Abstract
The host response to Mycobacterium tuberculosis includes macrophage activation, inflammation with increased immune effector cells, tissue necrosis and cavity formation, and fibrosis, distortion, and bronchiectasis. To evaluate the molecular basis of the immune response in the lungs of patients with active pulmonary tuberculosis (TB), we used bronchoalveolar lavage to obtain cells at the site of infection. Affymetrix Genechip micro-arrays and cDNA nylon filter microarrays interrogated gene expression in BAL cells from 11 healthy controls and 17 patients with active pulmonary TB. We found altered gene expression for 69 genes in TB versus normal controls that included cell surface markers, cytokines, chemokines, receptors, transcription factors, and complement components. In addition, TB BAL cell gene expression patternssegregated into 2 groups: one suggestive of a T helper type 1 (Th1) cellular immune response with increased STAT-4, IFN-γ receptor, and MIG expression with increased IFN-γ protein levels in BAL fluid; the other group displayed characteristics of Th2 immunity with increased STAT-6, CD81, and IL-10 receptor expression. We were able to demonstrate that a Th2 presentation could change to a Th1 pattern after anti-tuberculous treatment in one TB patient studied serially. These gene expression data support the conclusion that pulmonary TB produces a global change in the BAL cell transcriptome with manifestations of either Th1 or Th2 immunity.
Keywords: Human, tuberculosis, bronchoalveolar lavage, functional genomics, immunity
INTRODUCTION
Tuberculosis (TB) remains a common infectious disease, with nearly one third of the world’s population being infected by Mycobacterium tuberculosis (M tuberculosis) and 8 million new TB disease cases occurring each year. New York City experienced an epidemic of TB in 1992 when over 3800 cases were reported, and Directly Observed Therapy and TB Clinics have resulted in a 73% decline in the TB case rate since then (1). The incidence of TB remains high among the foreign born who represented 68% of reported active cases in New York City in 2004 (1). Understanding the host cellular immune response would be valuable for developing new treatments to improve clinical outcome, reduce the long duration of chemotherapy, and inhibit reactivation of latent infection.
M tuberculosis activates the cellular immune system. T helper type 1 (Th1) CD4+ lymphocytes secrete interferon-gamma (IFN-γ) and contribute to growth control of M tuberculosis infection in the lung (2). Interleukin (IL)-12 promotes IFN-γ production and Th1 development via signaling pathways that lead to signal transducer and activator of transcription-4 (STAT-4) activation (3). Studies using transgenic mice affecting IFN-γ, its receptor, signaling molecules, and downstream genes revealed extensive mycobacterial infection compared to their wildtype counterparts, demonstrating the importance of these genes in the control of M tuberculosis infection (4–7). We have previously shown that lymphocytes predominate in the bronchoalveolar lavage (BAL) from patients with minimally active pulmonary TB compared to extensive cavitary disease, and that these cells produce larger amounts of IFN-γ (8).
Furthermore, HIV co-infection is a major contributor to the reactivation of latent infection because of the defect in the cellular immune response. We have found that BAL cells from HIV-positive patients with active TB express less IFN-γ mRNA than immunologically competent patients with active TB, signifying the influence Th1 immunity has on M tuberculosis infection (9).
Several authors have detected elevated IL-4 levels at the site of disease in pulmonary TB patients, especially associated with increased tissue destruction and cavitation (10–13). IL-4 in the setting of intracellular infection such as TB, can suppress IFN-γ production and macrophage activation. Th2 cells produce the cytokines IL-4, -5, and –13, are associated with resistance to helminthic infections and are responsible for allergic illnesses. Ligation of IL-4 to its receptor leading to STAT-6 activation drives Th2 lymphocyte differentiation. Several other authors did not find an association between IL-4 or a Th2 response and advanced TB (14,15,16).
In order to describe the molecular basis of the lung TB host response, we studied gene expression in BAL cells obtained from pulmonary TB patients within the earliest phases of anti-mycobacterial treatment. We evaluated the genomic differences stimulated by M tuberculosis, and also describe two manifestations of active TB characterized by BAL cell gene expression patterns and cytokine production.
METHODS
Subjects
17 pulmonary TB patients and 11 healthy controls were recruited. All TB patients had active M tuberculosis pulmonary infection confirmed by sputum culture at presentation; 5 TB patients were resistant to isoniazid. All TB patients were HIV-1 negative and 7 were from Cape Town. All subjects signed informed consent approved by the NYU and University of Cape Town Institutional Review Boards.
Bronchoalveolar lavage
BAL was performed using a flexible bronchoscope and processed as described (17). TB patients had BAL performed in the radiographically involved and uninvolved lobes of the lung early in the initiation of conventional anti-tuberculous therapy. Four TB patients had a second BAL at 4. TB patients had BAL performed in the radiographically involved and uninvolved lobes of the lung within two weeks of the initiation of conventional anti-tuberculous therapy (N=25) and again four weeks after weeks after beginning anti-mycobacterial treatment. One of the TB patients had BAL performed after Directly Observed Therapy consisting of 6 months of appropriate anti-tuberculous medications, when there was clinical, radiographic, and mycobacterial evidence of cure. The BAL samples were processed within 30 minutes of acquisition and were filtered through two layers of sterile cotton gauze to remove mucus. Total cell count, cell viability with Trypan Blue, and BAL cell differentials (500 cells counted) were performed. Washed BAL cell pellets were immediately lysed in TRIzol Reagent (GIBCO-Life Technologies, Grand Island, NY) for RNA extraction. BAL fluid aliquots were frozen at −80°C for cytokine evaluation.
Cytokine levels
We used commercially available cytokine assay kits (R&D Systems, Minneapolis, MN, USA) to measure cytokine levels (IFN-γ, IL-4, and TNF-α) in theunconcentrated BAL fluid. We modified the IFN-γ commercial kit with a highly sensitive fluorometric detection technique (18,19). β-galactosidase-. β-galactosidase-conjugated streptavidin, which catalyzes the conversion of 4-methylumberlliferyl phosphate (MUG) to a fluorescent product (Molecular Probes, Eugene, OR), was used in place of horseradish peroxidase-conjugated streptavidin. Following overnight incubation, fluorescence intensities were measured on a Bio-Tek FL600 plate reader with 360/40 nm excitation and 460/40 nm emission (Bio-Tek Instruments, Inc., Winooski, VT, USA). Using this technique, the sensitivity of detection was increased 50- to 100-fold, which allowed the determination of IFN-γ levels ≤ 1 pg/ml.
Real-time quantitative RT-PCR
Total RNA was extracted from BAL cells in TRIzol Reagent, reverse transcribed using oligo-d(T), and first-strand PCR was performed following the manufacturer’s instructions for the SuperScript preamplification system (Life Technologies, Carlsbad, Calif.). PCR was carried out using oligonucleotide primers previously published (17).
RNA Labeling and Hybridization for cDNA filters
Ten of the BAL cell RNA samples were acquired prior to the availability of Affymetrix genechips and were radiolabeled and hybridized to high-density cDNA arrays (GF211, Research Genetics, Carlsbad, CA) described previously (17). These nylon arrays contain.
