Abstract
Conformational dynamics play a key role in enzyme catalysis. While protein motions have clear implications for ligand flux, a role for dynamics in the chemical step of enzyme catalysis has not been clearly established. We generated a mutant of E. coli dihydrofolate reductase (DHFR) that abrogates millisecond time scale fluctuations in the enzyme active site without perturbing its structural and electrostatic preorganization. Remarkably, this dynamic knockout severely impairs hydride transfer. Thus we have found a link between conformational fluctuations on the millisecond timescale and the chemical step of an enzymatic reaction, with broad implications for our understanding of enzyme mechanisms and for attempts to design novel protein catalysts.
Protein motions are critical for biological functions, but their precise role in enzyme catalysis remains unclear (1, 2). While there is convincing evidence that conformational fluctuations are essential for mediating substrate and cofactor binding as well as product release, and can be rate limiting for enzyme turnover (3–6), the importance of protein flexibility for progression along the chemical reaction coordinate remains a matter of debate (7, 8). In one view, electrostatic preorganization of the active site is considered to account fully for enzyme catalysis (9); on the other hand, hydrogen tunneling experiments suggest that a static model is inadequate and that fluctuations that reorganize the active site are required for the chemical step (7, 10, 11), which involves conformational sampling to facilitate hydrogen transfer as well as the transfer itself.
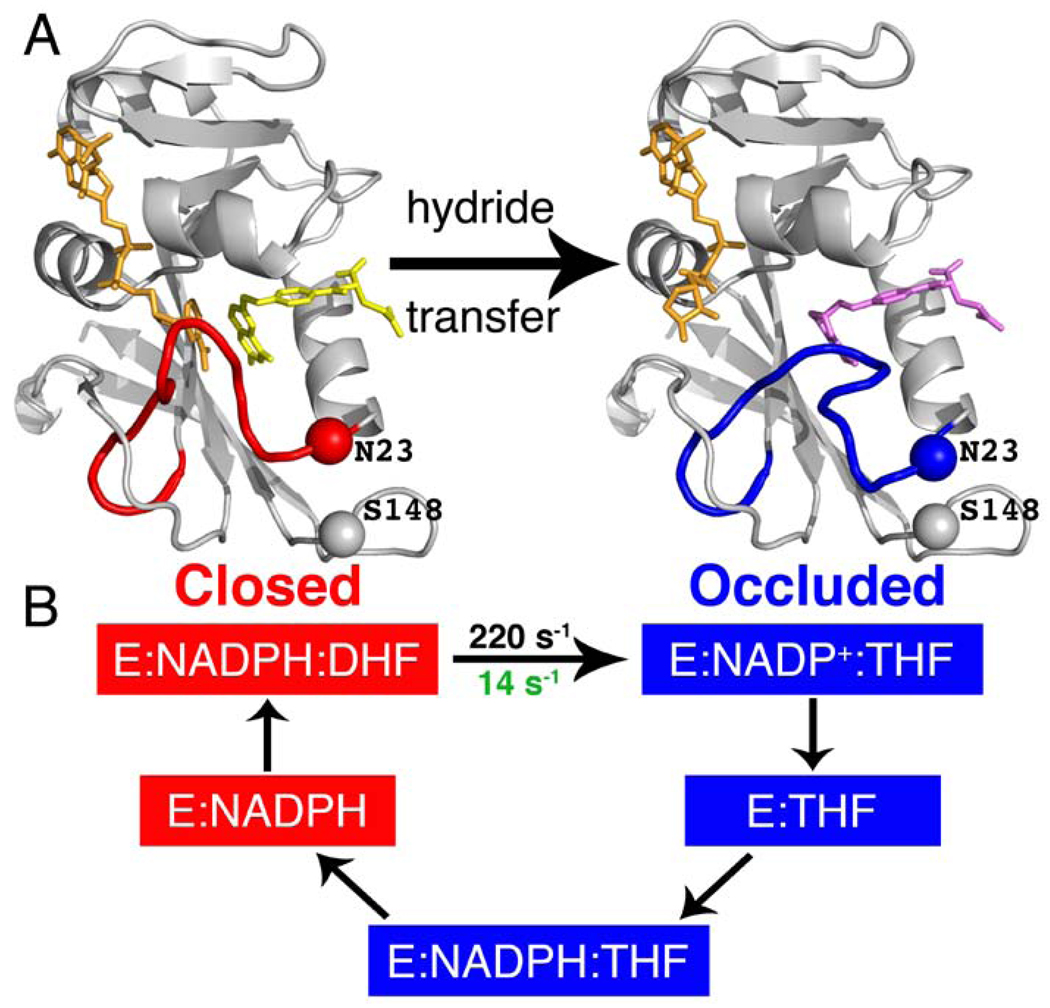
E. coli dihydrofolate reductase (ecDHFR) is a paradigm for understanding the relationship between structure, dynamics, and catalysis (4, 12–14). DHFR catalyzes the stereospecific reduction of dihydrofolate (DHF) to tetrahydrofolate (THF), using reduced nicotinamide adenine dinucleotide phosphate (NADPH) as cofactor. Five intermediates have been identified in the steady state catalytic cycle (7): E:NADPH, E:NADPH:DHF, E:NADP+:THF, E:THF, and E:NADPH:THF (Fig. 1). Large conformational changes are observed in the Met20 loop (residues 9–24) between the Michaelis complex E:NADPH:DHF (modeled by E:NADP+:folate (FOL) for structural studies) and the product complex, E:NADP+:THF (13). The Met20 loop adopts two dominant conformations: the ‘closed’ conformation, in which the loop packs tightly against the nicotinamide ring of the cofactor, and the ‘occluded’ conformation, in which the loop projects into the active site and sterically blocks the nicotinamide binding pocket. The holoenzyme (E:NADPH) and the model Michaelis complex (E:NADP+:FOL) are in the ‘closed’ conformation, while the three product complexes (E:NADP+:THF, E:THF, E:NADPH:THF) adopt the ‘occluded’ conformation (Fig. 1), which is stabilized by hydrogen bonds between Asn 23 and the backbone and side chain of Ser 148 (13, 15).
