Abstract
Although much is known about interleukin (IL)-1β and its role as a key mediator of cartilage destruction in osteoarthritis, only limited information is available on IL-1β signaling in chondrocyte dedifferentiation. Here, we have characterized the molecular mechanisms leading to the dedifferentiation of primary cultured articular chondrocytes by IL-1β treatment. IL-1β or lipopolysaccharide, but not phorbol 12-myristate 13-acetate, retinoic acid, or epidermal growth factor, induced nicotinamide phosphoribosyltransferase (NAMPT) expression, showing the association of inflammatory cytokines with NAMPT regulation. SIRT1, in turn, was activated NAMPT-dependently, without any alteration in the expression level. Activation or inhibition of SIRT1 oppositevely regulates IL-1β-mediated chondrocyte dedifferentiation, suggesting this protein as a key regulator of chondrocytes phenotype. SIRT1 activation promotes induction of ERK and p38 kinase activities, but not JNK, in response to IL-1β. Subsequently, ERK and p38 kinase activated by SIRT1 also induce SIRT1 activation, forming a positive feedback loop to sustain downstream signaling of these kinases. Moreover, we found that the SIRT1-ERK complex, but not SIRT1-p38, is engaged in IL-1β-induced chondrocyte dedifferentiation via a Sox-9-mediated mechanism. JNK is activated by IL-1β and modulates dedifferentiation of chondrocytes, but this pathway is independent on NAMPT-SIRT1 signaling. Based on these findings, we propose that IL-1β induces dedifferentiation of articular chondrocytes by up-regulation of SIRT1 activity enhanced by both NAMPT and ERK signaling.
Keywords: Cytokine, Differentiation, ERK, Histone Deacetylase, Signal Transduction, Dedifferentiation, MAPK, NAMPT, SIRT1, Chondrocytes
Introduction
Interleukin (IL)-1β is a proinflammatory cytokine protein that is processed to its active form by proteolytic enzymes such as caspase-1 (1). This cytokine plays a central role in connective tissue destruction and inflammation and is associated with a variety of cellular activities, including cell proliferation, differentiation, and apoptosis. For example, IL-1β in the inflamed joint disrupts the metabolic balance of chondrocytes, leading to a perturbation of the chondrocyte phenotype by reducing type II collagen expression (2). This event induces certain osteoarthritis (OA)2 degradative processes that are associated with the chondrocyte dedifferentiation produced by the onset of fibroblastic type I collagen expression. Mitogen-activated protein kinase (MAPK) is known as a mediator of proinflammatory cytokine-induced destruction of collagen and other extracellular matrix via the induction of matrix metalloproteinases (MMPs) and agrecanases (ADAMTS) (3 the transcription factor activator of protein-1 (AP-1) (4). Our previous report showed that c-Jun activation plays an important role in IL-1β-induced chondrocyte dedifferentiation via AP-1-mediated Sox-9 suppression (5). Other transcription factors, such as early growth response 1, epithelium-specific Ets transcription factor 1, and growth arrest and DNA damage-inducible protein 45β, inhibit transcription of cartilage matrix proteins such as type II collagen (6, 7). Some members of the Wnt family proteins, including Wnt-3a and Wnt-7a, are induced by IL-1β and are associated with cartilage destruction by the regulation of β-catenin signaling (8, 9). In addition, IL-1β inhibits the proliferation and differentiation of chondrocytes through a Sox-9-mediated mechanism (10). Although some molecules have been reported to function in association with chondrocyte dedifferentiation, the understanding of the role and molecular mechanisms of the other proteins in IL-1β-mediated cartilage destruction is at present quite limited.
Nicotinamide phosphoribosyltransferase (NAMPT), also known as visfatin or pre-B-cell colony-enhancing factor, is the rate-limiting enzyme that catalyzes the first step in the biosynthesis of nicotinamide adenine dinucleotide (NAD) from nicotinamide (NAM) (11). Recent studies have demonstrated that NAMPT-mediated NAD+ biosynthesis plays a crucial role in numerous physiological processes, including cell differentiation, the stress response, apoptosis, and organism longevity (12–15). NAMPT is up-regulated in human vascular smooth muscle cells during cell maturation and declines during cell aging (13). Caloric restriction in skeletal myoblasts stimulates NAMPT expression, leading to SIRT1-dependent impairment of myoblast differentiation (14). Neutrophil activation-mediated up-regulation of NAMPT acts as an anti-apoptotic factor for neutrophils in both clinical and experimental sepsis (16). The expression of NAMPT is increased in inflamed synovial tissue in mice and in the plasma and synovial fluid of patients with rheumatoid arthritis (17) or OA (18). In yeast, NAMPT plays an important role in regulating the activity of Sir2 (silent information regulator 2), which is an NAD-dependent deacetylase involved in chromatin silencing (19). In mammals, Sir2 comprises the seven homologues of SIRT1–7 belonging to the class III histone deacetylases; SIRT1 is the most closely related to yeast Sir2. Our previous report showed that ionizing radiation-mediated down-regulation of SIRT1 expression induces cellular senescence in rabbit articular chondrocytes, which in turn is associated with the development of OA (20). SIRT1 interacts with and deacetylates a large range of transcription factors, regulating target gene expression both negatively and positively (21, 22). For example, in human chondrocytes derived from OA patients, SIRT1 is recruited to the promoter of cartilage-specific genes through an interaction with the Sox-9 transcription factor, thereby targeting gene expression (22). Although histone deacetylases evidently affect chondrocyte biology, the exact function of NAMPT and SIRT1 in cartilage physiology remains largely unknown.
In this study, we investigated the role of NAMPT, the regulation of SIRT1 activity, and the interaction of these enzymes with MAPKs in cartilage metabolism, focusing on IL-1β-mediated dedifferentiation of chondrocytes. Here we discuss the potential contribution of NAMPT-dependent SIRT1 activity to the pathology of OA.
EXPERIMENTAL PROCEDURES
Primary Culture of Rabbit Articular Chondrocytes
Individual articular chondrocytes were isolated from knee joint cartilage slices of 2-week-old New Zealand White rabbits (Samtako, Osan, Korea) as described previously (20). Isolated single cells were resuspended in Dulbecco's modified Eagle's medium supplemented with 10% (v/v) fetal bovine serum, 50 μg/ml streptomycin, and 50 units/ml penicillin (Invitrogen). Cells were plated on culture dishes at a density of 5 × 104 cells/cm2 and maintained as monolayers at 37 °C in a humidified atmosphere of 5% CO2 in air. To induce dedifferentiation, passage 0 (P0) chondrocytes were serially subcultured up to P4 by plating cells each time at a density of 3 × 104 cells/cm2.
Treatment of Cells
For the experiment, 3-day cell cultures were treated with IL-1β (Calbiochem), lipopolysaccharide (LPS) (Sigma), phorbol 12-myristate 13-acetate (PMA) (Sigma), retinoic acid (RA) (Sigma), epidermal growth factor (EGF) (Invitrogen), transforming growth factor (TGF)-β1 (R&D Systems, Minneapolis, MN), or fibroblast growth factor (FGF) (R&D Systems) as indicated. For inhibitor or activator studies, the respective agents were added 1 h prior to IL-1β treatment. FK866 was used to inhibit NAMPT (Axon Medchem, Postbus, Netherlands). SB203580, PD98059, and SP600125 were used to inhibit p38, extracellular signal-regulated protein kinase (ERK), and c-Jun N-terminal kinase (JNK) (Calbiochem), respectively. We employed resveratrol to activate SIRT1 and NAM to inhibit SIRT1 (Sigma).
Cell Proliferation and Morphology
Chondrocytes were plated on culture dishes at a density of 5 × 104 cells/cm2. Cells were treated with IL-1β according to the designated experimental conditions. Cell proliferation was determined by direct counting using a hemocytometer, and cellular morphology was observed under microscopy.