Affymetrix genechips
For the newer Affymetrix GeneChips, total RNA was extracted from BAL cells in TRIzol Reagent (GIBCO-Life Technologies, Grand Island, NY). GeneChip Eukaryotic Poly-A RNA Control (Affymetrix, Santa Clara, CA) was added to each sample prior to extraction. Total RNA was purified with RNeasy Total RNA isolation kit (QIAGEN Sciences, Inc., Germantown, MD). Double-stranded cDNA was synthesized using T7-(dT)24 primer (5′-GGCCAGTGAATTGTAATACG ACTCACTATAGGGAGGCGG-(dT)24-3′) (Integrated DNA Technologies, Inc., Coralville, IA) and SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen, Carlsbad, CA). Biotin-labeled cRNA was produced in an in vitro transcription reaction (IVT) using the ENZO BioArray HighYield RNA Transcript Labeling Kit (Enzo Life Sciences, Farmingdale, NY). cRNA was purified, fragmented and hybridized to Affymetrix GeneChip Human Genome U133A, containing 22,283 probe sets. Chips were washed, stained with streptavidin phycoerythrin, hybridized, and scanned using the complete Affymetrix integrated GeneChip Instrument System including a GeneChip Fluidics Station and the Hewlett-Packard GeneArray Scanner (Affymetrix).
Affymetrix array data were normalized to reduce the biological and technical variability in measurements and to make arrays comparable to one another. We used the Robust Multi-Chip Analysis normalization method (RMA) which includes probe-level and chip-level (quantile) normalization (20,21). The. The normalization was performed in GeneTraffic, where data files were loaded onto the local GeneTraffic database with complete annotation based on MIAME standards for microarray experiments (Iobion Informatics, La Jolla, CA). A supervised cluster analysis was performed in GeneTraffic where the 69 genes differentially expressed between clusters in cDNA microarray experiments were selected for hierarchical clustering using Pearson correlation coefficient as a similarity metric and average linkage clustering for both genes and hybridization groups (22).
Statistical methods
Microarray data were normalized to remove variation and examined using clustering and discrimination methods to identify genes that were differentially expressed within each condition. In addition to normalizing the raw data from each array by median centering and log (base 2) transformed, we verified the results by quantile normalization. We used the Significance Analysis of Microarrays (SAM) procedure to estimate the False Discovery Rate (FDR) defined as the expected proportion of false discoveries among the tests (i.e. genes) that are declared significant (23,24). Cluster analysis was used to identify. We used the Significance Analysis of Microarrays (SAM) procedure to estimate the False Discovery Rate (FDR) defined as the expected proportion of false discoveries among the tests (i.e. genes) that are declared significant (18, 19). Cluster analysis was used to identify new classes among samples. The program TreeView was used for a visual representation of the results (Eisen Lab, Stanford University, CA) (22).
Descriptive statistics including means, SEM, and percentages were used to summarize the demographic variables, BAL cell differentials, ELISA and PCR data. A non-parametric, Mann-Whitney, two-tailed, unpaired t-test was utilized to evaluate these parameters. Linear regression analysis was performed for correlation between gene clusters and change over treatment periods.
RESULTS
Subject demographics and BAL cell differentials
The mean age of the TB subjects was 41 ± 3 (SEM) years old; 8 were of African descent (African or African-American), 6 were Asian, 2 were Hispanic, and 1 was white; 4 were known smokers; and the majority were male (Table 1). The mean age of the normal controls was 34 ± 2 years; 5 were African-American, 2 were Asian, 1 was Hispanic, and 3 were white; 6 were male; and 3 were smokers.
Table 1.
Patient Demographics and BAL cell differentials.
| Subject group (N) | Age (years) (mean ± SEM) | Male (N) | Smokers (N) | BAL (N) | BAL cell differential (mean %) | ||
|---|---|---|---|---|---|---|---|
| AM | L | PMN | |||||
| Controls (11) | 34 ± 2 | 6 | 3 | (17) | 92* | 7 | 1** |
|
| |||||||
| Active TB | 13 | 4 | IN (25) | 64 | 16 | 20 | |
| (17) | 41 ± 3 | UN (7) | 85 | 10 | 5 | ||
AM=alveolar macrophage, L=lymphocyte, PMN=polymorphonuclear neutrophil, IN=involved lobe, UN=uninvolved lobe.
p<0.001 compared to TB involved and uninvolved lobe.
p<0.001 compared to TB involved lobe.
BAL cell differentials revealed an increase in inflammatory cells (TB patients neutrophils were 5–20% versus controls 1%, p<0.01 and lymphocytes 10–16% in TB patients versus controls 7% Table 1). The results of chest radiographs for TB subjects showed focal infiltrates and nodules in 6 and multi-lobar disease in 11 (Table 2). The subjectsrecruited from Cape Town, South Africa (Table 2, 34–40) had multilobar infiltrates, nodules, and cavities, indicative of more advanced stage of active pulmonary TB.
Table 2.
Chest radiograph findings for TB subjects.
| Patient | Infiltrate | Nodules | Cavities | Number of involved lobes | Miliary | Cluster |
|---|---|---|---|---|---|---|
| TB A | Yes | 1 | 2 | |||
| TB B | Yes | 1 | 2 | |||
| TB C | Yes | 1 | 1 | |||
| TB D | Yes | Yes | 4 | 1 | ||
| TB E | Yes | 1 | 1 | |||
| TB F | Yes | Yes | Yes | 3 | 1 | |
| TB G | Yes | Yes | 2 | 1 | ||