Fig. 1.
Conformational changes that occur during the E. coli DHFR catalytic cycle. (A) Left, cartoon representation of E:NADP+:FOL crystal structure (1RX2, model for the Michaelis complex, E:NADPH:DHF) in the closed conformation. Right, crystal structure of E:NADP+:ddTHF (1RX4, model for the product complex) in the occluded conformation. NADP+ is shown in orange; FOL is shown in yellow and ddTHF in magenta. Red, Met20 loop in the closed conformation; blue, Met20 loop in the occluded conformation. The sites of mutation, N23 and S148 are shown as spheres. (B) Intermediates in the wild type E. coli DHFR catalytic cycle. Intermediates shown in red are in the closed conformation, and those in blue are in the occluded conformation. Prior to hydride transfer the Met20 loop is in the closed conformation, where it packs tightly against the nicotinamide ring of NADP+. Following hydride transfer, the Met20 loop adopts the occluded conformation, in which the nicotinamide ring of NADP+ is sterically hindered from binding in the active site. NADP+ undergoes a concurrent conformational change in which the nicotinamide ring is expelled from the binding pocket, initiating NADP+ release from the ternary product complex. The rate of hydride transfer in the wild type and N23PP/S148A mutant enzyme is indicated in black and green, respectively. Note that the mutation alters the pathway utilized for product and NADP+ release, as shown in Fig. S2.
In crystal structures of human and other vertebrate DHFRs, the Met20 loop is invariably closed (16). Alignment of the human and E. coli DHFR protein sequences suggests that the occluded conformation is destabilized in the human enzyme due to an inability to form the stabilizing hydrogen bond made by the S148 side chain in ecDHFR (Fig. S1). In addition, a polyproline sequence (PWPP) at the end of the ‘Met20’ loop of human DHFR may render the loop less flexible than in the E. coli enzyme. We hypothesized that a “dynamic knockout” mutant designed to impair the dynamics of the Met20 loop in ecDHFR would give direct information on the role of active-site loop fluctuations in catalysis by the E. coli enzyme. To this end, the ecDHFR mutant N23PP/S148A (Fig. 1) was engineered and characterized using X-ray crystallography, NMR spectroscopy and kinetic analysis (17).
