The expression, purification and crystallization of a PHD domain of human JARID1B are described.
Keywords: PHD domains, JARID1B, histone methylation
Abstract
Histone lysine methylation can be removed by proteins containing JmjC domains in a sequence- and methylation state-specific manner. JARID1B, a protein containing PHD and JmjC domains, is a histone demethylase specific for H3K4me2 and H3K4me3 which requires Fe(II) and α-ketoglutarate (α-KG) as cofactors to remove the methyl group. JARID1B has also been shown to play a critical role in the development of breast cancer. JARID1B contains JmjN, Arid and JmjC domains, a C5HC2 zinc-finger domain and three PHD domains. The first PHD domain (PHD1JARID1B; residues 306–360) is located at the N-terminus and is important for both histone demethylase activity and histone-tail recognition of JARID1B. Here, the expression, purification and crystallization of PHD1JARID1B is reported. A PHD1JARID1B crystal was grown by the hanging-drop vapour-diffusion method in reservoir solution consisting of 0.1 M HEPES pH 7.0, 2.2 M ammonium sulfate at 277 K. A zinc SAD data set was collected from a PHD1JARID1B crystal. The diffraction pattern of the PHD1JARID1B crystal extended to 1.65 Å resolution using synchrotron radiation. The crystal belonged to space group P43, with unit-cell parameters a = 51.7, b = 51.7, c = 36.2 Å.
1. Introduction
Histone methylation is recognized as an important modification that is linked to chromatin and transcription regulation as well as DNA-damage response. Five lysine (K) residues of histone H3 (H3K4, H3K9, H3K27, H3K36 and H3K79) and Lys20 of histone H4 (H4K20) have been identified to be sites for this modification (Margueron et al., 2005 ▶; Zhang & Reinberg, 2001 ▶). In general, methylation at H3K9, H3K27 and H4K20 is associated with transcription repression, and methylation at H3K4, H3K36 and H3K79 is associated with transcription activation (Zhang & Reinberg, 2001 ▶; Martin & Zhang, 2005 ▶). The first histone demethylase, LSD1 (lysine-specific demethylase 1), was identified in 2004 (Shi et al., 2004 ▶). To date, two distinct classes of demethylases have been characterized. The first class, represented by LSD1, use FAD as a cofactor through an amine-oxidation reaction to remove the lysine methyl group. The second class are the family of JmjC-domain-containing demethylases, which require FeII and α-ketoglutarate (α-KG) as cofactors in the demethylation process (Shi & Whetstine, 2007 ▶; Whetstine et al., 2006 ▶; Tsukada et al., 2006 ▶; Cloos et al., 2006 ▶; Chen et al., 2006 ▶).
Over 30 human JmjC-domain-containing proteins have been found through analysis of public protein-domain databases (Klose, Kallin et al., 2006 ▶; Shin & Janknecht, 2007 ▶; Xiang, Zhu, Han, Ye et al., 2007 ▶). Many of these proteins have been identified to be histone lysine demethylases (Lee, Villa et al., 2007 ▶; Lee, Norman et al., 2007 ▶; Xiang, Zhu, Han, Lin et al., 2007 ▶; Lan et al., 2007 ▶; Christensen et al., 2007 ▶; Cloos et al., 2006 ▶; Iwase et al., 2007 ▶; Klose, Yamane et al., 2006 ▶; Klose et al., 2007 ▶; Whetstine et al., 2006 ▶; Yamane et al., 2007 ▶; Agger et al., 2007 ▶). The structures of several representative JmjC-domain-containing histone lysine demethylase family members, including JMJD2A, PHF8, Kiaa1718 and JHDM1A, have been determined in the apo form or in complex with the substrate peptide and α-KG (Horton et al., 2010 ▶; Chen et al., 2006 ▶, 2007 ▶; Couture et al., 2007 ▶; Ng et al., 2007 ▶; Yang et al., 2010 ▶; Han et al., 2007 ▶). However, no structure has been reported for a member of the JARID1 subgroup which can specifically remove trimethylated histone H3K4.