Reverse Transcription-PCR
Total RNA was isolated using TRI reagent (Molecular Research Center, Cincinnati, OH). Reverse transcription reactions were performed using ImProm-II (Promega, Madison, WI) with rTaq polymerase (TaKaRa Bio Inc., Shiga, Japan) for 5 min at 70 °C for annealing and 60 min at 42 °C for extension of the first strand. The following primers and conditions were employed for PCR: type I collagen (COL1A1) (441-bp product, annealing temperature 60 °C, 27 cycles), sense 5′-GGC TTT CCT GGA GAG AAA GG-3′ and antisense 5′-ATA GAA CCA GCA GGG CCA GG-3′; type II collagen (COL2A1) (370-bp product, annealing temperature 60 °C, 20 cycles), sense 5′-GAC CCC ATG CAG TAC ATG CG-3′ and antisense 5′-AGC CGC CAT TGA TGG TCT CC-3′; Sox-9 (386-bp product, annealing temperature 62 °C, 27 cycles), sense 5′-GCG CGT GCA GCA CAA GAA GGA CCA CCC GGA TTA CAA GTA C-3′ and antisense 5′-CGA AGG TCT CGA TGT TGG AGA TGA CGT CGC TGC TCA GCT C-3′; glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (299-bp product, annealing temperature 50 °C, 21 cycles), sense 5′-TCA CCA TCT TCC AGG AGC GA-3′ and antisense 5′-CAC AAT GCC GAA GTG GTC GT-3′; SIRT1 (586-bp product, annealing temperature 61 °C, 30 cycles), sense 5′-TAG AGA ACC TTT GCC TCA TC-3′ and antisense 5′-AAA ATG TAA CGA TTT GGT GG-3′; NAMPT (174-bp product, annealing temperature 50 °C, 30 cycles), sense 5′-GCG GCA GAA GCC GAG TTC AAC-3′ and antisense 5′-CCC ATA AAA TAC TGT TTC CTC-3′; and IL-1β (500-bp product, annealing temperature 57 °C, 35 cycles), sense 5′-AGC TCA CCA GTG AGA TGA TG-3′ and antisense 5′-AGG TAT CTT GTC GTT ACT TTC-3′. The primers were designed based on the sequences of the human Sox-9, SIRT1, and NAMPT genes. Sequencing of the resulting PCR products for rabbit Sox-9, SIRT1, and NAMPT revealed that these gene fragments were 93.0, 88.0, and 92.0% homologous to their human counterparts, respectively (data not shown). Quantitative real-time PCR was performed using a Chromo4 cycler (Bio-Rad) and SYBR Premix Ex TaqTM (Takara Bio). All of the real-time PCR reactions were performed in triplicate, and the amplification signal from the target gene was normalized against that of GAPDH in the same reaction.
Reporter Gene Assay
Chondrocytes were transfected with a Sox-9 reporter gene with or without the indicated amount of several expression vectors using Lipofectamine PLUS reagent (Invitrogen) as described previously (5). After incubation of the transfected cells in complete medium for 12 h, the cells were treated with IL-1β, FK866, resveratrol, NAM, and/or MAPK inhibitors for an additional 36 h. Luciferase activity was determined with an assay kit from Promega and normalized against that of β-galactosidase.
Construction of the NAMPT Expression Vector and Transfection
The cDNA for wild-type NAMPT was RT-PCR-amplified from BEAS2B human bronchial epithelial cell line mRNA with the specific primers (sense 5′-GCCGCTAGCGGCCACCATGAATCCTGCGGCAGAAGCC-3′ and antisense 5′-CGGGATCCCGATGATGTGCTGCTTCCAGTTC-3′) designed to introduce NheI and BamHI restriction sequences at the 5′ and 3′ ends of the amplified fragment, respectively. The resulting cDNA was cloned into the NheI and BamHI sites of vector pAcGFP1-N1 (Clontech, Mountain View, CA). Chondrocytes from the day 2 cultures were transfected with pAcGFP1-N1 plasmids coding for wild-type NAMPT using Lipofectamine PLUS (Invitrogen) following the manufacturer's recommended procedure. Transfected cells were then left untreated or treated with the NAMPT inhibitor FK866 in complete medium for 36 h. Alternatively, chondrocytes from the day 2 cultures were infected with either control retrovirus or retrovirus containing wild-type SIRT1 or mutant SIRT-HY cDNA for 4 h as described in a previous report (20). After incubation of the infected cells in complete medium for 24 h, cells were left untreated or treated with 10 ng/ml IL-1β for an additional 36 h.
Knockdown of NAMPT by Small Interfering RNA (siRNA)
We examined two siRNA to silence NAMPT in chondrocytes and found that both caused effective knockdown of NAMPT. The siRNA-1 (5′-CAA ATT GGA TTG AGA CTA TTT-3′) and siRNA-2 (5′-GCA GAA CAC AGT ACC ATA ATT-3′) sequences were synthesized chemically according to the manufacturer's notes (Bioneer, Daejeon, Korea). A scrambled siRNA (5′-CCT ACG CCA CCA ATT TCG TTT-3′), used as a negative control, showed no significant homology to known gene sequences and did not regulate gene expression. Cells were transiently transfected with 50 nm siRNA in a serum-free medium for 5 h using Metafectene reagent (Biontex, Munich, Germany) according to the manufacturer's protocol. Cells were maintained for an additional 48 h, and the depletion of NAMPT was determined by Western blotting and quantitative RT-PCR analysis.
SIRT1 Deacetylase Activity Assay
SIRT1 activity was measured using a fluorometric SIRT1 assay kit (Sigma CS1040) according to the manufacturer's instructions, except that 1 μm trichostatin A was added to the reaction to block the class I, II, and IV histone deacetylases. Briefly, whole cell extracts were prepared with lysis buffer supplemented with 1 μm trichostatin A. 20 μl of the lysates were added to an opaque 96-well plate with 10 μl of acetylated Fluor-de-Lys substrate and 5 μl of NAD+. The extracts were then incubated for 20 min at room temperature, and the reactions were stopped by adding 5 μl of a developer solution supplemented with 1 μm trichostatin A. The fluorescence intensity at 450 nm (excitation 360 nm) was recorded and normalized to micrograms of protein. Experimental values are represented as a percentage of the control.
Determination of Apoptosis
Chondrocytes were plated on 35-mm dishes at a cell density of 4 × 105 cells and subjected to 10 ng/ml IL-1β for the indicated times. For quantification of cell death, cells were trypsinized, washed in phosphate-buffered saline (PBS), incubated in annexin V-FITC and propidium iodide according to the manufacturer's instructions (BD Biosciences), and then analyzed with a FACScan flow cytometer (BD Biosciences).
Immunofluorescence Confocal Microscopy
Chondrocytes were fixed with 3.5% paraformaldehyde in PBS for 10 min at room temperature. The cells were then permeabilized and blocked in PBS containing 0.1% Triton X-100 and 5% fetal calf serum for 30 min, respectively. The cells were washed with PBS three times and incubated for 1 h with the following primary antibodies: mouse monoclonal anti-type II collagen (Chemicon, Temecula, CA) or rabbit polyclonal anti-SIRT1 (Santa Cruz Biotechnology Inc., Santa Cruz, CA). The cells were washed and incubated with rhodamine- or fluorescein-conjugated secondary antibodies, washed again, and then observed using a Zeiss LSM 510 Meta confocal imaging system. Cell nuclei were identified by 4,6-diamidino-2-phenylindole (DAPI) staining.
GST Pulldown Assay
GST (Abcam, Cambridge, MA), GST-ERK2 (Millipore, Bedford, MA), or GST-pERK (Calbiochem) fusion protein was bound with glutathione-agarose beads (Sigma) in binding buffer (50 mm Tris-HCl, 100 mm NaCl, 1 mm EDTA, 1% Triton X-100, 1 mm PMSF, and 10% glycerol, pH 8.0) at 4 °C for 2 h. The bound glutathione-agarose beads were incubated with the active form of His-SIRT1 fusion protein (Abcam) for 12 h on a rotator in a cold room. The complex proteins were size-fractionated by electrophoresis and detected by Western blotting with anti-GST or anti-His antibody.
Western Blot Analysis
Chondrocytes were lysed in a buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% Nonidet P-40, and 0.1% SDS supplemented with protease inhibitors (10 μg/ml leupeptin, 10 μg/ml pepstatin A, 10 μg/ml aprotinin, and 1 mm 4-(2-aminoethyl)benzenesulfonyl fluoride) and phosphatase inhibitors (1 mm NaF and 1 mm Na3VO4). Proteins were fractionated by SDS-PAGE and transferred to a nitrocellulose membrane. Membranes were blocked with 5% nonfat dry milk in Tris-buffered saline and incubated with the primary antibodies for 1 h at room temperature. The following antibodies were used to detect proteins: mouse monoclonal anti-ERK (BD Biosciences); rabbit polyclonal anti-p38, -phospho-p38, -JNK, and -phospho-JNK (Cell Signaling Technology, Inc., Beverly, MA); rabbit polyclonal anti-acetyl-histone H3 (Millipore); mouse monoclonal anti-type II collagen (Chemicon); mouse monoclonal anti-β-actin (Sigma); mouse monoclonal anti-phospho-ERK and anti-phospho-c-Jun (Santa Cruz Biotechnology Inc.); and rabbit polyclonal anti-Sox-9, -SIRT1, -c-Jun, -NAMPT, -His, and -GST (Santa Cruz Biotechnology Inc.). Blots were developed with a peroxidase-conjugated secondary antibody and the ECL system (Amersham Biosciences).