To assess the effect of the mutations on ecDHFR activity, we carried out both steady state and pre-steady state kinetic measurements on N23PP/S148A DHFR. The overall turnover rate (kcat) is significantly lower than for the wild type enzyme (Table 1). For wild type ecDHFR, kcat is determined by dissociation of the THF product from the E:NADPH:THF complex (Fig.1) (10), whereas for the mutant enzyme, the magnitude of kcat is determined by dissociation of the NADP+ cofactor (Fig. S2). Thus, the reduced flexibility inherent in the design of N23PP/S148A ecDHFR imposes a change in the kinetic step that determines the steady state kcat. The hydride transfer step was monitored directly in pre-steady state kinetic experiments as described in supporting online material (Fig. S3). Remarkably, the rate of hydride transfer at pH 7.0 is 14 s−1 for the N23PP/S148A DHFR mutant, which is significantly slower than for wild-type ecDHFR (Table 1) (18). The measured rate for the chemical step has a primary kinetic isotope effect (KIE) of 3.0 (the same as the wild-type enzyme), consistent with the difference in zero point energies between the two hydrogen isotopes and confirming that we are, indeed, directly observing the rate of hydride transfer.
Table 1.
Kinetic parameters
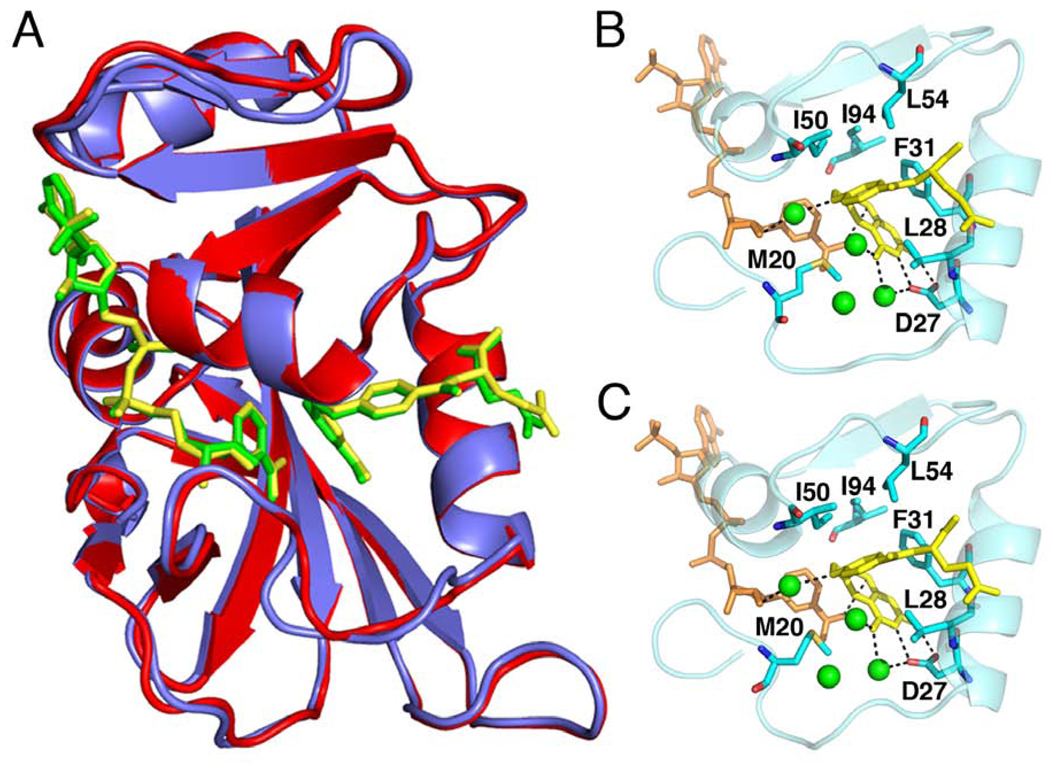
Structural differences between N23PP/S148A ecDHFR and wild-type ecDHFR were assessed by determining the crystal structure of the mutant enzyme in complex with NADP+ and folate and comparing it to the wild-type E:NADP+:FOL complex (13). To eliminate differences in refinement methods, the wild-type E:NADP+:FOL complex (13) was re-refined using the structure factors deposited in the PDB (code:1RX2), as described in supporting online material. The re-refined structure shows only very minor differences from the original 1RX2 coordinates. The N23PP/S148A ecDHFR at 1.6 Å resolution is almost identical to 1RX2-re-refined at 1.8 Å (13) (Fig. 2). Some minor adjustments at the end of the Met20 loop are observed, due to the proline insertion, but the loop is clearly in the closed conformation as in the wild-type E:NADP+:FOL complex. The ligand (Fig. S4) and active-site side chain conformations are maintained (Fig. 2), as are the hydrogen bonds (Table S2) and the locations of water molecules in the active site. The distance between hydride donor and acceptor atoms is slightly decreased (from 3.3 Å to 2.9 Å) in the N23PP/S148A mutant. This decrease cannot account for the impaired hydride transfer rate in the mutant, as a shorter distance would be expected to facilitate hydride transfer, not inhibit it. The similarity of the backbone conformations of the wild-type and N23PP/S148A E:NADP+:FOL complexes in solution is also illustrated by the near identity of 15N and 1H chemical shifts (Fig. S5).