| TB H | No | No | No | None | 1 | |
| 16 TB | 5 | Yes | 1 | |||
| 29 TB | Yes | 1 | 2 | |||
| 34 TB | Yes | Yes | Yes | 2–3 | 1 | |
| 35 TB | Yes | Yes | Yes | 2 | 1 | |
| 36 TB | Yes | Yes | Yes | 2–3 | 1 | |
| 37 TB | Yes | Yes | 2 | 1 | ||
| 38 TB | Yes | Yes | Yes | 3 | 1 | |
| 39 TB | Yes | Yes | Yes | 2–3 | 2 | |
| 40 TB | Yes | Yes | Yes | 2–3 | 2 |
Gene Expression in BAL Cells from TB patients
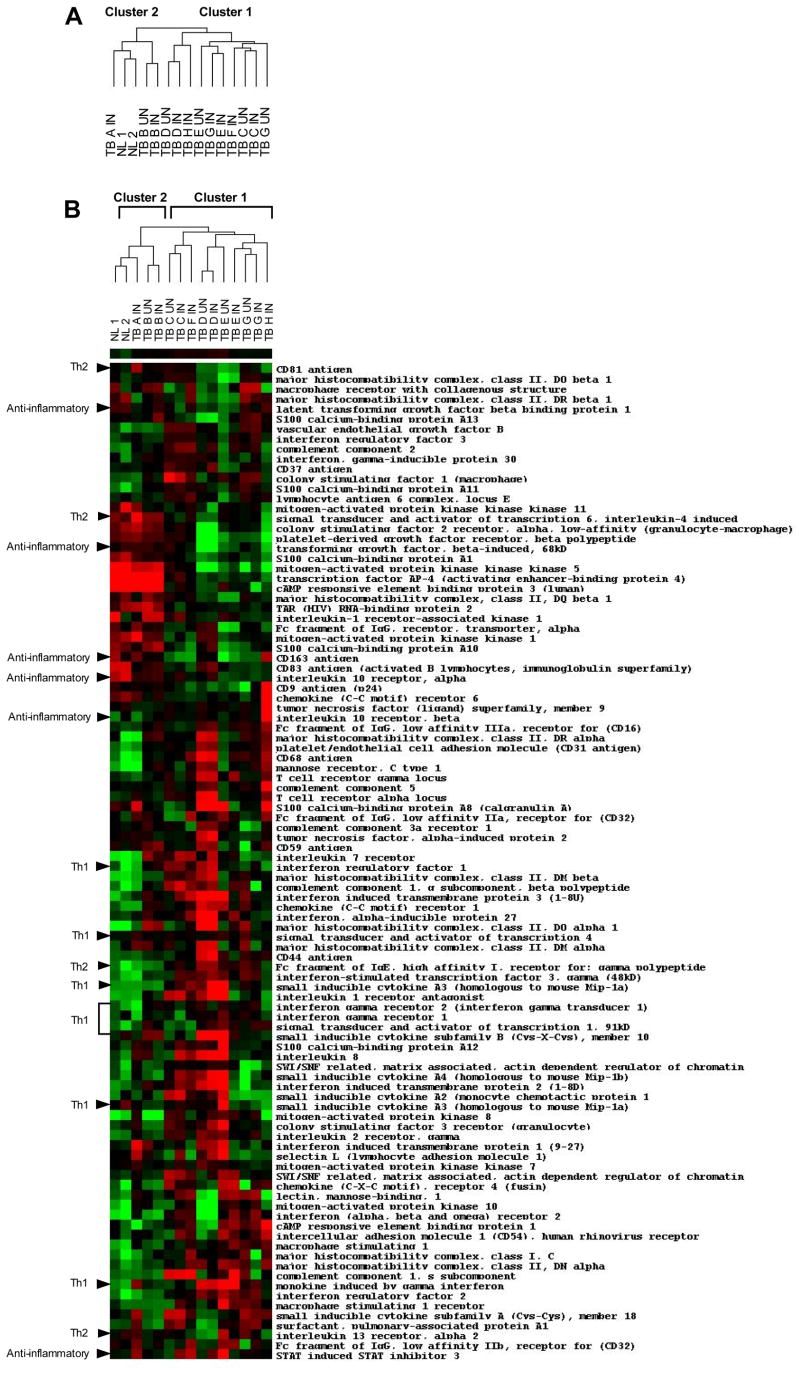
We first used nylon cDNA filters to assess gene expression in the BAL cells of 2 healthy volunteers and 8 pulmonary TB patients. We found that the pulmonary TB patients segregated into 2 groups using clustering algorithms (Figure 1A). One cluster contained 6 TB patients and the other contained 2 TB patients and the 2 control subjects. After quantile normalization, we found that 998 genes had a variance of at least 0.6 across all samples. Using Significance Analysis of Microarrays (SAM), which assesses the likelihood that an observed difference is due to chance by reporting a false discovery rate (FDR) rather than a p value, we found almost 3000 genes to be expressed differently between the 2 clusters at a FDR of less than 30%. The relationship between the individual samples and the differentially expressed genes were confirmed using two methods of normalization. An examination of the 998 genes (training set) differentiating the two clusters revealed 100 immunologically relevant genes (test set). The genes up-regulated in cluster 1 were Th1 related and the genes up-regulated in cluster 2 were Th2 related (Table 3). This clustering was unrelated to BAL cell differential or chest radiographic findings (Table 2). We constructed a new dendrogram from the test set of 100 genes (Figure 1B) that was similar to the dendrogram constructed from the entire set of genes (Figure 1A) confirming the test set of immunologically relevant genes corresponded with the much larger set of differentially regulated genes.
Figure 1.
BAL cells from TB patients and normal controls segregate into two groups of gene expression. (A) Dendrogram ofcDNA nylon filter microarray data from the BAL cells of 2 healthy volunteers (NL) and 8 TB patients (TB) (IN=involved lung segment, UN=uninvolved lung segment). Values of expression for all 4000 genes were normalized using the program Cluster and TreeView grouped each sample according to the pattern of gene regulation. TB BALcell mRNA gene expression patterns cluster into 2 groups. (B) Dendrogram of the same samples in Figure 1A for the test set of 100 immune response genes. The dendrogram is similar to the original dendrogram of all genes.
Table 3.
Test set of genes regulated in BAL cells from 8 TB subjects and 2 healthy subjects according to clustering in Figure 1.
| Up-regulated genes | Cluster 2 | Cluster 1 |
|---|---|---|
| Cell surface markers | CD9
CD37 CD81 (Th2) CD83 (anti-inflamm) CD163 (anti-inflamm) MARCO MHC class II DOβ1, DRβ1, DQβ1 TAR-RNA binding protein 2 Lymphocyte antigen 6 complex, antigen E Fc fragment of IgG, receptor, transporter, α |
CD16
CD31 CD32a & b CD44 CD59 CD68 MHC class II DRα, DMβ, DMα, DOα1, DNα MHC class I C Selectin L (lymphocyte adhesion molecule 1) Fc fragment of IgE (Th2) Mannose receptor C1 Mannose binding lectin TCR-α TCR-γ ICAM-1 |
| Transcription factors | IRF-3
STAT6 (Th2) MAPKKK-5 &11 MAPKK-1 Transcription factor AP-4 IRAK-1 CREB-3 |
IRF-1 (Th1)
IRF-2 ISGF-3 STAT1 (Th1) STAT4 (Th1) SOCS3 (anti-inflamm) MAPK-8 & 10 MAPKK-7 CREB-1 SWI/SNF related, matrix associated, actin dependent regulator of chromatin c2 & d2 |
| Cytokines, chemokines, and receptors | M-CSF
GM-CSF Rα VEGF B PDGF receptor beta polypeptide CCR-6 IL-10Rα (anti-inflamm) LTBP-1 (anti-inflamm) TGF-β induced 68kD (anti-inflamm) IFN-γ inducible protein 30 |
TNF superfamily member 9
TNF-α induced protein 2 IL-1RA IL-2Rγ IL-7R IL-8 IL-10Rβ (anti-inflamm) IL-13Rα2 (Th2) IFN-α inducible protein 27 IFN (type1) receptor 2 IFN-γ R1 & 2 (Th1) IFN induced transmembrane protein1, 2, & 3 G-CSF R MST1 MST1-R MIP-1αR (CCR-1) CCL-18, PARC CXCR-4, Fusin IP-10 (Th1) MCP-1 MIG (Th1) MIP-1α x2 (Th1) MIP-1β |
| Complement and receptors | Complement component 2 | Complement component 1q, β polypeptide
Complement component 1s Complement component 3a receptor 1 Complement component 5 |
| S100 calcium binding protein A1, A10, A11, A13 | Surfactant A1
S100 calcium binding protein A8, A12 |
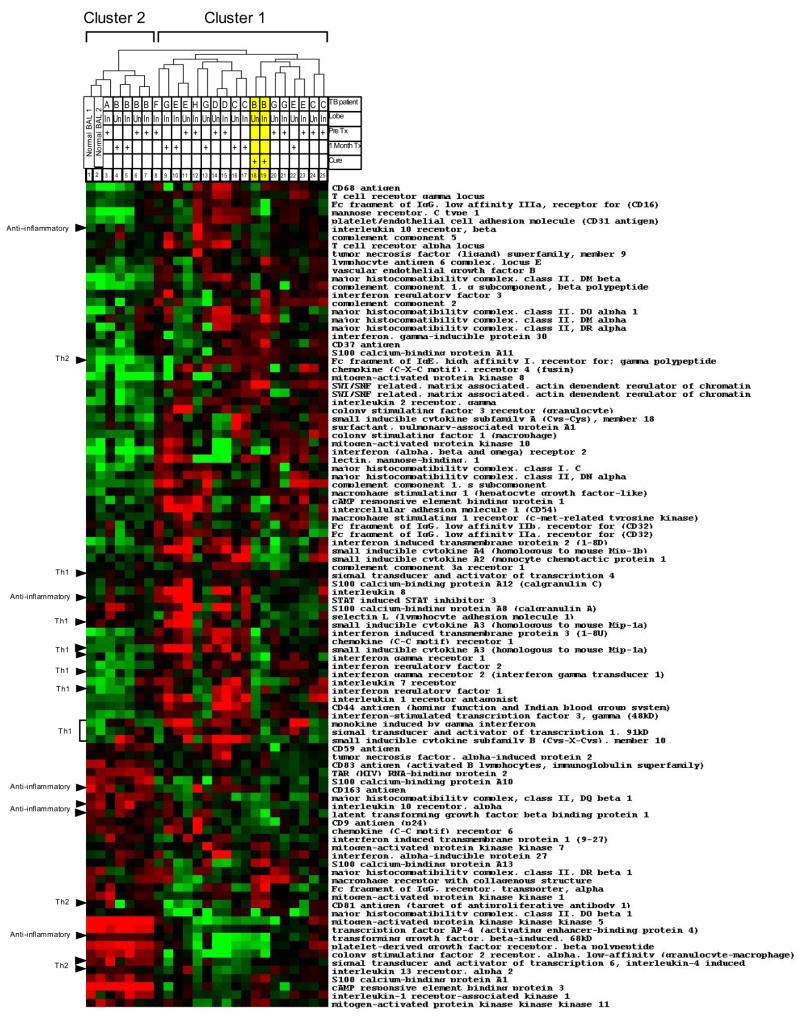
The analysis was repeated on an independent group of9 pulmonary TB patients and9 healthy volunteers usingthe more conventional Affymetrix oligonucleotide arrays. A dendrogram of the expression profile of the test set of immunologically relevant genes derived from the nylon cDNA array experiments showed segregation into two clusters and 69 genes were found to be differentially expressed by significance analysis (Figure 2). Again, clustering was independent of BAL cell differential and chest radiographic findings (Table 2). As in the nylon microarray data (Figure 1 and Table 2), STAT-4 was elevated in cluster 1 containing 6 TB patients. STAT-6 was elevated in cluster 2 which contained 3 TB patients and nine healthy controls. The test set confirmed the results obtained using cDNA arrays validating the clustering of TB BAL into Th1 and Th2 profiles.
Figure 2.