RESULTS
IL-1β Induces Dedifferentiation of Articular Chondrocytes without Cellular Senescence or Cell Death
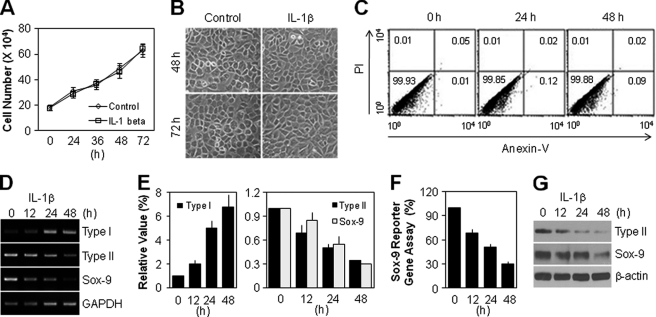
To determine whether IL-1β affects the pathophysiological processes of primary cultured rabbit articular chondrocytes, we assayed cell proliferation, morphological change, and apoptotic cell death after exposure to IL-1β. The total cell number by day 3 was almost the same in the control and IL-1β-treated groups at each time point (Fig. 1A). The cell size, a specific marker for senescent cells, was unchanged in the IL-1β-treated group compared with the control group at 48 and 72 h (Fig. 1B). Flow cytometry analysis revealed that IL-1β treatment did not induce chondrocyte apoptosis after 48 h culture (Fig. 1C). We next examined the phenotype of articular chondrocytes. As shown in Fig. 1D, RT-PCR analysis showed that IL-1β led to the induction of fibroblastic type I collagen expression and suppression of cartilage-specific type II collagen expression. IL-1β also inhibited Sox-9 expression, a major transcription factor of type II collagen. These results are consistent with the quantitative real-time PCR data, which showed a 4.9-fold increase in type I collagen, 2.0-fold decrease in type II collagen, and 1.8-fold decrease in Sox-9 at 24 h after IL-1β treatment (Fig. 1E). The reduced Sox-9 transcriptional activity was directly observed in IL-1β-treated cells as determined by Sox-9 reporter gene assay (Fig. 1F). As expected, Western blot analysis revealed that IL-1β treatment reduced the protein expression of type II collagen and Sox-9 in a time-dependent manner (Fig. 1G). Thus, these results indicate that IL-1β induces chondrocyte dedifferentiation without cellular senescence or apoptosis, which is an accepted function of IL-1β as one of the underlying causes of OA.
FIGURE 1.
IL-1β causes dedifferentiation of articular chondrocytes without apoptosis or cellular senescence-like phenotype. A and B, chondrocytes were left untreated (Control) or treated with 10 ng/ml IL-1β for the indicated periods. The total cell number was quantified by counting the surviving cells using trypan blue solution (A), and cell morphology was observed using light microscopy (B). C–G, chondrocytes were treated with 10 ng/ml IL-1β for the indicated periods. Cell death was detected using a FACScan flow cytometer, and the number in the corner of each quadrant is the percentage of total cells, referring to the percentage of annexin-V or/and propidium iodide (PI)-labeled cells (C). The transcript level of type I collagen, type II collagen, and Sox-9 was determined by conventional PCR (D) and quantitative real-time PCR (E) with GAPDH used as the loading control or internal control. The transcriptional activity of Sox-9 was determined with the reporter gene assay (F). The protein level of type II collagen and Sox-9 was determined by Western blotting (G). β-Actin was used as the loading control. Data are presented as the results of mean values with standard deviations (A, E, and F), a representative photomicrograph (B), or a typical experiment (C, D, and G) from at least four independent experiments.
IL-1β Up-regulates NAMPT Expression, Which Induces SIRT1 Activation in Articular Chondrocytes
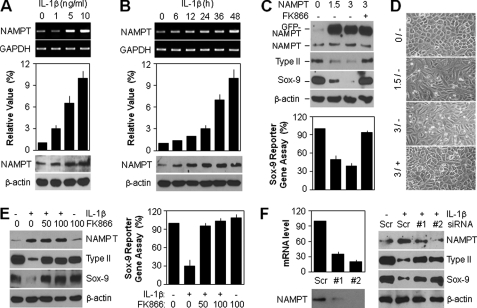
In an attempt to elucidate the role of NAMPT in chondrocyte dedifferentiation, we first determined the expression level of NAMPT after IL-1β treatment. As shown in Fig. 2A, various doses of IL-1β ranging from 0 to 10 ng/ml caused an increase in the transcript (top) and protein (bottom) levels of NAMPT in a concentration-dependent manner. Quantitative PCR analysis, which showed a 10.7-fold increase of NAMPT at 48 h in the 10 ng/ml IL-1β-treated group compared with the control group, was consistent with the conventional PCR data (Fig. 2A, middle). All of these phenomena were also observed to be time-dependent in response to 10 ng/ml IL-1β (Fig. 2B).
FIGURE 2.
IL-1β triggers induction of NAMPT expression, which promotes dedifferentiation of articular chondrocytes. A and B, chondrocytes were treated with the specified doses of IL-1β for 48 h (A) or 10 ng/ml IL-1β for the indicated periods (B). The transcript level of NAMPT was determined by conventional PCR (top) and quantitative real-time PCR (middle) with GAPDH used as the loading control or internal control. The protein level of NAMPT was determined by Western blotting (bottom) with β-actin used as the loading control. C and D, chondrocytes were transfected with the specified doses (μg) of NAMPT expression vector for 12 h and then left untreated (−) or treated (+) with 50 nm FK866 for an additional 36 h. The protein level of NAMPT, type II collagen, and Sox-9 was determined by Western blotting (C, top) with β-actin was used as the loading control. The transcriptional activity of Sox-9 was determined with the reporter gene assay (C, bottom), and cell morphology was observed using light microscopy (D). E, chondrocytes were pretreated with the specified doses (nm) of FK866 for 1 h and then left untreated (−) or treated (+) with 10 ng/ml IL-1β for 48 h. The protein level of NAMPT, type II collagen, and Sox-9 was determined by Western blotting (left) with β-actin used as the loading control. The transcriptional activity of Sox-9 was determined with the reporter gene assay (right). F, chondrocytes were transfected with scrambled siRNA (Scr) or NAMPT siRNAs (#1 and #2) in the absence (left) or presence (right) of 10 ng/ml IL-1β for 48 h. The expression level of NAMPT was determined by quantitative real-time PCR (top left) and Western blotting (bottom left) with GAPDH used as the internal control. The protein level of NAMPT, type II collagen, and Sox-9 was determined by Western blotting (right) with β-actin used as the loading control. Data are presented as the results of a typical experiment (A–C, E, and F) or a representative photomicrograph (D) from at least four independent experiments.
To examine whether NAMPT plays a crucial role in the regulation of Sox-9 and type II collagen, we directly induced chondrocytes to express ectopic wild-type GFP-tagged NAMPT in the absence or presence of its specific inhibitor, FK866. Treatment of 50 nm FK866 in chondrocytes did not show any change in phenotype, such as cell proliferation, cellular senescence, or apoptosis. Up to 72 h, the rate of cell proliferation or cell death was almost same between the control and FK866-treated groups. FK866 treatment did not induce senescence-associated-β-galactosidase activity and morphological change, the hallmarks of cellular senescence (supplemental Fig. 1). The overexpression of NAMPT reduced the Sox-9 and subsequent type II collagen expression levels, but FK866 treatment reversed the NAMPT-induced suppression of these proteins (Fig. 2C, top). Sox-9 reporter gene assay showed that NAMPT significantly reduced the transcriptional activity of Sox-9, an effect that was completely blocked by FK866 treatment (Fig. 2C, bottom). Ectopic overexpression of chondrocytes with NAMPT had a characteristic fibroblast-like morphology distinct from normal cells, whereas FK866 treatment markedly recovered the cell shape to normal (Fig. 2D). Consistent with the data from the direct gene experiments, the IL-1β-mediated induction of NAMPT expression also led to the suppression of Sox-9 and type II collagen, whereas pretreatment with FK866 fully recovered the phenomenon of chondrocytes induced by IL-1β (Fig. 2E, left). The reduction of Sox-9 transcription activity by IL-1β treatment was also significantly reversed in the FK866-treated groups, as determined by direct Sox-9 reporter gene assay (Fig. 2E, right). We also examined whether endogenous NAMPT could regulate chondrocyte dedifferentiation by using siRNA to knock down the endogenous NAMPT in chondrocytes. We found that two siRNAs caused effective knockdown of NAMPT expression as determined by Western blotting and quantitative RT-PCR analysis (Fig. 2F, left). Depletion of endogenous NAMPT recovered the expression level of type II collagen and Sox-9 proteins reduced by IL-1β treatment (Fig. 2F, right). These data collectively suggest that IL-1β-mediated chondrocyte dedifferentiation is attributable to the NAMPT-related signaling pathway in cells.