Fig. 2.
The three-dimensional structures of the E:NADP+:FOL complexes of the N23PP/S148A mutant and wild-type E. coli DHFR are almost identical. (A) Superposition of the crystal structures of wild-type E. coli DHFR (1RX2) and N23PP/S148A E. coli DHFR. Wild-type E. coli DHFR is shown in red, with yellow ligands and N23PP/S148A E. coli DHFR is shown in purple, with green ligands. (B) Active site configuration for wild-type ecDHFR (1RX2-re-refined). Folate is colored yellow, NADP+ orange, the waters are shown as green spheres, and key active-site residues are in blue sticks. (C) Active site configuration for N23PP/S148A ecDHFR. Colors are the same as for B. For clarity, only the major conformation of the glutamate moiety of folate is shown. The active site configurations are almost identical for wild type and N23PP/S148A ecDHFR, including placement of polar residues, key hydrophobic residues and waters, showing that the electrostatic nature of the active site is unchanged by the mutations.
The conservation of side chain conformations and water molecules in the active site of the wild type and mutant enzymes argues for a very similar electrostatic environment. However, a slight change in the Met20 side chain required further investigation. It has been proposed that the Met20 Sδ atom might assist catalysis by increasing the pKa of the substrate, thereby promoting protonation of the N5 atom of the pterin (19). The Met20 Sδ atom in N23PP/S148A E:NADP+:FOL is shifted ~0.3 Å relative to its position in the wild-type complex (20) (Fig. S6). We measured the pH-dependence of hydride transfer in the N23PP/S148A mutant to ensure that the apparent small difference in the position of the Met20 Sδ atom does not impair hydride transfer by decreasing the pKa for N5. The hydride transfer rate for N23PP/S148A ecDHFR decreases with increasing pH (21) with a pKa of 6.7 ± 0.1, which is the same, within experimental error, as the value for the wild-type enzyme (6.5 ± 0.1). If anything, the pKa is slightly higher for the mutant enzyme, which would facilitate hydride transfer rather than inhibit it. Thus, the X-ray and NMR data and the pKa measurements show that there are no significant differences in the structure or electrostatic environment of the active site between the E:NADP+:FOL complexes of wild-type and N23PP/S148A ecDHFR.