Heat map of Affymetrix GeneChip arrays hybridized with the RNA of BAL cells from additional 9 healthy volunteers and 9 TB patients using the test set of immune response genes (n=100) from the experiments in Figure 1. Black has an average expression level of that gene when compared to all subjects. Bright red represents a four-fold increase in expression above the mean and bright green represents a four-fold decrease in the expression from the average. BAL samples from 3 TB patients clustered with healthy volunteers and expressed Th2 related genes (cluster 2), whereas the remaining 6 TB patients have greater expression of Th1 related genes (cluster 1). Sixty-nine genes from the test set were significantly differentially expressed between these 2 groups (note several genes are duplicated on the map).
To assess the statistical significance of the difference between these two clusters we represented the data as a volcano plot (Figure 3A). The volcano plot used to identify significance and magnitude of change in expression of a set of genes between the groups in Figure 1. The plot displays the negative log of p-values from a t-test on the x-axis and the log2 of change between two conditions on the y-axis. The coordinates represent the magnitude of the difference in expression of that gene and its significance. Figure 3A demonstrates that there were many genes that were significantly different between the 2 groups and that this was not due to chance. The volcano plot of the test set of genes (Figure 3B) was similar in distribution to the graph in Figure 3A, verifying that the test set was characteristic of the larger group of genes. Thus, the clusters portrayed two different immunological states and a small group of genes was highly informative of global alterations of gene expression.
Figure 3.
Volcano plots. (A) Plot of the fold change against the p-value of all genes from cluster 1 compared to cluster 2 subjects indicating that there are many genes differentially regulated between clusters 1 and 2 that are significantly different. (B) The plot of 100 immunologically relevant genes has a similar pattern to plot A.
The TB patients in cluster 1 appeared to express genes important to a Th1 cellular immune response against pulmonary TB. Since IFN-γ is a central mediator of the Th1 cellular immune response, we measured the concentration of this cytokine in BAL fluid by high sensitivity ELISA. The BAL fluid of subjects in cluster 1 had higher levels of IFN-γ protein than the subjects in cluster 2 (Figure 4). There was no difference in IL-4 levels between the 2 groups (data not shown).
Figure 4.

Interferon-γ protein in BAL fluid. IFN-γ levels were measured by high sensitivity ELISA in theunconcentrated BALF from theTB patients in clusters 1 and 2 from Figure 1. Lower levels of IFN-γ were found in the BAL of patients in cluster 2 compared to cluster 1 (p=0.1).
Gene expression in pulmonary TB after treatment
We performed serial bronchoscopies on5 patients with pulmonary TBfrom the initial nylon microarray experiments to examine the genomic presentation at one month and at completion of treatment. Figure 5 shows the dendrogram of these BAL samples illustrating the clustering of the samples based on the similarity of their gene expression patterns. With this larger sample the gene expression profiles again clustered into 2 groups and the samples for TB patients A and B were still closely related to the 2 normal BAL samples (cluster 2) as in Figure 1. TB patient B started in cluster 2 and shifted to cluster 1 when cured of TBat 6 months (Figure 5, highlighted in yellow). In order to examine the statistical significance of the data presented in Figure 5 we graphed the ratio of regulated genes in BAL cells from the subjects in cluster 2 versus cluster 1 at diagnosis against the ratio of gene expression in BAL cells of TB patient (pt) B at diagnosis (dx) versus after completion of TB treatment. The significant correlation (linear regression: r2=0.3, p<0.0001, 95% confidence 0.44 to 0.84) confirmed the shift of BAL gene expression pattern after cure was consistent with a shift from cluster 2 to cluster 1. Therefore, it is possible to change from a Th2 gene expression pattern to a Th1 expression pattern with treatment of pulmonary TB.
Figure 5.
Serial BAL cell gene expression. BAL cell gene expression changed from Th2 to Th1 pattern after pulmonary TB was treated in TB (patient B). Dendrogram showing the clustering of GeneFilter RNA expression data from the BAL cells from 2 normal volunteers and 8 pulmonary TB patients from Figure 1. BAL was obtained from the radiographically involved lobe of the TB patients before and one month into treatment (n=4) with anti-tuberculous medications. The BAL from TB patients A and B cluster with the 2 healthy volunteers at diagnosis and have a Th2 pattern of immunity (as in Figure 1). The BAL from TB patient B obtained after completion of TB treatment (cure, highlighted in yellow) switched to cluster 1 with a Th1 gene expression pattern.
Genomics in the uninvolved lobe compared to the involved lobe
Interestingly, in figures 1 and 5 we noted that the involved and uninvolved BAL samples from the same TB patients clustered closely together. Comparing the gene expression data from each lung segment, we found only 7 genes to be significantly altered with a FDR less than 50% as determined by SAM.