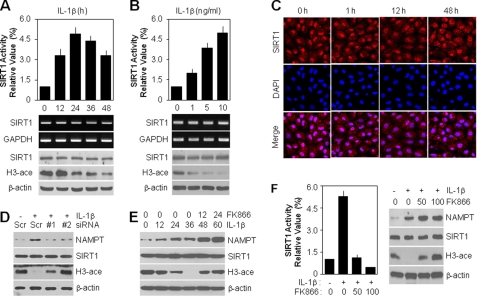
We further analyzed the activity and subcellular localization of intracellular SIRT1, the downstream target molecule in articular chondrocytes of the NAMPT-mediated NAD+ biosynthesis pathway. The cells displayed enhanced SIRT1 activity by 4.8-fold compared with the control group at 24 h after exposure to IL-1β, as determined by a SIRT1 deacetylase fluorometric assay kit (Fig. 3A, top). However, the transcript and protein levels of SIRT1 were unchanged under the condition of IL-1β-mediated chondrocyte dedifferentiation. Consistent with SIRT1 activity, the acetylation level of histone H3, a major deacetylation substrate of SIRT1, resulted in a time-dependent reduction by IL-1β treatment (Fig. 3A, middle and bottom). These phenomena were also dependent upon concentration, which ranged from 0 to 10 ng/ml (Fig. 3B). SIRT1 is known to localize in both the cytosol and the nucleus, and this subcellular localization was unchanged at the early or late time point after IL-1β treatment (Fig. 3C). To understand whether IL-1β-mediated SIRT1 activation is due to NAMPT, we examined the effect of direct loss of function of endogenous NAMPT using siRNAs. As shown in Fig. 3D, down-regulation of endogenous NAMPT reduced SIRT1 activity, based on the recovery of histone H3 acetylation in siRNA-transfected groups. We next examined SIRT1 activity in cells treated with FK866 to inhibit NAMPT activity chemically. NAMPT expression was increased time-dependently with IL-1β treatment and promoted the basal level of SIRT1 activation in a NAMPT-dependent manner. FK866 also time-dependently inhibited SIRT1 activity increased by NAMPT without a decrease in NAMPT protein level (Fig. 3E). We further directly analyzed SIRT1 activity to confirm the effect of NAMPT on SIRT1 activation more clearly in cells treated with IL-1β alone or a combination of IL-1β and FK866. The potently increased SIRT1 activity induced by IL-1β was completely blocked by FK866 in a dose-dependent fashion, as determined by either a direct SIRT1 deacetylase fluorometric assay kit (Fig. 3F, left) or an indirect Western blot analysis, which detected the acetylation level of histone H3 (Fig. 3F, right).
FIGURE 3.
IL-1β induces NAMPT-dependent SIRT1 activity but not expression level in articular chondrocytes. A and B, chondrocytes were treated with 10 ng/ml IL-1β for the indicated periods (A) or with specified doses of IL-1β for 24 h (B). IL-1β-mediated SIRT1 activity was determined using a SIRT1 fluorometric assay kit (top). The transcript level of SIRT1 was determined by conventional PCR (middle), and the protein level of SIRT1 and acetylation of histone H3 (H3-ace) were determined by Western blotting (bottom) with GAPDH and β-actin used as loading controls, respectively. C, chondrocytes were treated with 10 ng/ml IL-1β for the indicated periods. Distribution of SIRT1 was determined by immunofluorescence confocal microscopy. Cells were identified by DAPI staining of the nuclei. D, chondrocytes were transfected with scrambled siRNA (Scr) or NAMPT siRNAs (#1 and #2) in the absence (−) or presence (+) of 10 ng/ml IL-1β for 48 h. Protein levels of NAMPT and SIRT1 and acetylation of histone H3 were determined by Western blotting with β-actin used as the loading control. E, chondrocytes were treated with 10 ng/ml IL-1β alone for 0–36 h, and then cells incubated for 36 h were treated with 50 nm FK866 for an additional 12 or 24 h. Protein levels of NAMPT and SIRT1 and acetylation of histone H3 were determined by Western blotting with β-actin used as the loading control. F, chondrocytes were pretreated with the specified doses (nm) of FK866 for 1 h and then left untreated (−) or treated (+) with 10 ng/ml IL-1β for 24 h. SIRT1 activity was determined using a SIRT1 fluorometric assay kit (left). Protein levels of NAMPT and SIRT1 and acetylation of histone H3 were determined by Western blotting (right) with β-actin used as the loading control. Data are presented as the results of a typical experiment (A, B, and D–F) or a representative photomicrograph (C) from at least four independent experiments.
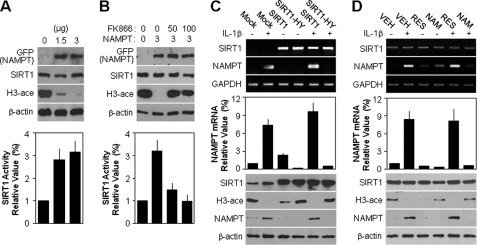
To better understand the relationship between SIRT1 activity and NAMPT expression, we transfected chondrocytes with wild-type NAMPT expression vector. As shown by the acetylation level of histone H3, overexpression of wild-type NAMPT was associated with an induction of SIRT1 activity without any evident alteration of SIRT1 expression (Fig. 4A, top). Consistent with this result, SIRT1 activity was enhanced by 3.2-fold in cells transfected with 3 μg of NAMPT compared with the control group, as determined by a SIRT1 deacetylase fluorometric assay kit (Fig. 4A, bottom). SIRT1 activity induced by ectopic overexpression of NAMPT was significantly inhibited by FK866 treatment in a dose-dependent manner as determined by Western blotting (Fig. 4B, top) and a SIRT1 deacetylase fluorometric assay kit (Fig. 4B, bottom), indicating the NAMPT-dependent positive activation of SIRT1. Next, we investigated whether SIRT1 forms a positive feedback loop by regulating NAMPT expression. For this experiment, chondrocytes were infected with a recombinant retrovirus encoding the wild-type or deacetylase-dead mutant form of dominant-negative SIRT1, SIRT1-HY. As shown in Fig. 4C, ectopic overexpression of SIRT1 or SIRT1-HY was confirmed by conventional PCR (top) and Western blot analysis, and their activities were indirectly assayed by evaluating the acetylation level of histone H3 (bottom). Surprisingly, SIRT1-HY overexpression markedly reduced the IL-1β-induced transcript level, as determined by conventional PCR (Fig. 4C, top) and quantitative PCR (middle), and the protein level of NAMPT, as determined by Western blotting (bottom). Consistent with direct SIRT1-HY gene regulation, indirect inhibition of SIRT1 activity with the chemical inhibitor NAM was also blocked at the NAMPT transcript (Fig. 4D, top and middle) and protein (bottom) levels induced by IL-1β. These results support the formation of a NAMPT-SIRT1 positive feedback loop to sustain downstream signaling.
FIGURE 4.
NAMPT-dependent SIRT1 activity promotes induction of NAMPT expression to form a positive feedback loop in articular chondrocytes. A and B, chondrocytes were transfected with the specified doses (μg) of NAMPT expression vector for 36 h (A) or for 5 h and then treated with the specified doses (nm) of FK866 for an additional 36 h (B). Protein levels of GFP-tagged NAMPT and SIRT1 and acetylation of histone H3 (H3-ace) were detected by Western blotting (top) with β-actin used as the loading control. SIRT1 activity was determined using a SIRT1 fluorometric assay kit (bottom). C, chondrocytes were infected with control retrovirus (Mock) or retrovirus containing wild-type (SIRT1) or dominant-negative SIRT1 (SIRT1-HY) cDNA. Infected cells were cultured in complete medium for 24 h and then left untreated (−) or treated (+) with 10 ng/ml IL-1β for an additional 36 h. Transcript levels of SIRT1 and NAMPT were determined by conventional PCR (top) and quantitative real-time PCR (middle) with GAPDH used as the loading control or internal control. Protein levels of SIRT1 and NAMPT and acetylation of histone H3 were detected by Western blotting (bottom) with β-actin used as the loading control. D, chondrocytes were pretreated with vehicle alone (VEH), 50 μm resveratrol (RES), or 1 mm NAM for 1 h and then left untreated (−) or treated (+) with 10 ng/ml IL-1β for an additional 36 h. All experiments were performed as described in B. Data are presented as the results of a typical experiment from at least four independent experiments.