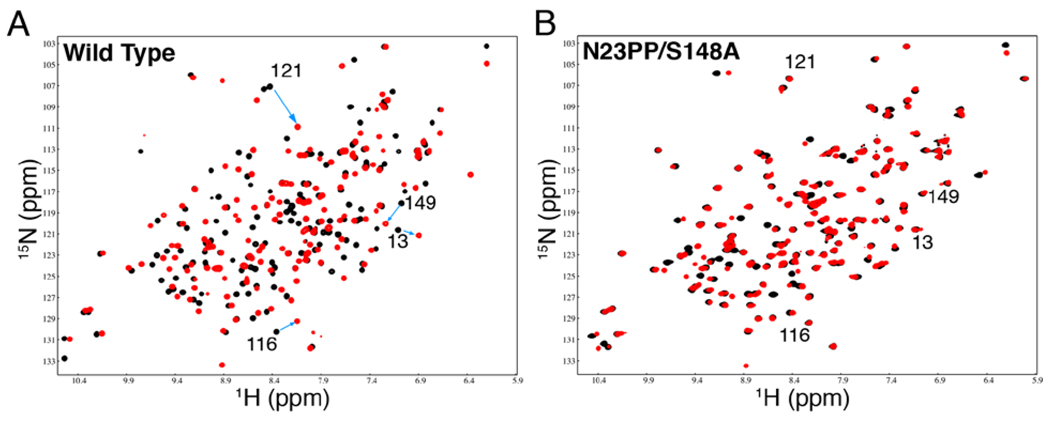
In wild-type ecDHFR, the Met20 loop adopts the closed ground state conformation in the Michaelis model complex, E:NADP+:FOL, and the occluded ground state conformation in the product ternary complex, E:NADP+:THF (13, 15, 22). Previous NMR studies identified marker resonances, mostly in the Met20 and FG loops, for which 15N and 1H chemical shifts differ significantly between the closed and occluded conformations (22). Unlike the wild-type enzyme, the E:NADP+:FOL and E:NADP+:THF complexes of the N23PP/S148A mutant enzyme have almost identical 1H and 15N chemical shifts for most residues, including those in the Met20, FG, and GH loops (Fig. 3 and Fig. S7), showing that the mutant enzyme remains in the closed conformation across the chemical step. The occluded conformation itself is not relevant to hydride transfer, which takes place within an ensemble of states in which the active site loops are closed (13). However, the fact that the mutant enzyme is unable to adopt an occluded conformation suggested that the flexibility of the active site loops may be significantly dampened and prompted us to examine the dynamics of the N23PP/S148A Michaelis model complex (E:NADP+:FOL)..
Fig. 3.
The Met20 loop of N23PP/S148A E. coli DHFR remains in the closed position across the chemical step. (A) 1H-15N HSQC spectra of wild-type E:NADP+:FOL (model Michaelis complex, black) in the closed conformation and E:NADP+:THF (product complex, red) in the occluded conformation. Large chemical shift differences are observed, particularly for residues in regions that undergo the closed-to-occluded structural change. Chemical shift changes from closed (black) to occluded (red) are indicated by arrows for several residues in the active site loops. (B) 1H-15N HSQC spectra of N23PP/S148A E:NADP+:FOL (model Michaelis complex, black) and E:NADP+:THF (product complex, red). The 1H-15N HSQC spectra for the complexes of the mutant enzyme are similar; the cross peaks corresponding to the active site loops do not shift, and appear in the position corresponding to the closed conformation for both complexes. A quantitative chemical shift analysis is presented in Fig. S8.