To evaluate inflammation in the involved lung segment, TNF-α was assayed by ELISA., TNF-α levels were higher in the involved lung segments compared the uninvolved lung segments, but did not reach significance (p=0.1) (figure 7A). TNF-α levels were higher in the involved lung segments compared the uninvolved lung segments in the BAL fluid, but did not reach significance (p=0.1). The mRNA levels for TNF-α measured by microarray was similar for involved and uninvolved lung segments. We also evaluated gene expression of the chemokine IFN-γ inducible protein 10 kD (IP-10) by real-time quantitative RT-PCR from mRNA extracted from BAL cells from healthy controls, and the uninvolved and involved lobes of TB patients. Values were corrected for GAPDH expression. IP-10 expression was elevated in both the radiographically involved and uninvolved lobes of the lung in patients with pulmonary tuberculosis compared to normal controls (p<0.05). There was no significant correlation between IP-10 levels and BAL lymphocyte count.
DISCUSSION
We compared BAL cell gene expression between pulmonary TB patients and normal controls finding 69 genes associated with immune activation and inflammation using 2 different microarray techniques. In addition, we found two BAL cell gene expression patterns in pulmonary TB: one presentation had up-regulation of genes involved in the Th2 immune response and the other a Th1 immune response. Interestingly, this pattern of gene expression could change from a Th2 to a Th1 presentation over time with anti-tuberculosis treatment. We found regulation of multiple cytokines, chemokines and their receptors, signaling molecules, and surface antigens in BAL cells from subjects with M tuberculosis infection.
In a study perfomed on murine macrophages exposed to IFN-γ or live M tuberculosis, Ehrt et al observed differential regulation of 25% of genes (2664 genes). They found five times as many genes related to immunity and inflammation were induced than suppressed. Highly induced genes included: MHC class II antigen, monokine induced by IFN-γ (MIG), RANTES, secretory leukocyte protease inhibitor (SLPI), macrophage inflammatory protein-1β (MIP-1β), nitric oxide synthase 2 (NOS2), IP-10, and serum amyloid A (26). Keller et al found consistent up-regulation of G-CSF expression in infected macrophages from mice susceptible to TB compared to macrophages from TB resistant species (27). The authors surmised that macrophages from susceptible mice favor an inflammatory response that predominantly recruits cells involved in tissue destruction (granulocytes) rather than antibacterial protection. Ragno et al and Wang and colleagues also found induction of cytokines, chemokines, ribosomal proteins, and STAT-1 in human monocyte derived macrophages exposed to live M tuberculosis in vitro (28,29). Compared to Ehrt’s murine macrophage model, only 10% of the genes were similarly regulated.
Grassi and colleagues utilized cDNA microarray technology to evaluated 847 immune-inflammatory related genes in the BAL cells from patients with pulmonary TB (30). They identified 54 genes of limited variance within the group that were up-regulated (17) or down regulated (4) compared to non-TB patients, and the 33 remaining genes were similarly modulated in both groups. They found up-regulation of ICAM-1, IRF-1, STAT-1, IL-1β, CD64, CCR4, IL-8, MIP-1α, MIP-1β, MCP-1, IFN-γR2, and GM-CSF receptor in the BAL of TB subjects. Despite the increased transcription of many IFN-γ related genes, the investigators were unable to detect increase in IFN-γ mRNA transcription with a sensitive quantitative RT-PCR assay. However, they were able to correlate increased expression of IFN-γ and TNF-α with increasing tissue damage as scored on chest radiographs (30).
Only 10% of the genes reported were similar to the murine macrophage model (21, 24).
In comparison to these microarray studies, we found 2 genomic presentations of pulmonary TB in humans. Cluster 1 pulmonary TB patients expressed predominantly Th1 related genes and functionally produced more IFN-γ. The BAL cells had increased expression of STAT-4, a transcription factor required for Th1 cells to activate IFN-γ transcription after IL-12 stimulation; IFN-γ receptor 1 and 2, their signaling molecules STAT-1 and IRF-1, and the chemokines IP-10, monokine induced by gamma interferon (MIG) and MIP-1α responsible for Th1 cell migration and differentiation. Cluster 1 had up-regulation of ICAM-1, inflammatory. Cluster 1 had up-regulation of ICAM-1, cytokines, chemokines and their receptors, as well as genes responsible for antigen presentation. This correlated well with the higher levels of IFN-γ found in the BAL fluid, which we have observed previously (8). We might expect less chest radiographic involvement in the Th1 cluster if these mediators were better able to control TB infection, but we found a Th1 pattern in advanced radiographic TB presentations as well.
Cluster 2 pulmonary TB patients had elevation of STAT-6, an essential mediator of the Th2 phenotype. There was also an increase in IL-13 receptor expression, which in conjunction with the IL-4 receptor, utilizes STAT-6 for signaling. Furthermore, there was an increase in CD81 expression, which is also involved in Th2 polarization, suggesting that some normal individuals as well as patients with pulmonary TB may have Th2 immunity. In addition, these subjects had increased expression of genes involved in suppressing inflammation: IL-10 receptor, TGF-β related genes, and CD163. IL-10, produced early in Th2 differentiation, appears to be an important inhibitor of the host immune response to TB (31). Peripheral blood mononuclear cells (PBMC) and mononuclear cells from infected body fluids from anergic patients produce more IL-10 and less IFN-γ after PPD stimulation (32,33). M tuberculosis has been shown to induce IL-10 in human macrophages, inducing inhibitory transcription factors thereby restricting inflammatory cytokine production (34). Transforming growth factor beta (TGF-β) is another anti-inflammatory cytokine produced by monocytes and dendritic cells after mycobacterial infection and like IL-10, is capable of inhibiting IFN-γ and proinflammatory cytokine production, antigen presentation, and cellular activation (35). Despite the anti-inflammatory effects of IL-10 and TGF-β, we found evidence for macrophage activation in cluster 2 subjects with increased expression of GM-CSF receptor, dendritic cell markers CD83 and S100, and MHC class II molecules in BAL cells.
Leprosy is another mycobacterial disease known to present with either a Th1 or Th2 phenotype; analogous gene expression profiling in lepromatous versus tuberculoid skin lesions has been reported (36–38). Not only did the 2 classes of skin lesions in leprosy cluster separately, but the expression profiles also revealed. Not only did the 2 classes cluster separately, but the expression profiles also revealed marked differences in genes related to the immune response and were consistent with the previously shown Th1/Th2 dichotomy. In fact, lepromatous lesions had increased expression of TGF-β, LTBP, IL-10, CD47, CD83, and STAT-6, as we found in our cluster 2 subjects, implicating a Th2 phenotype.