SIRT1 Acts as a Key Mediator in IL-1β-induced Dedifferentiation of Articular Chondrocytes
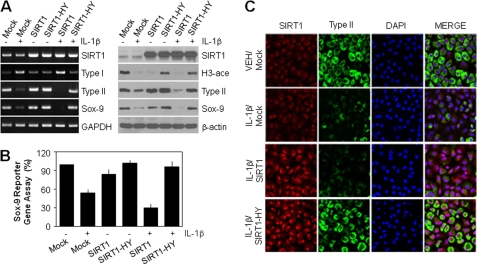
To examine whether SIRT1 activation is required for IL-1β-induced chondrocyte dedifferentiation, we directly infected cells with a recombinant retrovirus encoding SIRT1 or SIRT1-HY. Cells expressing SIRT1-HY in combination with IL-1β displayed inhibition of SIRT1 activity, based on the induction of histone H3 acetylation (Fig. 5A, right). IL-1β-mediated suppression of Sox-9 and type II collagen was markedly recovered in cells expressing SIRT1-HY at the transcript (Fig. 5A, left) and protein (right) levels. Treatment of cells with IL-1β led to an increased transcript level of type I collagen, which was markedly inhibited by the ectopic expression of SIRT1-HY (Fig. 5A, left). In the Sox-9 reporter gene assay, reduced Sox-9 transcriptional activity was observed in cells treated either with IL-1β alone or in combination with SIRT1 overexpression. However, SIRT1-HY-infected cells together with IL-1β treatment led to a marked increase in the transcriptional activity of Sox-9 compared with the IL-1β-treated group (Fig. 5B). The effect of SIRT1 on chondrocyte dedifferentiation was further evaluated under immunofluorescence microscopy. Double staining for type II collagen and SIRT1 in chondrocytes showed that cells expressing ectopic SIRT1 together with IL-1β treatment were negative for type II collagen staining, whereas SIRT1-HY expression reversed the IL-1β-induced suppression of type II collagen expression (Fig. 5C).
FIGURE 5.
SIRT1 acts as a key mediator in IL-1β-mediated dedifferentiation of articular chondrocytes. A and B, chondrocytes were infected with control retrovirus (Mock) or with retrovirus containing wild-type (SIRT1) or dominant-negative SIRT1 (SIRT1-HY) cDNA. Infected cells were cultured in complete medium for 24 h and then and left untreated (−) or treated (+) with 10 ng/ml IL-1β for an additional 36 h. Transcript levels of SIRT1, type I collagen, type II collagen, and Sox-9 were determined by conventional PCR (A, left) with GAPDH used as the loading control. Protein levels of SIRT1, type II collagen, and Sox-9 and acetylation of histone H3 were detected by Western blotting (A, right) with β-actin used as the loading control. The transcriptional activity of Sox-9 was determined with the reporter gene assay (B). C, chondrocytes were infected with control retrovirus or with retrovirus containing wild-type or dominant-negative SIRT1 cDNA as described in A. Infected cells were cultured in complete medium for 24 h and then left untreated (VEH) or treated with 10 ng/ml IL-1β for an additional 36 h. Expressions of SIRT1 and type II collagen were determined by double immunostaining confocal fluorescence microscopy. Cells were identified by DAPI staining of the nuclei. Data are presented as the results of a typical experiment (A), mean values with standard deviations (B), or a representative photomicrograph (C) from at least four independent experiments.
The role of SIRT1 was further investigated using the chemical activator resveratrol or the inhibitor NAM. Although the SIRT1 expression level was unchanged after resveratrol or NAM treatment in the absence or presence of IL-1β (Fig. 6A), NAM inhibited SIRT1 activity, based on the increase in the acetylation level of histone H3 (Fig. 6A, right). Treatment of cells with NAM for 1 h prior to IL-1β treatment led to a significant increase in the transcript (Fig. 6A, left) and protein (Fig. 6A, right) levels of type II collagen and Sox-9, which were markedly decreased by IL-1β treatment. In the Sox-9 reporter gene assay, a combination of the NAM and IL-1β led to a significant increase in transcriptional activity compared with the IL-1β-treated group, but this was not the case with a combination of resveratrol and IL-1β (Fig. 6B). Consistent with direct SIRT1-HY gene expression, treatment of cells with a combination of NAM and IL-1β revealed that the number of type II collagen positive cells increased to 90% of the initial number evaluated following IL-1β but not in case of a combination of resveratrol and IL-1β (Fig. 6C). These results collectively suggest that SIRT1 activation is sufficient to cause abrogation of type II collagen expression in response to IL-1β.
FIGURE 6.
Chemical activation and inhibition of SIRT1 have opposite effects on IL-1β-mediated dedifferentiation of articular chondrocytes. A and B, chondrocytes were pretreated with vehicle alone (VEH), 50 μm resveratrol (RES), or 1 mm NAM for 1 h and then left untreated (−) or treated (+) with 10 ng/ml IL-1β. After 48 h, transcript levels of SIRT1, type I collagen, type II collagen, and Sox-9 were determined by conventional PCR (A, left) with GAPDH used as the loading control. Protein levels of SIRT1, type II collagen, and Sox-9 and acetylation of histone H3 (H3-ace) were detected by Western blotting (A, right) with β-actin used as the loading control. The transcriptional activity of Sox-9 was determined with the reporter gene assay (B). C, chondrocytes were pretreated with vehicle alone, 50 μm resveratrol, or 1 mm NAM for 1 h and then left untreated (VEH) or treated with 10 ng/ml IL-1β for an additional 48 h. Expressions of SIRT1 and type II collagen were determined by double immunostaining confocal fluorescence microscopy. Cells were identified by DAPI staining of nuclei. Data are presented as the results of a typical experiment (A), mean values with standard deviations (B), or a representative photomicrograph (C) from at least four independent experiments.
NAMPT-SIRT1 Signaling Activates ERK and p38 Kinase, but SIRT1-ERK Complex Participates in IL-1β-induced Chondrocyte Dedifferentiation
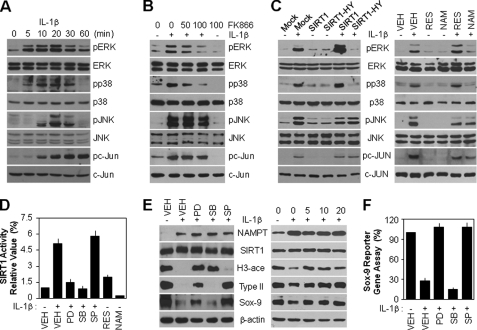
We examined whether IL-1β-mediated MAPK activation is associated with the NAMPT-SIRT1 signaling pathway in articular chondrocytes. As shown in Fig. 7A, IL-1β promoted a transient increase in the phosphorylation of ERK, p38 kinase, and JNK, leading to a peak in their activities at 20 min after IL-1β treatment. In addition, phosphorylation of c-Jun, a downstream target molecule of JNK, was continuous for up to 1 h after treatment. The total cellular MAPK and c-Jun protein levels remained unchanged. Next, chondrocytes were preincubated with FK866 prior to IL-1β treatment. This compound significantly inhibited ERK and p38 kinase activated by IL-1β in a dose-dependent manner, indicating that these processes occur downstream of NAMPT signaling; however, FK866 did not block JNK and c-Jun activation (Fig. 7B). To establish the linkage between SIRT1 and MAPK activation in IL-1β-treated articular chondrocytes, SIRT1 was stimulated or inhibited in cells either by using genes directly or pharmacological agents indirectly. The stimulation or inhibition of SIRT1 with either wild type or SIRT1-HY in the absence of IL-1β did not affect MAPK and c-Jun activation compared with the control group (Fig. 7C, left); this phenomenon was also observed in the experiments using resveratrol or NAM (Fig. 7C, right). However, the overexpression of SIRT1-HY completely blocked IL-1β-induced ERK and p38 kinase activation, but not that of JNK and c-Jun (Fig. 7C, left), with concomitant suppression of the same MAPK activity after NAM treatment (Fig. 7C, right).
FIGURE 7.
NAMPT-SIRT1 signaling boosts both ERK and p38 kinase activations, but only ERK is associated with IL-1β-mediated dedifferentiation of articular chondrocytes. A, chondrocytes were treated with 10 ng/ml IL-1β for the indicated periods. Expression and phosphorylation levels of ERK, p38, JNK, and c-Jun were determined by Western blotting. B, chondrocytes were pretreated with the specified doses (nm) of FK866 for 1 h and then left untreated (−) or treated (+) with 10 ng/ml IL-1β for 20 min (for ERK, p38, and JNK detection) or 60 min (for c-Jun detection). Expression and phosphorylation levels of ERK, p38, JNK, and c-Jun were determined by Western blotting. C, chondrocytes were infected with control retrovirus (Mock) or retrovirus containing wild-type (SIRT1) or dominant-negative SIRT1 (SIRT1-HY) cDNA. Infected cells were cultured in complete medium for 24 h and then left untreated (−) or treated (+) with 10 ng/ml IL-1β for an additional 20 min (left). Chondrocytes were pretreated with vehicle alone (VEH), 50 μm resveratrol (RES), or 1 mm NAM for 1 h and then left untreated (−) or treated (+) with 10 ng/ml IL-1β for an additional 20 min (right). Expression and phosphorylation levels of ERK, p38, JNK, and c-Jun were determined by Western blotting. D, chondrocytes were pretreated with vehicle alone, 20 μm SB203580 (SB), 20 μm PD98059 (PD), 20 μm SP600125 (SP), 50 μm resveratrol as a positive control, or 1 mm NAM as negative control for 1 h and then left untreated (−) or treated (+) with 10 ng/ml IL-1β for 36 h. SIRT1 activity was determined using a SIRT1 fluorometric assay kit. E and F, chondrocytes were left untreated (−) or treated (+) with 10 ng/ml IL-1β for 36 h in the absence (VEH) or presence of 20 μm SB203580, 20 μm PD98059, or 20 μm SP600125 (E (left) and F) or in the specified doses (μm) of PD98059 (E, right). Protein levels of NAMPT, SIRT1, type II collagen, and Sox-9 and acetylation of histone H3 were determined by Western blotting, with β-actin as the loading control (E). The transcriptional activity of Sox-9 was determined with the reporter gene assay (F). Data are presented as the results of a typical experiment (A–C and E) or mean values with standard deviations (D and F) from at least four independent experiments.