JARID1-subgroup members are highly conserved from yeast to humans and contain a similar domain architecture comprising JmjN, Arid and JmjC domains, a C5HC2 zinc-finger domain and two or three PHD domains (designated PHD1, PHD2 and PHD3 from the N-terminus to the C-terminus). There are four JARID1-group members in mammals, JARID1A (RBP2), JARID1B (PLU-1), JARID1C (SMCX) and JARID1D (SMCY), which have all been identified to be demethylases for trimethylated and dimethylated H3K4 (Iwase et al., 2007 ▶; Christensen et al., 2007 ▶; Klose et al., 2007 ▶; Tahiliani et al., 2007 ▶; Lee, Norman et al., 2007 ▶; Yamane et al., 2007 ▶). The four proteins are all important transcriptional corepressors since they remove the transcription-activating mark trimethylated H3K4. Studies of human malignancies show that JARID1B plays a critical role in the development of breast cancer (Lu et al., 1999 ▶; Barrett et al., 2002 ▶; Yamane et al., 2007 ▶).
Among the recognizable domains in JARID1B, PHD domains are predicted to be involved in histone-tail recognition. JARID1B contains three PHD domains: PHD1 is the first PHD domain, located between the Arid and JmjC domains, while the PHD2 and PHD3 domains are located at the C-terminus. Recent studies of Lid, a homologue of JARID1B in Drosophila, found that PHD1 is required for demethylase activity towards H3K4me3, while PHD2 and PHD3 are not (Li et al., 2010 ▶). Interestingly, unbiased peptide screening for PHD1 interaction indicates that PHD1 can specifically bind to histone H3, either unmodified or with K9 methylation, suggesting that PHD1 plays an important role in the nucleosome-localization and demethylase activity of JARID1B (Li et al., 2010 ▶). Given the highly conserved sequence homology of PHD1 of Lid and JARID1B (58% identity, 80% homology), PHD1JARID1B may also be important for the function of JARID1B. We have found that PHD1JARID1B indeed plays an important role in histone-tail recognition and histone demethylase activity (unpublished data). Here, we report the expression, purification and crystallization of PHD1JARID1B. The structure will reveal the mechanism of histone-tail recognition by PHD1JARID1B.
2. Materials and methods
2.1. Protein expression and purification
The ORF of PHD1JARID1B (residues 306–360 of JARID1B; NP_006609.3; Homo sapiens) was engineered into modified pGEX-6P-1 vector (GE Healthcare) using BamHI and XhoI restriction sites. The construct was verified by DNA sequencing and the plasmid was transformed into Escherichia coli strain BL21 (DE3) competent cells. The transformants were grown at 310 K to an OD600 of 1.0 in Luria broth medium containing 100 µg ml−1 ampicillin and were induced by the addition of 0.2 mM isopropyl β-d-1-thiogalactopyranoside. PHD1JARID1B was overexpressed as a fusion protein with a GST tag at the N-terminus. After a further 12 h incubation at 288 K, the cells were pelleted and resuspended in lysis buffer consisting of 25 mM Tris pH 8.0, 150 mM NaCl supplemented with DNAse and protease inhibitors. Cells were lysed on ice using a French press and the solution was clarified by centrifugation at 15 000 rev min−1 for 30 min at 277 K. The supernatant was applied onto a Glutathione Sepharose 4B column (1 ml resin per column; GE Healthcare) equilibrated with lysis buffer. After washing with buffer containing 25 mM Tris pH 8.0, 150 mM NaCl, the fusion protein was digested on the column with PreScission (3C) protease overnight at 277 K. The molecular weight of PHD1JARID1B is 6.1 kDa including the additional Gly-Pro-Leu-Gly-Ser sequence from 3C cleavage. The eluted protein was then loaded onto an Source 15Q anion-exchange column (GE Healthcare) and eluted with a linear gradient of 0–0.5 M NaCl at a flow rate of 10 ml min−1. The peak fractions were collected and further purified by gel-filtration chromatography on a Superdex 200 10/300 GL column (GE Healthcare) with buffer consisting of 10 mM Tris pH 8.0, 50 mM NaCl and 3 mM DTT. PHD1JARID1B-containing fractions were concentrated to 40 mg ml−1 using an ultracentrifugal filter tube (Millipore) and used for crystallization.