Carr-Purcell-Meiboom-Gill (CPMG)-based R2 relaxation dispersion NMR experiments were used to monitor the sampling of higher energy, lowly populated conformational substates that are associated with millisecond timescale structural fluctuations (23, 24). Previous studies of wild-type ecDHFR revealed millisecond timescale motions in the active site and C-terminal region in all five intermediate complexes of the catalytic cycle (4, 25). Surprisingly, relaxation dispersion experiments on the E:NADP+:FOL complex of N23PP/S148A DHFR showed no dispersion for any active site residues (with the sole exception of Ala 9), indicating that fluctuations on the millisecond timescale are abrogated in the mutant enzyme complex (Fig. 4). Residues associated with the C-terminus, however, undergo fluctuations on the millisecond timescale similar to those in the wild-type enzyme, confirming that the mutations affect dynamics only in the active site. R1, R2 and heteronuclear NOE experiments show that the Met20 loop of N23PP/S148A in the E:NADP+:FOL complex is rather rigid on the picosecond to nanosecond timescale, as in the wild-type enzyme (26) (Fig. S8). Several residues exhibiting exchange contributions from millisecond timescale conformational fluctuations in the active site loops have elevated R2 values in wild-type ecDHFR, while the values for these residues remain close to average for the N23PP/S148A mutant, suggesting that the mutations have not simply moved the motions to a faster, microsecond timescale. No minor resonances, which might be expected if there was a small population (~3% or higher) of an alternative Met20 loop conformation in slow exchange with the closed ground state, are observed in the 15N-HSQC spectrum of N23PP/S148A E:NADP+:FOL. Thus, we conclude that the millisecond time scale fluctuations in active site of the wild-type enzyme have been abolished in the N23PP/S148A mutant.
Fig. 4.
Millisecond timescale dynamics in the active site loops of the E:NADP+:FOL complex are impaired by the N23PP/S148A mutation. (A) Representative 15N R2 relaxation dispersion curves for wild type (black), N23PP/S148A (red), N23PP (blue) and S148A (magenta) E:NADP+:FOL. Dispersion data were collected at pH 7.6 and at 301 K. Top, active site loop residues (11, 95, 121), for which the dispersion observed in wild type E:NADP+:FOL is not observed in either N23PP/S148A or N23PP E:NADP+:FOL. S148A retains dispersion for residue 11 and other active site residues, shown in Fig. S11. Bottom, C-terminal associated residues (129, 131, 133), for which dispersion is observed for the wild type and mutant proteins. Residues which display 15N relaxation dispersion in the E:NADP+:FOL complex are mapped onto the structure for wild type (B), N23PP/S148A (C) and S148A (D). 1RX2 coordinates are used for representations in B and D. Red spheres indicate active site-associated residues for which R2 dispersion is observed. Blue spheres indicate C-terminal-associated residues for which R2 dispersion is observed. NADP+ is shown as orange sticks, FOL as yellow sticks. For clarity, only the major conformation of the glutamate moiety of folate is shown.
To obtain further insights into the effects of the N23PP/S148A mutations, we generated two additional ecDHFR mutants, N23PP and S148A. The N23PP mutation is, in itself, sufficient to abrogate the active-site flexibility required for efficient catalysis. The hydride transfer rate for the N23PP mutant (14 ± 1 s−1) is the same as for N23PP/S148A, while NMR experiments show that the Met20 loop remains in the closed conformation across the hydride transfer step, and that the millisecond time scale fluctuations in the active site are quenched in the E:NADP+:FOL complex of N23PP DHFR (Table 1, Fig. 4, Fig.S9). An analysis of S148A ecDHFR reveals that the decreased hydride transfer rate does not simply arise from impairment of the closed to occluded transition in the mutant enzymes. S148A ecDHFR also remains in the closed ground state conformation across the hydride transfer step (Fig. S10), yet its hydride transfer rate (157 s−1) is decreased only slightly from that of the wild-type enzyme (27). R2 dispersion experiments on the S148A E:NADP+:FOL complex show that, while the millisecond timescale dynamics are dampened compared to wild-type, several active-site residues do retain flexibility, sampling higher energy conformational substates that do not correspond to the occluded state (Fig. 4 and Fig. S11). We conclude that, unlike the N23PP and N23PP/S148A mutants, the S148A mutant retains residual motions in the active site, which are not associated with the closed-occluded transition, but which likely play a role in promoting hydride transfer.