To further ascertain whether SIRT1 activity is dependent on MAPK activation, we pretreated cells with specific MAPK inhibitors prior to IL-1β treatment. Inhibition of ERK with PD98059 or of p38 kinase with SB203580 completely blocked IL-1β-induced SIRT1 activity as determined by a SIRT1 fluorometric assay. Inhibition of JNK with SP600125 had no effect on SIRT1 activity, indicating that the JNK pathway is not associated with IL-1β-induced SIRT1 activity (Fig. 7D). Consistent with these findings, the pretreatment of cells with PD98059 or SB203580, but not SP600125, led to a potent increase in the acetylation level of histone H3 after IL-1β treatment with no alteration in SIRT1 protein expression (Fig. 7E, left). These results strongly support the formation of either a SIRT1-ERK or SIRT1-p38 kinase positive feedback loop to sustain downstream signaling. Moreover, only SIRT1-mediated ERK activation, but not SIRT1-p38 kinase, was involved in the regulation of type II collagen and Sox-9 expression in response to IL-1β. However, the JNK pathway was associated with the regulation of type II collagen expression in a SIRT1-independent manner (Fig. 7E, left). We also confirmed that dose-dependent inhibition of ERK inhibited SIRT1 activity based on the rescued level of the acetylation of histone H3 decreased by IL-1β, subsequently resulting in an increase in the expression of type II collagen and Sox-9 (Fig. 7E, right). A Sox-9 reporter gene assay also showed that reduced Sox-9 transcriptional activity by IL-1β was rescued by the inhibition of ERK and JNK, consistent with the Western blot (Fig. 7F). Thus, these results indicate that IL-1β induces chondrocyte dedifferentiation, which is mediated by the SIRT1-dependent ERK signaling and SIRT1-independent JNK signaling pathways.
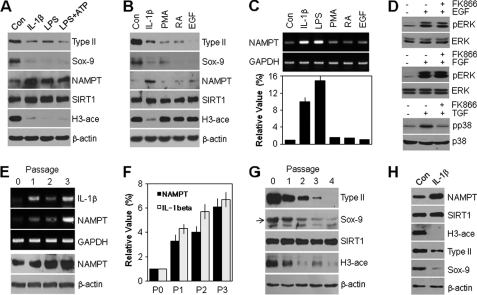
To determine whether NAMPT-dependent SIRT1 activation is associated exclusively with the response to IL-1β, cells were incubated with four additional agents, LPS, PMA, RA, and EGF, previously reported to induce dedifferentiation of chondrocytes (23–25). We found that only LPS, known to induce IL-1β and other cytokines, led to an induction of the NAMPT protein level and subsequent promotion of SIRT1 activity without any alteration of the SIRT1 protein level, based on the reduction of the acetylation level of histone H3 (Fig. 8A). However, NAMPT expression and SIRT1 activity did not change in response to PMA, RA, or EGF, although each of these agents down-regulated the protein level of type II collagen and Sox-9 (Fig. 8B). In addition, the transcription level of NAMPT was markedly increased in the IL-1β- and LPS-treated groups but not in the PMA-, RA-, or EGF-treated group, as determined by conventional PCR (Fig. 8C, top) and quantitative PCR (bottom). These results demonstrate that chondrocyte dedifferentiation via the NAMPT-dependent SIRT1 signaling pathway was limited to inflammatory cytokine stimuli such as LPS and IL-1β. We further examined the relationship of SIRT1 signaling with cytokines, focusing on ERK and p38 kinase activities. As shown in Fig. 8D, EGF and FGF markedly induced ERK activity, but inhibition of SIRT1 by pretreatment with FK866 did not block their activity. TGF-β1, one of the cytokines, significantly led to induction of NAMPT expression (data not shown) and p38 kinase activity, and FK866 completely inhibited the activation of p38 kinase promoted by TGF-β1. These results indicate that ERK or p38 kinase activation by SIRT1 is due to the cytokine-mediated signaling pathway. We confirmed the roles of these pathways using the dedifferentiation of articular chondrocytes promoted by monolayer subculture system. Subculturing of the chondrocytes serially from passage 0 to 4 resulted in a flattened and fibroblast-like morphology (data not shown). Notably, the transcript level of IL-1β increased during the dedifferentiation in the serial subculture in a similar manner to NAMPT mRNA expression, as determined by conventional PCR (Fig. 8E, top). Quantitative PCR showed that the cells displayed enhanced expression of IL-1β and NAMPT by 5.7- and 4.0-fold, respectively, at passage 2 (Fig. 8F), with concomitant accumulation of the NAMPT protein, as detected by Western blotting (Fig. 8E, bottom). Consistent with NAMPT expression, the increase in SIRT1 activity was observed throughout the culture period, as shown by the acetylation level of histone H3 (Fig. 8G). As expected, the expression of type II collagen and Sox-9 was high at P0, began to decrease at P1, and was almost undetectable at P3 and thereafter, confirming the typical chondrocyte dedifferentiation induced by monolayer culture system (Fig. 8G). We finally examined whether the effects of IL-1β on NAMPT and SIRT-1 require continuous or repeated stimulation with IL-1β for dedifferentiation. For this experiment, chondrocytes were treated with 10 ng/ml IL-1β for 4 h; then IL-1β was removed from the culture medium, and the chondrocytes were cultured continuously in normal culture medium for an additional 48 h. As shown in Fig. 8H, NAMPT expression and SIRT1 activity were induced at 48 h, which in turn down-regulated type II collagen and Sox-9 expression, although chondrocytes were cultured without stimulation from IL-1β after the initial 4 h. Taken together, these results indicate that continuously accumulated NAMPT and SIRT1 signaling by IL-1β affects cartilage-specific gene expression in chondrocytes.
FIGURE 8.
NAMPT-SIRT1-mediated dedifferentiation of articular chondrocytes is responsible for only IL-1β, but not PMA, RA, or EGF reagents. A–C, chondrocytes were treated with vehicle alone (Con), 10 ng/ml IL-1β, 10 ng/ml LPS alone or in combination with 5 mm ATP, 10 nm PMA, 1 μm RA, or 10 ng/ml EGF for 48 h as designed. Protein levels of type II collagen, Sox-9, NAMPT, and SIRT1 and acetylation of histone H3 (H3-ace) were detected by Western blotting (A and B) with β-actin used as the loading control. The transcript level of NAMPT was determined by conventional PCR (C, top) and quantitative real-time PCR (C, bottom) with GAPDH used as the loading control or internal control. D, chondrocytes were pretreated with 50 nm FK866 to inhibit NAMPT-dependent SIRT1 activity for 1 h and then treated with 10 ng/ml EGF, 10 ng/ml FGF, or 5 ng/ml TGF-β for 20 min. Expression and phosphorylation levels of ERK and p38 were determined by Western blotting. E–G, chondrocytes were serially subcultured as monolayers up to passage 3 (E and F) or 4 (G). Transcript levels of NAMPT and IL-1β were determined by conventional PCR (E, top) and quantitative real-time PCR (F) with GAPDH used as the loading control or internal control. Protein levels of NAMPT, type II collagen, Sox-9, and SIRT1 and acetylation of histone H3 were detected by Western blotting (E, bottom, and G) with β-actin used as the loading control. H, chondrocytes were left untreated (Con) or treated with 10 ng/ml IL-1β for 4 h; then the IL-1β was removed by exchanging the medium, and the chondrocytes were cultured continuously in normal culture medium for an additional 48 h. Protein levels of NAMPT, SIRT1, type II collagen, and Sox-9 and acetylation of histone H3 were detected by Western blotting with β-actin used as the loading control. Data are presented as the results of a typical experiment from at least four independent experiments.