2.2. Protein crystallization
Initial crystallization trials were performed using Crystal Screen, Index, SaltRX and PEG/Ion kits from Hampton Research and Wizard I and II kits from Emerald BioSystems at 277 K. These initial screens were set up using the hanging-drop vapour-diffusion method by mixing 1 µl protein solution and 1 µl reservoir solution. The initial conditions yielded clusters of needle-shaped crystals of about 0.02 mm in width. The crystal conditons were further optimized by variation of the pH, protein concentration, precipitants and additives. Approximately 200 conditions were set up to optimize crystallization.
2.3. Data collection and processing
All crystals were mounted in nylon loops and flash-cooled in liquid nitrogen using the reservoir buffer as cryoprotectant. Data collection was carried out on BL17U at Shanghai Synchrotron Radiation Facility (SSRF, People’s Republic of China) using a MAR CCD MX-225 detector. The wavelength of the radiation was 1.2823 Å and the crystal-to-detector distance was 90 mm. The exposure time for each frame was 1 s with 1° oscillation and 180 frames were collected. The data were indexed, integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
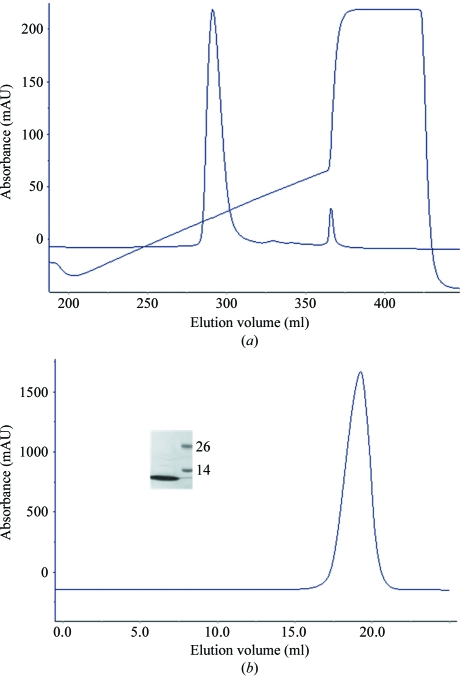
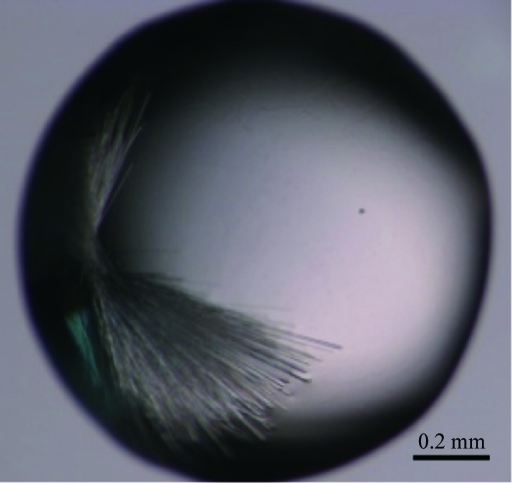
Recombinant PHD1JARID1B protein was expressed, purified to homogeneity (Fig. 1 ▶) and used for crystallization. The purified PHD1JARID1B protein was estimated to have a purity of >95% (inset in Fig. 1 ▶ b). Initial screening was performed at 277 K and crystals appeared in several conditions as clusters of needle-shaped crystals of about 0.02 mm in width. Most of the initial crystallization conditions contained ammonium sulfate, which was chosen to be the essential component for further optimization. After a further optimization screen of solution buffer pH, precipitants and additives, we obtained crystals that were suitable for diffraction. The crystals (Fig. 2 ▶) used for data collection were obtained by mixing 1 µl protein solution (40 mg ml−1) with 1 µl reservoir solution consisting of 0.1 M HEPES pH 7.0, 2.2 M ammonium sulfate at 277 K. The crystals appeared after four weeks.