There is growing evidence that Th1/Th2 balance is important in controlling pulmonary TB. BAL cells but not PBMC from patients with active pulmonary TB had more IFN-γ producing lymphocytes than IL-4 and IL-10 producing lymphocytes in response to in vitro stimulation with PPD (39). Other studies found PPD. Other studies found PPD specific T cells from the peripheral blood (PB) or TB infected body fluids at diagnosis had a Th0 (naïve T helper cell) cytokine profile with production of both IFN-γ and IL-4 (40). After 6 months of effective anti-tuberculous treatment, most PPD-responsive T cell clones showed a Th1 profile (IFN-γ only) (40). In contrast, in treatment failure increased numbers of IL-4 producing Th2 clones were noted in areas of disease (40,41). Mazzarella et al also found increased IL-4 production by BAL lymphocytes in cavitary TB samples and increased IFN-γ production in noncavitary TB BAL (13). Recent studies by Dheda and colleagues examined the role of IL-4δ2, the splice variant and antagonist of IL-4, in pulmonary TB (42). They found increased levels of both IL-4 and IL-4δ2 mRNA in the BAL of TB patients. Advanced radiologic disease correlated with the IL-4/IFN-γ ratio and after 6 months of anti-tuberculous treatment, IL-4 expression remained the same, yet there was increased IL-4δ2 and IFN-γ expression in BAL. Thus, IL-4δ2 expression may promote a more effective immune response in the lung by antagonizing an ineffective Th2 response in TB (42).
An appropriate lymphocytic immune response appears most effective in controlling active M tuberculosis infection. Yet macrophages remain the predominant alveolar cell and are required for antigen presentation, lymphocyte activation, and type 1 vs. 2 polarization. Dheda et al found that IL-4δ2 was also expressed in the non-T cell fraction of TB BAL (42). Moreover, Verreck and colleagues found that human macrophages polarized in the presence of GM-CSF secreted high levels of IL-23, a heterodimer sharing the same IL-12 p40 chain and similar Th1 cytokine function, after mycobacterial activation (43). These cells efficiently stimulated Th1 cells by up-regulating their antigen presenting and co-stimulatory cell surface molecule expression. Only after the addition of IFN-γ did these macrophages produce IL-12. In contrast, human macrophages polarized with M-CSF produced IL-10 and MCP-1, which stimulates Th2 polarization, and supported intracellular M tuberculosis overgrowth. In addition, these macrophages down regulated cell surface expression of HLA-DR, CD86, and CD40, potentially promoting chronic mycobacterial infection (43). Since there was no difference in the number of BAL lymphocytes between the 2 clusters of TB patients in our study, and the majority of BAL cells are macrophages, the differences in immune gene regulation may be due to differences in the response of AM to infection.
The development of a Th2 as opposed to a Th1 response to M tuberculosis infection at presentation may be genetically predetermined, or it may be in response to other environmental stimuli such asthma or parasitic or helminthic infections predominant in developing countries (44). A diminished Th1 response and shift of the Th2 response to a Th1 response after successful treatment of pulmonary TB has been noted in African and U.S. immigrant TB patients (15,45). We found a similar shift from a Th2 gene expression pattern in the BAL from a TB patient at diagnosis to a Th1 genomic profile after completion of anti-tuberculosis medications and cure of pulmonary TB.
Gene expression patterns and the clinical characteristics of disease may not only depend upon human susceptibility, but on the virulence of the infecting organism. Human AM infected with a virulent strain of M tuberculosis (H37Rv) altered the expression of greater than 400 genes compared to only 17 in AM infected with an avirulent strain (H37Ra) (46). The genes induced by H37Rv were predominantly those characteristic of a Th1 immune response. Spira et al found consistent down regulation of pro-apoptotic genes in human AM after in vitro infection with H37Rv (47). H37Ra infection led to a gene expression pattern favoring macrophage apoptosis. Moreover, mammalian gene expression patterns vary in relation to the physiological states (e.g., rapid growth phase) of the tubercle bacillus (48).
We found similar gene expression in BAL cells from radiographically involved and uninvolved lobes probably reflecting subclinical M tuberculosis infection in radiographically uninvolved lobes. TNF-α levels were higher in the BAL fluid from involved lobes, and we previously noted increases in TNF-α in BAL cell 24-hour supernatants from involved sites (25). The chest radiograph may be an inaccurate measure of tuberculous involvement. Since the genomic profiles were similar, the disparity between gene regulation and inflammation may also be secondary to post-transcriptional effects. Keller et al considered this when studying gene expression patterns in different mouse strains that were resistant or susceptible to M tuberculosis infection (27). Infected macrophages from resistant mouse strains (C57BL/6 and BALB/c) produced more TNF-α and IL-10 compared to macrophages from susceptible strains (DBA/2 and CBA/J). Yet the difference in protein levels were not reflected at the mRNA level. Clarification would require assessment of gene transcription and translation and could result in the discovery of. Clarification would require assessment of gene transcription and translation and could result in the discovery of a potential immune mediator of disease.
There are significant limitations to our study. We used 2 different microarray platforms, BAL includes a mixed population of cells, and we had a limited sample size. The genomics presented in this report can only describe steady state mRNA levels in BAL cells; the mRNA observations present a static picture. Cytokine and cell differential data are presented more as confirmation of sample origin (involved versus uninvolved segments of the lung) rather than determining or distinguishing the phenotype of the genomic profiles (cluster 1 versus cluster 2 TB patients). The functional consequences of the mRNA data are beyond the scope of this investigation. BAL cells are a mixed population of leukocytes, but the predominant cell type are macrophages with only a 15% difference between involved and uninvolved lung segments (Table 1). Our results likely represent the effect of regulated gene expression in macrophages. The mRNA expression from lymphocytes or PMN did not appear to alter gene expression profiles using the multiple modes of detection (nylon filter array, genechip array, and RT-PCR). It is possible that the cytokine differences between involved and uninvolved lung segments came from these minor cell populations. Nevertheless, the observations were robust. Smoking was not a factor since most of our study subjects were nonsmokers or ex-smokers. There were 3 smoker controls and 4 TB patients. Changes in sensitivity and cross hybridization artifacts between the two platforms did not materially alter the results. We demonstrated 2 gene expression profiles in independent TB populations in the U.S. and South Africa with both cDNA and Affymetrix platforms.
Figure 6.
Graph of ratio of regulated genes in BAL cells from the subjects in cluster 2 versus cluster 1 at diagnosis (figure 1) compared to the ratio of gene expression in BAL cells of TB patient (pt) B at diagnosis (dx) versus after TB was completely treated, showing a significant correlation. This confirms the shift of BAL gene expression pattern after treatment in patient B is consistent with a change from cluster 2 to cluster 1 or a Th2 to a Th1 pattern of gene expression.
Acknowledgments
This work was supported by NIH/NCRR grants M01 RR00096 and HL-59832, HL-57879, HL-68517, NYC Speakers Award to BR, and Uehara Memorial Foundation Award to YH.
We would like to thank Patricia Soteropoulos, PhD and Anthony Galante from the Center for Applied Genomics at the International Center for Public Health, Newark, NJ for performing the Affymetrix GeneChip hybridizations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.The New York City Department of Health and Mental Hygiene. Tuberculosis cases down 73% citywide since the peak of the NYC epidemic. 2005 Press Release # 024-05 ed. http://www.ci.nyc.ny.us/html/doh/html/pr/pr024-05.shtml.