DISCUSSION
Although IL-1β is known to induce cell death in mouse chondrocyte-like ATDC5 cells (26) and clinical OA cartilage specimens (27), it is still controversial as to whether IL-1β induces chondrocyte death. In the present study, we analyzed the effects of IL-1β on primary cultured rabbit articular chondrocytes and the elements in the IL-1β-related dedifferentiation signaling pathway.
We showed that rabbit articular chondrocytes did not exhibit cell death or cellular senescence following IL-1β treatment. However, they did exhibit an increase in dedifferentiation due to IL-1β-mediated induction of NAMPT expression in a time-dependent manner, as deduced from an NAMPT inhibition experiment in which the loss of chondrocyte phenotype generated by IL-1β treatment was completely restored (Fig. 2). Our data showed that NAMPT expression was serially up-regulated up to 48 h in a time-dependent manner following IL-1β treatment. Under the same condition, SIRT1 activity peaked at 24 h and then decreased slowly up to 48 h, which was still much higher than in the control group. Despite the decrease in SIRT1 activity, the expression level of type II collagen and Sox-9 was down-regulated continuously at 36 and 48 h. In the subculture system, chondrocyte dedifferentiation was highly induced in P3 or P4 by suppressing type II collagen expression, although the greatest -fold change in NAMPT and IL-1β occurred from P0 to P1 (Fig. 8). Therefore, we suggest that once chondrocytes were exposed to IL-1β, cartilage-specific gene expression, such as type II collagen and Sox-9, was down-regulated via continuously accumulated NAMPT and SIRT1 signaling. Recently, the effect of NAMPT on the inflammatory and immune responses has attracted considerable attention. For example, IL-6 is reported to suppress the mRNA expression of NAMPT via ERK activation in 3T3-L1 adipocytes (28), whereas NAMPT is up-regulated by IL-1β in neutrophils and functions as a novel inhibitor of apoptosis following exposure to inflammatory stimuli (16). Our result showing IL-1β-dependent NAMPT overproduction in chondrocytes is similar to the data on neutrophils except for the NAMPT function, although the cell lines used are different. One study also reported that the plasma concentration of NAMPT is increased in patients with rheumatoid arthritis caused by a chronic and systemic inflammatory disorder (17). However, the effect of NAMPT on cartilage physiology still remains largely unknown. Our investigation on the role of NAMPT in cartilage destruction shows that IL-1β-mediated NAMPT overexpression is sufficient to induce chondrocyte dedifferentiation via down-regulation of Sox-9, a major transcription factor that regulates the type II collagen (COL2A1) level. This mechanism was confirmed using the NAMPT-specific inhibitor FK866, which blocked the IL-1β-induced alteration of chondrocyte phenotype through a restoration of Sox-9 activity. Therefore, we consider the chondrocyte dedifferentiation induced by NAMPT overproduction to be associated with the Sox-9-mediated signaling pathway in response to IL-1β.
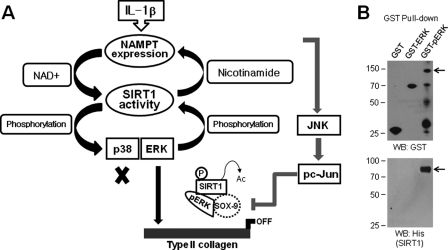
It is established that NAMPT functions as an intra- and extracellular NAD biosynthetic enzyme and that SIRT1, as the NAD-consuming enzyme, is required in several important signal transduction pathways in mammalian cells (29). In this study, SIRT1 activation was dependent on NAMPT in IL-1β-treated articular chondrocytes, and activated SIRT1 subsequently was associated with the induction of NAMPT transcription and translation levels (Fig. 4), establishing a positive feedback system for the sustained activation of downstream signaling. Ramsey et al. (30) reported a similar feedback loop for the circadian clock gene, a downstream target gene of SIRT1, in the liver and white adipose tissue. Interestingly, we found that SIRT1 activation was also always accompanied by the activation of the MAPKs ERK and p38, but not JNK, in IL-1β-treated chondrocytes. Treatment of cells with PD98059 for ERK inhibition or SB203580 for p38 kinase inhibition did not have any effect on NAMPT expression but completely blocked SIRT1 activity (Fig. 7). This means that the activation of these two MAPKs is a prerequisite for SIRT1 activation, thus establishing a second positive feedback loop for the activation of SIRT1 and the sustained signaling cascade of MAPKs downstream. At present, the mechanism of SIRT1 activation is not well understood, but it is known that SIRT1 activity is up-regulated by sumoylation at lysine 734 via the attachment of small ubiquitin-related modifiers (31). In addition, phosphorylation of this protein also promotes its enzymatic activity. For example, SIRT1 forms a complex with cyclin-dependent kinase 1 and is phosphorylated at threonine 530 and serine 540 as the targets of this kinase (32). This is the first detailed report on SIRT1 phosphorylation by ERK or p38 kinase; the endogenous mechanisms that regulate SIRT1 activity will require further examination. Based on the findings described above, SIRT1 is considered an efficient second intracellular mediator in IL-1β-treated chondrocytes, acting either downstream of NAMPT signaling or upstream of ERK and p38 kinase (Fig. 9A). The findings collectively suggest that SIRT1-mediated regulation of MAPK proteins may help explain the diversity of SIRT1-dependent cellular responses.
FIGURE 9.
Schematic summary of a signaling pathway underlying IL-1β-induced dedifferentiation of primary cultured articular chondrocytes. A, schematic summary of SIRT1 function either as downstream of NAMPT signaling or as upstream of ERK and p38 kinase. B, 0.25 μg of GST, 0.5 μg of GST-ERK2, or 2.0 μg of GST-pERK fusion proteins were initially bound with glutathione-agarose beads for 2 h, and then each complex was incubated with the active form of 0.5 μg of His-SIRT1 fusion proteins for 12 h. The protein complexes were detected by Western blotting with anti-GST (top) or anti-His antibody (bottom).
Because both SIRT1 and MAPK regulate a very broad and complex array of physiological processes, it is to be expected that the activation of SIRT1 and MAPKs induced by IL-1β would exert an effect on the normal chondrocyte phenotype. IL-1β in fact induces the dedifferentiation of chondrocytes, which is characterized by a suppression of cartilage-specific type II collagen expression and the onset of type I collagen expression (Fig. 1). As depicted in Fig. 5, dominant-negative SIRT1 blocked the down-regulation of the transcript and protein levels of type II collagen and Sox-9 in response to IL-1β. These findings suggest that SIRT1 is sufficient to drive chondrocyte dedifferentiation and thus plays an important role in the development of OA. It is reported that ERK is involved in the regulation of chondrogenesis and the maintenance of the differentiated chondrocyte phenotype in a PKC-dependent or -independent manner (33). In this study, we found that activation of SIRT1 forms a complex with activated ERK or p38 kinase as determined by GST pulldown assay (Fig. 9B), but only the SIRT1-ERK complex participates in the loss of the differentiated phenotype. Although we initially defined SIRT1 as an ERK regulator in IL-1β-treated chondrocytes, the ERK signaling cascade in chondrocyte dedifferentiation remains largely unknown. On the other hand, the SIRT1-p38 kinase complex did not affect the type II collagen and Sox-9 levels, indicating that it is not related to the regulation of chondrocyte phenotype and, instead, may be associated with the regulation of other physiological processes such as inflammation. We reported previously that the binding of p38 kinase to SIRT1 leads to an induction of SIRT1 protein degradation, which acts as a key regulatory mechanism in the induction of cellular senescence in irradiated articular chondrocytes (20). Therefore, we suppose that SIRT1 protein homeostasis is essential for the maintenance of the differentiated chondrocyte phenotype. JNK was activated by IL-1β but had no effect on its activity by SIRT1 in our experiment. Furthermore, the IL-1β-mediated decrease in type II collagen and Sox-9 expression was affected by the enhanced JNK activity, indicating an association of the NAMPT-SIRT1-independent JNK signaling pathway with chondrocyte dedifferentiation (Fig. 9). Dvir-Ginzberg et al. (22) report that elevation of the SIRT1 protein level in human chondrocytes derived from OA patients leads to a dramatic increase in cartilage-specific gene expression, and therefore, they suggest that SIRT1 and NAMPT may provide a positive role in human cartilage. This is somewhat inconsistent with our results, which were obtained during the cytokine-induced dedifferentiation process of normal rabbit articular chondrocytes. Our results showed that NAMPT and SIRT1 activations significantly lowered Sox-9 and type II collagen expressions and thus have a negative function in the maintenance of normal chondrocyte phenotype. The difference in SIRT1 function for regulating gene expression may be because of dissimilarities in the species, experimental conditions, and OA inducer. These results support the notion that SIRT1 plays a role in the coordination of gene expression and cellular responses in both normal and pathophysiological settings.