Figure 1.
PHD1JARID1B protein purification. (a) Ion-exchange (Source 15Q; GE Healthcare) profile of PHD1JARID1B protein. (b) Gel-filtration (Superdex 200 10/300 GL; GE Healthcare) profile of PHD1JARID1B protein. Inset: one of the peak fractions was analyzed by SDS–PAGE with Coomassie Blue staining. The left lane contained the peak fraction and molecular weights are indicated in kDa in the right lane.
Figure 2.
Crystals of PHD1JARID1B grown in 0.1 M HEPES pH 7.0, 2.2 M ammonium sulfate at 277 K.
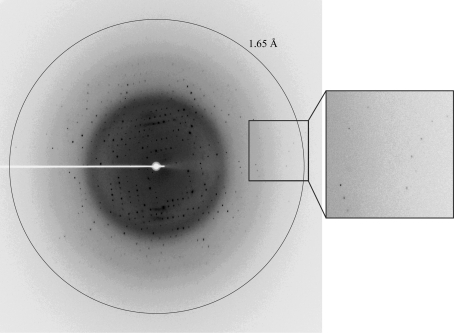
All PHD domains contain two or three Zn atoms according to their primary sequences. In order to solve the phase problem, anomalous data at a wavelength of 1.2823 Å were collected for zinc SAD (Fig. 3 ▶). The diffraction extended to 1.65 Å resolution. The crystals belonged to space group P43, with unit-cell parameters a = 51.7, b = 51.7, c = 36.2 Å. After initial data processing, two molecules were predicted to be present in the asymmetric unit. The estimated solvent content was 38% and the Matthews coefficient was 1.99 Å3 Da−1. Based on sequence analysis, PHD1JARID1B contains two zinc ions in each monomer. Data-collection statistics are summarized in Table 1 ▶. The PHD domain of SmcY protein (PDB entry 2e6r; S. Kadirvel, F. He, Y. Muto, M. Inoue, T. Kigawa, M. Shirouzu, T. Terada & S. Yokoyama, unpublished work) shows 81% sequence similarity and 57% identity to PHD1JARID1B. Therefore, it should be possible to solve the PHD1JARID1B structure by molecular replacement.
Figure 3.
X-ray diffraction pattern of the PHD1JARID1B crystal. The ring indicates a resolution of 1.65 Å. An enlargement of the region in the box is shown on the right.
Table 1. Diffraction data statistics for PHD1JARID1B .
Values in parentheses are for the highest resolution shell.
| Wavelength (Å) | 1.2823 |
| Resolution (Å) | 50.00–1.65 (1.71–1.65) |
| Space group | P43 |
| Unit-cell parameters (Å) | a = 51.7, b = 51.7, c = 36.2 |
| Completeness (%) | 96.7 (80.5) |
| Rmerge† (%) | 7.8 (37.4) |
| 〈I/σ(I)〉 | 46.9 (3.1) |
| Multiplicity | 6.8 (4.7) |
| Total No. of reflections | 77112 (4441) |
| No. of unique reflections | 11340 (945) |
R
merge = 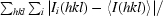
 , where I
i(hkl) is the ith measurement of the intensity of reflection hkl and 〈I(hkl)〉 is the mean intensity of reflection hkl.
, where I
i(hkl) is the ith measurement of the intensity of reflection hkl and 〈I(hkl)〉 is the mean intensity of reflection hkl.