Considering that the E:NADP+:FOL complexes, which are models for the Michaelis complex, have virtually identical active site structures in both the N23PP/S148A mutant and wild-type enzyme, it is most unlikely that hydride transfer is impaired in the mutant because of differences in the structural and electrostatic environment of the active site. While the ground state structures are almost indistinguishable, the wild-type and mutant enzymes differ substantially in their ability to sample alternative ground state active site conformations and access higher energy conformational substates through millisecond time scale fluctuations. The ground state conformational change between the closed Michaelis complex and the occluded product complex in wild-type ecDHFR is not observed for any of the three mutants, since the active site loops are restrained in the closed conformation. The millisecond timescale motions in the active site that allow the wild-type enzyme to sample higher energy substates are eliminated in the N23PP/S148A and N32PP mutants, while the S148A mutant still retains some flexibility, and has a correspondingly higher rate of hydride transfer. We, therefore, conclude that the significantly decreased rate of hydride transfer in the N23PP/S148A and N32PP mutant enzymes does not result from changes in structural or electrostatic preorganization of the active site, but from impaired flexibility.
Molecular simulations consistently show that Met20 loop residues move closer to the cofactor on progressing from reactant to transition state. These residues are predicted to play an important role in electrostatic stabilization of the transition state and in promoting protonation of the N5 atom of the pterin moiety of the substrate (19, 28, 29). This movement is likely required to tightly close and compact the active site, to correctly orient and constrain the substrate and cofactor, and to promote hydride transfer by reducing the distance between the donor and acceptor atoms. Although the active site of the N23PP/S148A mutant is fully preorganized in the ground state, millisecond timescale fluctuations of the active site are restricted such that the enzyme cannot efficiently sample higher energy conformational substates that are conducive to formation of the transition state; impairment of conformational fluctuations in the active site leads to strong inhibition of the chemical step. Fine-tuning of conformational flexibility is likely a general phenomenon that can be harnessed in engineering of efficient protein catalysts.
Supplementary Material
References and Notes
- 1.Hammes-Schiffer S, Benkovic SJ. Annu. Rev. Biochem. 2006;75:519. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 2.Henzler-Wildman K, Kern D. Nature. 2007;450:964. doi: 10.1038/nature06522. [DOI] [PubMed] [Google Scholar]
- 3.Wolf-Watz M, et al. Nat Struct Mol Biol. 2004;11:945. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- 4.Boehr DD, McElheny D, Dyson HJ, Wright PE. Science. 2006;313:1638. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 5.Doucet N, Watt ED, Loria JP. Biochemistry. 2009;48:7160. doi: 10.1021/bi900830g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watt ED, Shimada H, Kovrigin EL, Loria JP. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11981. doi: 10.1073/pnas.0702551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagel ZD, Klinman JP. Nat Chem Biol. 2009;5:543. doi: 10.1038/nchembio.204. [DOI] [PubMed] [Google Scholar]
- 8.Kamerlin SC, Warshel A. Proteins. 2010;78:1339. doi: 10.1002/prot.22654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pisliakov AV, Cao J, Kamerlin SC, Warshel A. Proc. Natl. Acad. Sci. U. S. A. 2009;106:17359. doi: 10.1073/pnas.0909150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang ZX, Lee T, Resing KA, Ahn NG, Klinman JP. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9556. doi: 10.1073/pnas.0403337101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benkovic SJ, Hammes-Schiffer S. Science. 2003;301:1196. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 12.Fierke CA, Johnson KA, Benkovic SJ. Biochemistry. 1987;26:4085. doi: 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- 13.Sawaya MR, Kraut J. Biochemistry. 1997;36:586. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 14.Schnell JR, Dyson HJ, Wright PE. Annu. Rev. Biophys. Biomol. Struct. 2004;33:119. doi: 10.1146/annurev.biophys.33.110502.133613. [DOI] [PubMed] [Google Scholar]
- 15.Venkitakrishnan RP, et al. Biochemistry. 2004;43:16046. doi: 10.1021/bi048119y. [DOI] [PubMed] [Google Scholar]
- 16.Davies JF, 2nd, et al. Biochemistry. 1990;29:9467. doi: 10.1021/bi00492a021. [DOI] [PubMed] [Google Scholar]
- 17.Materials and methods are available as supporting material on Science Online
- 18.Cameron CE, Benkovic SJ. Biochemistry. 1997;36:15792. doi: 10.1021/bi9716231. [DOI] [PubMed] [Google Scholar]
- 19.Khavrutskii IV, Price DJ, Lee J, Brooks CL., 3rd Protein Sci. 2007;16:1087. doi: 10.1110/ps.062724307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.The terminal methyl of M20 is not clearly defined by the electron density in 1RX2 and in 1RX2-rerefined, and is likely flexible. In addition, density for water 47 is observed in 1RX2-rerefined, but was not built into 1RX2. A discussion on building M20 and water47 into 1RX2-rerefined is included in the Supporting Online Material.