- 2.Schluger NW, Rom WN. The host immune response to tuberculosis. Am J Respir Crit Care Med. 1998;157(3 Pt 1):679–91. doi: 10.1164/ajrccm.157.3.9708002. [DOI] [PubMed] [Google Scholar]
- 3.Rengarajan J, Szabo SJ, Glimcher LH. Transcriptional regulation of Th1/Th2 polarization. Immunol Today. 2000;21(10):479–83. doi: 10.1016/s0167-5699(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 4.Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon gamma in resistance to Mycobacterium tuberculosis infection. J Exp Med. 1993;178(6):2249–54. doi: 10.1084/jem.178.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamijo R, Le J, Shapiro D, Havell EA, Huang S, Aguet M, Bosland M, Vilcek J. Mice that lack the interferon-gamma receptor have profoundly altered responses to infection with Bacillus Calmette-Guerin and subsequent challenge with lipopolysaccharide. J Exp Med. 1993;178(4):1435–40. doi: 10.1084/jem.178.4.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamijo R, Harada H, Matsuyama T, Bosland M, Gerecitano J, Shapiro D, Le J, Koh SI, Kimura T, Green SJ, et al. Requirement for transcription factor IRF-1 in NO synthase induction in macrophages. Science. 1994;263(5153):1612–5. doi: 10.1126/science.7510419. [DOI] [PubMed] [Google Scholar]
- 7.MacMicking JD, North RJ, LaCourse R, Mudgett JS, Shah SK, Nathan CF. Identification of nitric oxide synthase as a protective locus against tuberculosis. Proc Natl Acad Sci U S A. 1997;94(10):5243–8. doi: 10.1073/pnas.94.10.5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Condos R, Rom WN, Liu YM, Schluger NW. Local immune responses correlate with presentation and outcome in tuberculosis. Am J Respir Crit Care Med. 1998;157(3 Pt 1):729–35. doi: 10.1164/ajrccm.157.3.9705044. [DOI] [PubMed] [Google Scholar]
- 9.Law KF, Jagirdar J, Weiden MD, Bodkin M, Rom WN. Tuberculosis in HIV-positive patients: cellular response and immune activation in the lung. Am J Respir Crit Care Med. 1996;153(4 Pt 1):1377–84. doi: 10.1164/ajrccm.153.4.8616569. [DOI] [PubMed] [Google Scholar]
- 10.van Crevel R, Ottenhoff TH, van Der Meer JW. Innate Immunity to Mycobacterium tuberculosis. Clin Microbiol Rev. 2002;15(2):294–309. doi: 10.1128/CMR.15.2.294-309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sousa AO, Salem JI, Lee FK, Vercosa MC, Cruaud P, Bloom BR, Lagrange PH, David HL. An epidemic of tuberculosis with a high rate of tuberculin anergy among a population previously unexposed to tuberculosis, the Yanomami Indians of the Brazilian Amazon. Proc Natl Acad Sci U S A. 1997;94(24):13227–32. doi: 10.1073/pnas.94.24.13227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raju B, Hoshino Y, Kuwabara K, Belitskaya I, Prabhakar S, Canova A, Gold JA, Condos R, Pine RI, Brown S, Rom WN, Weiden MD. Aerosolized gamma interferon (IFN-gamma) induces expression of the genes encoding the IFN-gamma-inducible 10-kilodalton protein but not inducible nitric oxide synthase in the lung during tuberculosis. Infect Immun. 2004;72(3):1275–83. doi: 10.1128/IAI.72.3.1275-1283.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gee KR, Sun WC, Bhalgat MK, Upson RH, Klaubert DH, Latham KA, Haugland RP. Fluorogenic substrates based on fluorinated umbelliferones for continuous assays of phosphatases and beta-galactosidases. Anal Biochem. 1999;273(1):41–8. doi: 10.1006/abio.1999.4202. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez CR, Fei DT, Keyt B, Baly DL. A sensitive fluorometric enzyme-linked immunosorbent assay that measures vascular endothelial growth factor165 in human plasma. J Immunol Methods. 1998;219(1–2):45–55. doi: 10.1016/s0022-1759(98)00131-8. [DOI] [PubMed] [Google Scholar]
- 15.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19(2):185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 16.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 17.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95(25):14863–8. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benyamini Y, Hochberg Y. Controlling the false discovery rate: a powerful and practical approach to multiple testing. J R Statist Soc B. 1995;57:289–300. [Google Scholar]
- 19.Storey J. A direct approach to false discovery rates. Journal of the Royal Statistical Society, Series B. 2002;64:479–498. [Google Scholar]
- 20.Gaede KI, Mamat U, Muller-Quernheim J. Differential gene expression pattern in alveolar macrophages of patients with sarcoidosis and tuberculosis. J Mol Med. 2004;82(3):206–10. doi: 10.1007/s00109-003-0511-2. [DOI] [PubMed] [Google Scholar]
- 21.Ehrt S, Schnappinger D, Bekiranov S, Drenkow J, Shi S, Gingeras TR, Gaasterland T, Schoolnik G, Nathan C. Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J Exp Med. 2001;194(8):1123–40. doi: 10.1084/jem.194.8.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller C, Lauber J, Blumenthal A, Buer J, Ehlers S. Resistance and susceptibility to tuberculosis analysed at the transcriptome level: lessons from mouse macrophages. Tuberculosis (Edinb) 2004;84(3–4):144–58. doi: 10.1016/j.tube.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 23.Ragno S, Romano M, Howell S, Pappin DJ, Jenner PJ, Colston MJ. Changes in gene expression in macrophages infected with Mycobacterium tuberculosis: a combined transcriptomic and proteomic approach. Immunology. 2001;104(1):99–108. doi: 10.1046/j.0019-2805.2001.01274.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang JP, Rought SE, Corbeil J, Guiney DG. Gene expression profiling detects patterns of human macrophage responses following Mycobacterium tuberculosis infection. FEMS Immunol Med Microbiol. 2003;39(2):163–72. doi: 10.1016/S0928-8244(03)00223-2. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Broser M, Cohen H, Bodkin M, Law K, Reibman J, Rom WN. Enhanced interleukin-8 release and gene expression in macrophages after exposure to Mycobacterium tuberculosis and its components. J Clin Invest. 1995;95(2):586–92. doi: 10.1172/JCI117702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang JC, Wysocki A, Tchou-Wong KM, Moskowitz N, Zhang Y, Rom WN. Effect of Mycobacterium tuberculosis and its components on macrophages and the release of matrix metalloproteinases. Thorax. 1996;51(3):306–11. doi: 10.1136/thx.51.3.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cliff JM, I, Andrade N, Mistry R, Clayton CL, Lennon MG, Lewis AP, Duncan K, Lukey PT, Dockrell HM. Differential gene expression identifies novel markers of CD4+ and CD8+ T cell activation following stimulation by Mycobacterium tuberculosis. J Immunol. 2004;173(1):485–93. doi: 10.4049/jimmunol.173.1.485. [DOI] [PubMed] [Google Scholar]
