated time points after exposure to IR; representative images (left) and quantitative analysis (right) are shown. Each value represents the mean ± S.E. of three independent experiments using Student's t test. B, U2OS cells were transfected with the indicated siRNAs and exposed to IR at the indicated doses. Fourteen days later, cells were stained with crystal violet. Colonies containing more than 50 cells were counted. Each value represents the mean ± S.D. of three independent experiments. C, a proposed model of DNA damage signaling involving RNF2-BMI1-mediated H2AX monoubiquitination and phosphorylation. The recruitment of the RNF2-BMI1 complex at the damaged site mediates monoubiquitination of H2AX, which facilitates the recruitment of p-ATM, γ-H2AX formation, and in turn recruiting downstream DNA damage responsive proteins such as MDC1 to the damaged loci to activate the DNA damage signaling pathway.
DISCUSSION
Recent studies have illustrated the diversity of post-translational modifications on histone variant H2AX, which provides fine-tuned regulatory mechanisms for DDR and DNA repair (15, 17). Among them, several reports have highlighted DNA damage-dependent polyubiquitination of H2AX as a new mechanism in regulating the structural hierarchy of DDR. However, the potential function of H2AX modification by a single ubiquitin moiety, monoubiquitination, in DDR remains largely unknown. As shown in our proposed model (Fig. 4C), our study provided detailed mechanistic evidence to support the role of RNF2-BMI1 in DSB-associated DDR via regulating H2AX monoubiquitination (Lys120), which is involved in the recruitment of p-ATM and the subsequent phosphorylation (Ser139) of H2AX. RNF2-BMI1-mediated H2AX monoubiquitination is a very upstream event required for γ-H2AX formation and subsequent recruitment of early DDR proteins such as MDC1. In addition, as a consequence of impaired recruitment of early DDR proteins, we indeed found that depletion of BMI1 and RNF2 significantly inhibits the recruitment of BRCA1 to the DNA lesions, which is a downstream molecule of the ATM signaling pathway and plays a critical role in HR repair.
During the revision of our manuscript, Facchino et al. (25), recently reported that BMI1 depletion impaired the phosphorylation of H2AX and the recruitment of p-ATM to the damaged sites after irradiation in controlling DSBs by studying a different cell type. Their results were consistent with our findings. More importantly, in addition to these interesting observations, our study identifies a unique ubiquitination mechanism in controlling the initiation of DNA damage signaling, which is distinct from the ubiquitin signaling induced by RNF8 and RNF168 in DDR. As shown in our results, RNF2-BMI1 primarily regulates monoubiquitination of H2AX in response to DNA damage, whereas RNF8 and RNF168 are known to mediate DNA damage-induced polyubiquitination of H2AX. Thereby, these structurally discernable ubiquitin signals generated by different E3 ligases potentially provide a diversity of regulatory mechanisms at different stages of DDR. It has been shown that RNF8 binds to MDC1, which is recruited to DSBs dependent on activation of γ-H2AX. RNF8 promotes a series of ubiquitination events to recruit BRCA1 to DSBs and also provides ubiquitin marks of H2AX and H2A to recruit a second E3 ligase RNF168 to amplify ubiquitin signals. In contrast, our findings indicate that RNF2-BMI1 initiates H2AX monoubiquitination. This unique H2AX modification precedes and initiates the recruitment of ATM, MDC1, and BRCA1 to DNA damage sites.