- 21.At basic pHs, the hydride transfer rate (khyd) is decreased to 3.3 s-1 for N23PP/S148A due to the limited sources of protons for the hydride transfer step. The pKa values are derived from fitting the pH Vs. khyd profile as described in the supplementary material
- 22.Osborne MJ, Venkitakrishnan RP, Dyson HJ, Wright PE. Protein Sci. 2003;12:2230. doi: 10.1110/ps.03219603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palmer AG, 3rd, Kroenke CD, Loria JP. Methods Enzymol. 2001;339:204. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- 24.Mittermaier A, Kay LE. Science. 2006;312:224. doi: 10.1126/science.1124964. [DOI] [PubMed] [Google Scholar]
- 25.McElheny D, Schnell JR, Lansing JC, Dyson HJ, Wright PE. Proc. Natl. Acad. Sci. U. S. A. 2005;102:5032. doi: 10.1073/pnas.0500699102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Osborne MJ, Schnell J, Benkovic SJ, Dyson HJ, Wright PE. Biochemistry. 2001;40:9846. doi: 10.1021/bi010621k. [DOI] [PubMed] [Google Scholar]
- 27.Miller GP, Wahnon DC, Benkovic SJ. Biochemistry. 2001;40:867. doi: 10.1021/bi001608n. [DOI] [PubMed] [Google Scholar]
- 28.Agarwal PK, Billeter SR, Rajagopalan PT, Benkovic SJ, Hammes-Schiffer S. Proc. Natl. Acad. Sci. U. S. A. 2002;99:2794. doi: 10.1073/pnas.052005999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garcia-Viloca M, Truhlar DG, Gao J. Biochemistry. 2003;42:13558. doi: 10.1021/bi034824f. [DOI] [PubMed] [Google Scholar]
- 30.The authors acknowledge L. Tuttle for invaluable discussions, S. Bae for NMR data on the S148A mutant, H. Tien and D. Marciano of the Robotics Core at the Joint Center for Structural Genomics (JCSG) for automated crystal screening, and X. Dai for technical assistance with crystallographic data collection. This work was supported by the National Institutes of Health Grant GM75995 and the Skaggs Institute of Chemical Biology. The JCSG is supported by NIH National Institute of General Medical Sciences (NIGMS) (U54 GM094586). DCE is supported by a predoctoral fellowship from the Achievement Rewards for College Scientists Foundation, and Grant GM080209 from the NIH Molecular Evolution Training Program. The GM/CA CAT 23-ID-D has been funded in whole or in part with federal funds from National Cancer Institute (Y1-CO-1020) and NIGMS (Y1-GM-1104). Use of the Advanced Photon Source (APS) was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract no. DE-AC02-06CH11357. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIGMS or the NIH. Coordinates and structure factors of N23PP/S148A ecDHFR and 1RX2-re-refined are deposited in the Protein Data Bank (PDB code 3QL0 and 3QL3). Author contributions: GB and PEW designed the research. GB performed NMR experiments. GB and DCE performed crystallography experiments and structure refinement. IAW supervised structure refinement. JL and JG performed kinetic experiments and analysis. GB, PEW and SJB analysed and interpreted the data. GB, PEW and JL wrote the manuscript. IAW, HJD and SJB edited the manuscript.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.