One of the interesting findings here is that NAMPT-SIRT1 signaling appears to mediate the dedifferentiation of chondrocytes via a restricted pathway. As shown in Fig. 8, IL-1β, one of the proinflammatory cytokines that plays a pivotal role in the pathogenesis of arthritis, specifically enhances NAMPT expression and subsequently activates SIRT1 protein. These phenomena were also observed during LPS- and LPS/ATP-induced chondrocyte dedifferentiation. LPS plays a key role as an immunological stimulator and drives the cells to release inflammatory cytokines such as IL-6, IL-8, and IL-10, which are responsible for the progression of inflammatory reactions (34). Furthermore, a serial monolayer culture system is a well defined procedure to demonstrate the loss of chondrocyte phenotype (5). Consistent with the down-regulation of type II collagen, the monolayer subculture resulted in the overproduction of IL-1β expression in a passage-dependent manner. We consider this the source of the increased expression of NAMPT and, in turn, the activation of SIRT1 in the subculture model system. However, other dedifferentiation-inducing stimulators, including PMA as an activator of protein kinase C, RA as a metabolite of vitamin A, and EGF as a growth factor, did not affect the NAMPT level or SIRT1 activity, although all of these agents significantly promote chondrocyte dedifferentiation in various ways. Therefore, NAMPT-SIRT1 signaling appears to be responsible for the chondrocyte dedifferentiation caused by direct inflammatory cytokine activity.
In summary, these results collectively indicate that the regulation of SIRT1 activity is an essential event in the regulation of IL-1β-mediated chondrocyte dedifferentiation. SIRT1 activation is performed using double feedback loop systems, which are associated with NAMPT as the upstream regulator and with MAPKs as the targets. Finally, the interaction of SIRT1 and ERK, but not SIRT1 and p38 kinase, led to the loss of chondrocyte phenotype in a Sox-9 signaling mechanism-dependent manner. Therefore, inhibition of the NAMPT or SIRT1 pathway appears to be an effective approach to arresting inflammatory cytokine-induced tissue damage and may be applicable to the treatment of OA.
Supplementary Material
This work was supported by the Nuclear Research and Development Program of the National Research Foundation of Korea and in part by Korea Research Foundation Grant KRF-2008-313-C00260.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- OA
- osteoarthritis
- NAM
- nicotinamide
- NAMPT
- nicotinamide phosphoribosyltransferase
- P
- passage (e.g. P0)
- PMA
- phorbol 12-myristate 13-acetate
- RA
- retinoic acid.
REFERENCES
- 1. Martinon F., Tschopp J. (2004) Cell 117, 561–674 [DOI] [PubMed] [Google Scholar]
- 2. Sandell L. J., Aigner T. (2001) Arthritis Res. 3, 107–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Goldring M. B., Otero M., Tsuchimochi K., Ijiri K., Li Y. (2008) Ann. Rheum. Dis. 67, iii75–iii82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jacques C., Gosset M., Berenbaum F., Gabay C. (2006) Vitam. Horm. 74, 371–403 [DOI] [PubMed] [Google Scholar]
- 5. Hwang S. G., Yu S. S., Poo H., Chun J. S. (2005) J. Biol. Chem. 280, 29780–29787 [DOI] [PubMed] [Google Scholar]
- 6. Peng H., Tan L., Osaki M., Zhan Y., Ijiri K., Tsuchimochi K., Otero M., Wang H., Choy B. K., Grall F. T., Gu X., Libermann T. A., Oettgen P., Goldring M. B. (2008) J. Cell. Physiol. 215, 562–573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ijiri K., Zerbini L. F., Peng H., Correa R. G., Lu B., Walsh N., Zhao Y., Taniguchi N., Huang X. L., Otu H., Wang H., Wang J. F., Komiya S., Ducy P., Rahman M. U., Flavell R. A., Gravallese E. M., Oettgen P., Libermann T. A., Goldring M. B. (2005) J. Biol. Chem. 280, 38544–38555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hwang S. G., Ryu J. H., Kim I. C., Jho E. H., Jung H. C., Kim K., Kim S. J., Chun J. S. (2004) J. Biol. Chem. 279, 26597–26604 [DOI] [PubMed] [Google Scholar]
- 9. Hwang S. G., Yu S. S., Lee S. W., Chun J. S. (2005) FEBS Lett. 579, 4837–4842 [DOI] [PubMed] [Google Scholar]
- 10. Murakami S., Lefebvre V., de Crombrugghe B. (2000) J. Biol. Chem. 275, 3687–3692 [DOI] [PubMed] [Google Scholar]
- 11. Wang T., Zhang X., Bheda P., Revollo J. R., Imai S., Wolberger C. (2006) Nat. Struct. Mol. Biol. 13, 661–662 [DOI] [PubMed] [Google Scholar]
- 12. Revollo J. R., Grimm A. A., Imai S. (2004) J. Biol. Chem. 279, 50754–50763 [DOI] [PubMed] [Google Scholar]
- 13. van der Veer E., Ho C., O'Neil C., Barbosa N., Scott R., Cregan S. P., Pickering J. G. (2007) J. Biol. Chem. 282, 10841–10845 [DOI] [PubMed] [Google Scholar]
- 14. Fulco M., Cen Y., Zhao P., Hoffman E. P., McBurney M. W., Sauve A. A., Sartorelli V. (2008) Dev. Cell 14, 661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yang H., Yang T., Baur J. A., Perez E., Matsui T., Carmona J. J., Lamming D. W., Souza-Pinto N. C., Bohr V. A., Rosenzweig A., de Cabo R., Sauve A. A., Sinclair D. A. (2007) Cell 130, 1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jia S. H., Li Y., Parodo J., Kapus A., Fan L., Rotstein O. D., Marshall J. C. (2004) J. Clin. Invest. 113, 1318–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Otero M., Lago R., Gomez R., Lago F., Dieguez C., Gómez-Reino J. J., Gualillo O. (2006) Ann. Rheum. Dis. 65, 1198–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Presle N., Pottie P., Dumond H., Guillaume C., Lapicque F., Pallu S., Mainard D., Netter P., Terlain B. (2006) Osteoarthr. Cartil. 14, 690–695 [DOI] [PubMed] [Google Scholar]
- 19. Anderson R. M., Bitterman K. J., Wood J. G., Medvedik O., Sinclair D. A. (2003) Nature 423, 181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hong E. H., Lee S. J., Kim J. S., Lee K. H., Um H. D., Kim J. H., Kim S. J., Kim J. I., Hwang S. G. (2010) J. Biol. Chem. 285, 1283–1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang T., Kraus W. L. (2010) Biochim. Biophys. Acta 1804, 1666–1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dvir-Ginzberg M., Gagarina V., Lee E. J., Hall D. J. (2008) J. Biol. Chem. 283, 36300–36310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bobacz K., Sunk I. G., Hofstaetter J. G., Amoyo L., Toma C. D., Akira S., Weichhart T., Saemann M., Smolen J. S. (2007) Arthritis Rheum. 56, 1880–1893 [DOI] [PubMed] [Google Scholar]
- 24. Klooster A. R., Bernier S. M. (2005) Arthritis Res. Ther. 7, 127–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ryu J. H., Kim S. J., Kim S. H., Oh C. D., Hwang S. G., Chun C. H., Oh S. H., Seong J. K., Huh T. L., Chun J. S. (2002) Development 129, 5541–5550 [DOI] [PubMed] [Google Scholar]
- 26. Yasuhara R., Miyamoto Y., Akaike T., Akuta T., Nakamura M., Takami M., Morimura N., Yasu K., Kamijo R. (2005) Biochem. J. 389, 315–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Héraud F., Héraud A., Harmand M. F. (2000) Ann. Rheum. Dis. 59, 959–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kralisch S., Klein J., Lossner U., Bluher M., Paschke R., Stumvoll M., Fasshauer M. (2005) Am. J. Physiol. Endocrinol. Metab. 289, E586–E590 [DOI] [PubMed] [Google Scholar]
- 29. Garten A., Petzold S., Körner A., Imai S., Kiess W. (2009) Trends Endocrinol. Metab. 20, 130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramsey K. M., Yoshino J., Brace C. S., Abrassart D., Kobayashi Y., Marcheva B., Chong J. L., Buhr E. D., Lee C., Takahashi J. D., Imai S., Bass J. (2009) Science 324, 651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang Y., Fu W., Chen J., Olashaw N., Zhang X., Nicosia S. V., Bhalla K., Bai W. (2007) Nat. Cell Biol. 9, 1253–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sasaki T., Maier B., Koclega K. D., Chruszcz M., Gluba W., Stukenberg P. T., Minor W., Scrable H. (2008) PLoS ONE 3, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yoon Y. M., Kim S. J., Oh C. D., Ju J. W., Song W. K., Yoo Y. J., Huh T. L., Chun J. S. (2002) J. Biol. Chem. 277, 8412–8420 [DOI] [PubMed] [Google Scholar]
- 34. Levy O. (2005) J. Endotoxin Res. 11, 113–116 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.