It is well established that H2AX is phosphorylated by ATM, ATR, and DNA-PK kinases and acts as a platform to recruit and retain repair proteins to DNA lesions. In agreement with this notion, the mutant H2AX K120R lacking the monoubiquitination site at Lys120 significantly impairs ATM recruitment and consequently the formation of γ-H2AX. Collectively our data indicate that RNF2-BMI1 plays a distinct role at an early stage of DDR by initiating monoubiquitination of H2AX at Lys120. It is worth noticing that Ismail et al. (26), and Alajez et al. (27), also reported that the RNF2-BMI1 complex mediates the monoubiquitination of H2AX. However, both of them focused on that BMI1 knockdown led to decreased 53BP1 and BRCA1 foci formation at relative late time points after IR, which occur downstream of γ-H2AX formation. By carefully dissecting DDR in RNF2-BMI-depleted cells at a very early stage, we observed that the RNF2-BMI1 complex was involved in regulating the activation of H2AX. With additional mechanistic studies, we further revealed that monoubiquitination of H2AX specifically occurs at Lys119/Lys120, which has not been reported from other groups. It is well known that the overlapping functions of ATM and DNA-PK play an important role in maintaining genomic stability after exposure to a DNA-damaged agent. In the conditions of impaired ATM function, which activates H2AX at an initial stage of DDR, the compensatory effect of DNA-PK could contribute to H2AX phosphorylation at late time points after IR. This compensatory effect from DNA-PK can be induced 30 min after IR. As shown in supplemental Fig. S12, phosphorylation of H2AX in RNF2-deficient cells was only delayed without treatment with the DNA-PK inhibitor, but it was completely abolished in the presence of the DNA-PK inhibitor. This phenomenon could explain why other groups did not observe impaired H2AX phosphorylation in BMI1-depleted cells at 1 h or even later time points after exposure to IR. In summary, our work provides illuminating evidence of how ubiquitination and phosphorylation interact in coordinating the complex cellular system to maintain genomic stability at different stages of DDR.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S12.
- DSB
- double-strand break
- IR
- ionizing radiation
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1. Paull T. T., Rogakou E. P., Yamazaki V., Kirchgessner C. U., Gellert M., Bonner W. M. (2000) Curr. Biol. 10, 886–895 [DOI] [PubMed] [Google Scholar]
- 2. Downs J. A., Nussenzweig M. C., Nussenzweig A. (2007) Nature 447, 951–958 [DOI] [PubMed] [Google Scholar]
- 3. Rogakou E. P., Boon C., Redon C., Bonner W. M. (1999) J. Cell Biol. 146, 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Harper J. W., Elledge S. J. (2007) Mol. Cell 28, 739–745 [DOI] [PubMed] [Google Scholar]
- 5. Jackson S. P., Bartek J. (2009) Nature 461, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fernandez-Capetillo O., Lee A., Nussenzweig M., Nussenzweig A. (2004) DNA Repair 3, 959–967 [DOI] [PubMed] [Google Scholar]
- 7. Stucki M., Jackson S. P. (2006) DNA Repair 5, 534–543 [DOI] [PubMed] [Google Scholar]
- 8. Stewart G. S., Wang B., Bignell C. R., Taylor A. M., Elledge S. J. (2003) Nature 421, 961–966 [DOI] [PubMed] [Google Scholar]
- 9. Rai R., Dai H., Multani A. S., Li K., Chin K., Gray J., Lahad J. P., Liang J., Mills G. B., Meric-Bernstam F., Lin S. Y. (2006) Cancer Cell 10, 145–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xie A., Hartlerode A., Stucki M., Odate S., Puget N., Kwok A., Nagaraju G., Yan C., Alt F. W., Chen J., Jackson S. P., Scully R. (2007) Mol. Cell 28, 1045–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bergink S., Jentsch S. (2009) Nature 458, 461–467 [DOI] [PubMed] [Google Scholar]
- 12. Lukas J., Bartek J. (2009) Nature 458, 581–583 [DOI] [PubMed] [Google Scholar]
- 13. Messick T. E., Greenberg R. A. (2009) J. Cell Biol. 187, 319–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stewart G. S., Panier S., Townsend K., Al-Hakim A. K., Kolas N. K., Miller E. S., Nakada S., Ylanko J., Olivarius S., Mendez M., Oldreive C., Wildenhain J., Tagliaferro A., Pelletier L., Taubenheim N., Durandy A., Byrd P. J., Stankovic T., Taylor A. M., Durocher D. (2009) Cell 136, 420–434 [DOI] [PubMed] [Google Scholar]