The crystal structure of PHD1JARID1B will deepen our understanding of the molecular mechanism of histone-tail recognition by JARID1B.
Acknowledgments
We thank all staff members of beamline BL17U at SSRF (People’s Republic of China) for their help with data collection. This work was supported by grants from the National Natural Science Foundation of China (31000325), the Shanghai Municipal Natural Science Foundation (10ZR1402800), the Ministry of Science and Technology of China (2009ZX09503-006) and the Shanghai Leading Academic Discipline Project (B111).
References
- Agger, K., Cloos, P. A., Christensen, J., Pasini, D., Rose, S., Rappsilber, J., Issaeva, I., Canaani, E., Salcini, A. E. & Helin, K. (2007). Nature (London), 449, 731–734. [DOI] [PubMed]
- Barrett, A., Madsen, B., Copier, J., Lu, P. J., Cooper, L., Scibetta, A. G., Burchell, J. & Taylor-Papadimitriou, J. (2002). Int. J. Cancer, 101, 581–588. [DOI] [PubMed]
- Chen, Z., Zang, J., Kappler, J., Hong, X., Crawford, F., Wang, Q., Lan, F., Jiang, C., Whetstine, J., Dai, S., Hansen, K., Shi, Y. & Zhang, G. (2007). Proc. Natl Acad. Sci. USA, 104, 10818–10823. [DOI] [PMC free article] [PubMed]
- Chen, Z., Zang, J., Whetstine, J., Hong, X., Davrazou, F., Kutateladze, T. G., Simpson, M., Mao, Q., Pan, C.-H., Dai, S., Hagman, J., Hansen, K., Shi, Y. & Zhang, G. (2006). Cell, 125, 691–702. [DOI] [PubMed]
- Christensen, J., Agger, K., Cloos, P. A., Pasini, D., Rose, S., Sennels, L., Rappsilber, J., Hansen, K. H., Salcini, A. E. & Helin, K. (2007). Cell, 128, 1063–1076. [DOI] [PubMed]
- Cloos, P. A., Christensen, J., Agger, K., Maiolica, A., Rappsilber, J., Antal, T., Hansen, K. H. & Helin, K. (2006). Nature (London), 442, 307–311. [DOI] [PubMed]
- Couture, J.-F., Collazo, E., Ortiz-Tello, P. A., Brunzelle, J. S. & Trievel, R. C. (2007). Nature Struct. Mol. Biol. 14, 689–695. [DOI] [PubMed]
- Han, Z., Liu, P., Gu, L., Zhang, Y., Li, H., Chen, S. & Chai, J. (2007). Frontier Sci. 1, 52–61.
- Horton, J. R., Upadhyay, A. K., Qi, H. H., Zhang, X., Shi, Y. & Cheng, X. (2010). Nature Struct. Mol. Biol. 17, 38–43. [DOI] [PMC free article] [PubMed]
- Iwase, S., Lan, F., Bayliss, P., de la Torre-Ubieta, L., Huarte, M., Qi, H. H., Whetstine, J. R., Bonni, A., Roberts, T. M. & Shi, Y. (2007). Cell, 128, 1077–1088. [DOI] [PubMed]
- Klose, R. J., Kallin, E. M. & Zhang, Y. (2006). Nature Rev. Genet. 7, 715–727. [DOI] [PubMed]
- Klose, R. J., Yamane, K., Bae, Y., Zhang, D., Erdjument-Bromage, H., Tempst, P., Wong, J. & Zhang, Y. (2006). Nature (London), 442, 312–316. [DOI] [PubMed]