- 28.Spira A, Carroll JD, Liu G, Aziz Z, Shah V, Kornfeld H, Keane J. Apoptosis genes in human alveolar macrophages infected with virulent or attenuated Mycobacterium tuberculosis: a pivotal role for tumor necrosis factor. Am J Respir Cell Mol Biol. 2003;29(5):545–51. doi: 10.1165/rcmb.2002-0310OC. [DOI] [PubMed] [Google Scholar]
- 29.Glimcher LH, Murphy KM. Lineage commitment in the immune system: the T helper lymphocyte grows up. Genes Dev. 2000;14(14):1693–711. [PubMed] [Google Scholar]
- 30.Condos R, Raju B, Canova A, Zhao BY, Weiden M, Rom WN, Pine R. Recombinant gamma interferon stimulates signal transduction and gene expression in alveolar macrophages in vitro and in tuberculosis patients. Infect Immun. 2003;71(4):2058–64. doi: 10.1128/IAI.71.4.2058-2064.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saukkonen JJ, Bazydlo B, Thomas M, Strieter RM, Keane J, Kornfeld H. Beta-chemokines are induced by Mycobacterium tuberculosis and inhibit its growth. Infect Immun. 2002;70(4):1684–93. doi: 10.1128/IAI.70.4.1684-1693.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sauty A, Dziejman M, Taha RA, Iarossi AS, Neote K, Garcia-Zepeda EA, Hamid Q, Luster AD. The T cell-specific CXC chemokines IP-10, Mig, and I-TAC are expressed by activated human bronchial epithelial cells. J Immunol. 1999;162(6):3549–58. [PubMed] [Google Scholar]
- 33.Law K, Weiden M, Harkin T, Tchou-Wong K, Chi C, Rom WN. Increased release of interleukin-1 beta, interleukin-6, and tumor necrosis factor-alpha by bronchoalveolar cells lavaged from involved sites in pulmonary tuberculosis. Am J Respir Crit Care Med. 1996;153(2):799–804. doi: 10.1164/ajrccm.153.2.8564135. [DOI] [PubMed] [Google Scholar]
- 34.Hershey GK. IL-13 receptors and signaling pathways: an evolving web. J Allergy Clin Immunol. 2003;111(4):677–90. doi: 10.1067/mai.2003.1333. quiz 691. [DOI] [PubMed] [Google Scholar]
- 35.Deng J, Dekruyff RH, Freeman GJ, Umetsu DT, Levy S. Critical role of CD81 in cognate T-B cell interactions leading to Th2 responses. Int Immunol. 2002;14(5):513–23. doi: 10.1093/intimm/14.5.513. [DOI] [PubMed] [Google Scholar]
- 36.Hogger P, Sorg C. Soluble CD163 inhibits phorbol ester-induced lymphocyte proliferation. Biochem Biophys Res Commun. 2001;288(4):841–3. doi: 10.1006/bbrc.2001.5845. [DOI] [PubMed] [Google Scholar]
- 37.Pettersen RD, Hestdal K, Olafsen MK, Lie SO, Lindberg FP. CD47 signals T cell death. J Immunol. 1999;162(12):7031–40. [PubMed] [Google Scholar]
- 38.Cousins DJ, Lee TH, Staynov DZ. Cytokine coexpression during human Th1/Th2 cell differentiation: direct evidence for coordinated expression of Th2 cytokines. J Immunol. 2002;169(5):2498–506. doi: 10.4049/jimmunol.169.5.2498. [DOI] [PubMed] [Google Scholar]
- 39.Boussiotis VA, Tsai EY, Yunis EJ, Thim S, Delgado JC, Dascher CC, Berezovskaya A, Rousset D, Reynes JM, Goldfeld AE. IL-10-producing T cells suppress immune responses in anergic tuberculosis patients. J Clin Invest. 2000;105(9):1317–25. doi: 10.1172/JCI9918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Delgado JC, Tsai EY, Thim S, Baena A, Boussiotis VA, Reynes JM, Sath S, Grosjean P, Yunis EJ, Goldfeld AE. Antigen-specific and persistent tuberculin anergy in a cohort of pulmonary tuberculosis patients from rural Cambodia. Proc Natl Acad Sci U S A. 2002;99(11):7576–81. doi: 10.1073/pnas.062056099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka N, Hoshino Y, Gold J, Hoshino S, Martiniuk F, Kurata T, Pine R, Levy D, Rom WN, Weiden M. Interleukin-10 induces inhibitory C/EBPbeta through STAT-3 and represses HIV-1 transcription in macrophages. Am J Respir Cell Mol Biol. 2005;33(4):406–11. doi: 10.1165/rcmb.2005-0140OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toossi Z, Ellner JJ. The role of TGF beta in the pathogenesis of human tuberculosis. Clin Immunol Immunopathol. 1998;87(2):107–14. doi: 10.1006/clin.1998.4528. [DOI] [PubMed] [Google Scholar]
- 43.Salgame P, Abrams JS, Clayberger C, Goldstein H, Convit J, Modlin RL, Bloom BR. Differing lymphokine profiles of functional subsets of human CD4 and CD8 T cell clones. Science. 1991;254(5029):279–82. doi: 10.1126/science.254.5029.279. [DOI] [PubMed] [Google Scholar]
- 44.Yamamura M, Uyemura K, Deans RJ, Weinberg K, Rea TH, Bloom BR, Modlin RL. Defining protective responses to pathogens: cytokine profiles in leprosy lesions. Science. 1991;254(5029):277–9. doi: 10.1126/science.254.5029.277. [DOI] [PubMed] [Google Scholar]
- 45.Bleharski JR, Li H, Meinken C, Graeber TG, Ochoa MT, Yamamura M, Burdick A, Sarno EN, Wagner M, Rollinghoff M, Rea TH, Colonna M, Stenger S, Bloom BR, Eisenberg D, Modlin RL. Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science. 2003;301(5639):1527–30. doi: 10.1126/science.1087785. [DOI] [PubMed] [Google Scholar]
- 46.Schwander SK, Torres M, Sada E, Carranza C, Ramos E, Tary-Lehmann M, Wallis RS, Sierra J, Rich EA. Enhanced responses to Mycobacterium tuberculosis antigens by human alveolar lymphocytes during active pulmonary tuberculosis. J Infect Dis. 1998;178(5):1434–45. doi: 10.1086/314454. [DOI] [PubMed] [Google Scholar]
- 47.Dieli F, Singh M, Spallek R, Romano A, Titone L, Sireci G, Friscia G, Di Sano C, Santini D, Salerno A, Ivanyi J. Change of Th0 to Th1 cell-cytokine profile following tuberculosis chemotherapy. Scand J Immunol. 2000;52(1):96–102. doi: 10.1046/j.1365-3083.2000.00744.x. [DOI] [PubMed] [Google Scholar]
- 48.Marchant A, Amedei A, Azzurri A, Vekemans J, Benagiano M, Tamburini C, Lienhardt C, Corrah T, McAdam KP, Romagnani S, D’Elios MM, Del Prete G. Polarization of PPD-specific T-cell response of patients with tuberculosis from Th0 to Th1 profile after successful antimycobacterial therapy or in vitro conditioning with interferon-alpha or interleukin-12. Am J Respir Cell Mol Biol. 2001;24(2):187–94. doi: 10.1165/ajrcmb.24.2.4274. [DOI] [PubMed] [Google Scholar]
- 49.Lienhardt C, Azzurri A, Amedei A, Fielding K, Sillah J, Sow OY, Bah B, Benagiano M, Diallo A, Manetti R, Manneh K, Gustafson P, Bennett S, D’Elios MM, McAdam K, Del Prete G. Active tuberculosis in Africa is associated with reduced Th1 and increased Th2 activity in vivo. Eur J Immunol. 2002;32(6):1605–13. doi: 10.1002/1521-4141(200206)32:6<1605::AID-IMMU1605>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 50.Zhang M, Lin Y, Iyer DV, Gong J, Abrams JS, Barnes PF. T-cell cytokine responses in human infection with Mycobacterium tuberculosis. Infect Immun. 1995;63(8):3231–4. doi: 10.1128/iai.63.8.3231-3234.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]