- 15. Mailand N., Bekker-Jensen S., Faustrup H., Melander F., Bartek J., Lukas C., Lukas J. (2007) Cell 131, 887–900 [DOI] [PubMed] [Google Scholar]
- 16. Doil C., Mailand N., Bekker-Jensen S., Menard P., Larsen D. H., Pepperkok R., Ellenberg J., Panier S., Durocher D., Bartek J., Lukas J., Lukas C. (2009) Cell 136, 435–446 [DOI] [PubMed] [Google Scholar]
- 17. Huen M. S., Grant R., Manke I., Minn K., Yu X., Yaffe M. B., Chen J. (2007) Cell 131, 901–914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kolas N. K., Chapman J. R., Nakada S., Ylanko J., Chahwan R., Sweeney F. D., Panier S., Mendez M., Wildenhain J., Thomson T. M., Pelletier L., Jackson S. P., Durocher D. (2007) Science 318, 1637–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang B., Elledge S. J. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 20759–20763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bonner W. M., Redon C. E., Dickey J. S., Nakamura A. J., Sedelnikova O. A., Solier S., Pommier Y. (2008) Nat. Rev. Cancer 8, 957–967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bassing C. H., Chua K. F., Sekiguchi J., Suh H., Whitlow S. R., Fleming J. C., Monroe B. C., Ciccone D. N., Yan C., Vlasakova K., Livingston D. M., Ferguson D. O., Scully R., Alt F. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 8173–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Celeste A., Petersen S., Romanienko P. J., Fernandez-Capetillo O., Chen H. T., Sedelnikova O. A., Reina-San-Martin B., Coppola V., Meffre E., Difilippantonio M. J., Redon C., Pilch D. R., Olaru A., Eckhaus M., Camerini-Otero R. D., Tessarollo L., Livak F., Manova K., Bonner W. M., Nussenzweig M. C., Nussenzweig A. (2002) Science 296, 922–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Celeste A., Fernandez-Capetillo O., Kruhlak M. J., Pilch D. R., Staudt D. W., Lee A., Bonner R. F., Bonner W. M., Nussenzweig A. (2003) Nat. Cell Biol. 5, 675–679 [DOI] [PubMed] [Google Scholar]
- 24. Wang H., Wang L., Erdjument-Bromage H., Vidal M., Tempst P., Jones R. S., Zhang Y. (2004) Nature 431, 873–878 [DOI] [PubMed] [Google Scholar]
- 25. Facchino S., Abdouh M., Chatoo W., Bernier G. (2010) J. Neurosci. 30, 10096–10111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ismail I. H., Andrin C., McDonald D., Hendzel M. J. (2010) J. Cell Biol. 191, 45–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Alajez N. M., Shi W., Hui A. B., Yue S., Ng R., Lo K. W., Bastianutto C., O'Sullivan B., Gullane P., Liu F. F. (2009) Cell Death Differ. 16, 1469–1479 [DOI] [PubMed] [Google Scholar]
- 28. Buchwald G., van der Stoop P., Weichenrieder O., Perrakis A., van Lohuizen M., Sixma T. K. (2006) EMBO J. 25, 2465–2474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bergink S., Salomons F. A., Hoogstraten D., Groothuis T. A., de Waard H., Wu J., Yuan L., Citterio E., Houtsmuller A. B., Neefjes J., Hoeijmakers J. H., Vermeulen W., Dantuma N. P. (2006) Genes Dev. 20, 1343–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Celeste A., Difilippantonio S., Difilippantonio M. J., Fernandez-Capetillo O., Pilch D. R., Sedelnikova O. A., Eckhaus M., Ried T., Bonner W. M., Nussenzweig A. (2003) Cell 114, 371–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bassing C. H., Suh H., Ferguson D. O., Chua K. F., Manis J., Eckersdorff M., Gleason M., Bronson R., Lee C., Alt F. W. (2003) Cell 114, 359–370 [DOI] [PubMed] [Google Scholar]
- 32. Stucki M., Clapperton J. A., Mohammad D., Yaffe M. B., Smerdon S. J., Jackson S. P. (2005) Cell 123, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 33. Stiff T., O'Driscoll M., Rief N., Iwabuchi K., Löbrich M., Jeggo P. A. (2004) Cancer Res. 64, 2390–2396 [DOI] [PubMed] [Google Scholar]
- 34. Uziel T., Lerenthal Y., Moyal L., Andegeko Y., Mittelman L., Shiloh Y. (2003) EMBO J. 22, 5612–5621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Voncken J. W., Roelen B. A., Roefs M., de Vries S., Verhoeven E., Marino S., Deschamps J., van Lohuizen M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2468–2473 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.