- Klose, R. J., Yan, Q., Tothova, Z., Yamane, K., Erdjument-Bromage, H., Tempst, P., Gilliland, D. G., Zhang, Y. & Kaelin, W. G. (2007). Cell, 128, 889–900. [DOI] [PubMed]
- Lan, F., Bayliss, P. E., Rinn, J. L., Whetstine, J. R., Wang, J. K., Chen, S., Iwase, S., Alpatov, R., Issaeva, I., Canaani, E., Roberts, T. M., Chang, H. Y. & Shi, Y. (2007). Nature (London), 449, 689–694. [DOI] [PubMed]
- Lee, M. G., Norman, J., Shilatifard, A. & Shiekhattar, R. (2007). Cell, 128, 877–887. [DOI] [PubMed]
- Lee, M. G., Villa, R., Trojer, P., Norman, J., Yan, K. P., Reinberg, D., Di Croce, L. & Shiekhattar, R. (2007). Science, 318, 447–450. [DOI] [PubMed]
- Li, L., Greer, C., Eisenman, R. N. & Secombe, J. (2010). PLoS Genet. 6, e1001221. [DOI] [PMC free article] [PubMed]
- Lu, P. J., Sundquist, K., Baeckstrom, D., Poulsom, R., Hanby, A., Meier-Ewert, S., Jones, T., Mitchell, M., Pitha-Rowe, P., Freemont, P. & Taylor-Papadimitriou, J. (1999). J. Biol. Chem. 274, 15633–15645. [DOI] [PubMed]
- Margueron, R., Trojer, P. & Reinberg, D. (2005). Curr. Opin. Genet. Dev. 15, 163–176. [DOI] [PubMed]
- Martin, C. & Zhang, Y. (2005). Nature Rev. 6, 838–849. [DOI] [PubMed]
- Ng, S. S. et al. (2007). Nature (London), 448, 87–91.
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Shi, Y., Lan, F., Matson, C., Mulligan, P., Whetstine, J. R., Cole, P. A., Casero, R. A. & Shi, Y. (2004). Cell, 119, 941–953. [DOI] [PubMed]
- Shi, Y. & Whetstine, J. R. (2007). Mol. Cell, 25, 1–14. [DOI] [PubMed]
- Shin, S. & Janknecht, R. (2007). Biochem. Biophys. Res. Commun. 353, 973–977. [DOI] [PubMed]
- Tahiliani, M., Mei, P., Fang, R., Leonor, T., Rutenberg, M., Shimizu, F., Li, J., Rao, A. & Shi, Y. (2007). Nature (London), 447, 601–605. [DOI] [PubMed]
- Tsukada, Y., Fang, J., Erdjument-Bromage, H., Warren, M. E., Borchers, C. H., Tempst, P. & Zhang, Y. (2006). Nature (London), 439, 811–816. [DOI] [PubMed]
- Whetstine, J. R., Nottke, A., Lan, F., Huarte, M., Smolikov, S., Chen, Z., Spooner, E., Li, E., Zhang, G., Colaiacovo, M. & Shi, Y. (2006). Cell, 125, 467–481. [DOI] [PubMed]
- Xiang, Y., Zhu, Z., Han, G., Lin, H., Xu, L. & Chen, C. D. (2007). Cell Res. 17, 850–857. [DOI] [PubMed]
- Xiang, Y., Zhu, Z., Han, G., Ye, X., Xu, B., Peng, Z., Ma, Y., Yu, Y., Lin, H., Chen, A. P. & Chen, C. D. (2007). Proc. Natl Acad. Sci. USA, 104, 19226–19231. [DOI] [PMC free article] [PubMed]
- Yamane, K., Tateishi, K., Klose, R. J., Fang, J., Fabrizio, L. A., Erdjument-Bromage, H., Taylor-Papadimitriou, J., Tempst, P. & Zhang, Y. (2007). Mol. Cell, 25, 801–812. [DOI] [PubMed]
- Yang, Y. et al. (2010). Cell Res. 20, 886–898. [DOI] [PubMed]
- Zhang, Y. & Reinberg, D. (2001). Genes Dev. 15, 2343–2360. [DOI] [PubMed]