Abstract
The use of assisted reproduction treatment, especially intracytoplasmic sperm injection (ICSI), is now linked to a range of adverse consequences, the aetiology of which remains largely undefined. The objective was to determine differences in gene expression of blastocysts generated by ICSI as well as ICSI with artificial oocyte activation (ICSI-A) versus the less manipulative IVF, providing fundamental genetic information that can be used to aid in the diagnosis or treatment of those adversely affected by assisted reproduction treatment, as well as stimulate research to further refine these techniques. Murine blastocysts were generated by ICSI, ICSI-A and IVF, and processed for a microarray-based analysis of gene expression. Ten blastocysts were pooled for each procedure and three independent replicates generated. The data were then processed to determine differential gene expression and to identify biological pathways affected by the procedures. In blastocysts derived by ICSI versus IVF, the expression of 197 genes differed (P < 0.01). In blastocysts derived by ICSI-A versus IVF and ICSI-A versus ICSI, the expression of 132 and 65 genes differed respectively (P < 0.01). Procedural-induced changes in genes regulating specific biological pathways revealed some consistency to known adverse consequences. Detailed investigation of procedure-specific dysfunction is therefore warranted.
Keywords: activation, blastocyst, gene expression, ICSI, IVF, mouse
Introduction
The use of assisted reproductive technology has increased dramatically over the last 30 years, providing an unprecedented opportunity for infertile couples to conceive a child. Unfortunately, the use of techniques such as intracytoplasmic sperm injection (ICSI) has become increasingly linked to adverse consequences that can affect both the mother and her child. In the mother, this includes an increased incidence of placental abruption, pre-eclampsia and stillbirths (Blumenfeld et al., 1992; Aytoz et al., 1998; Devroey and Van Steirteghem, 2004; Katalinic et al., 2004; Lucifero et al., 2004; Osmanagaoglu et al., 2004; Unger et al., 2004; Bonduelle et al., 2005; Lidegaard et al., 2005; Pinborg et al., 2005; Woldringh, 2005; Buckett et al., 2008; Arav et al., 2010; Poret et al., 2010). In children, dysfunction can range from the development of tumours and carcinomas (White et al., 1990; Toren et al., 1995; Odone-Filho et al., 2002; Moll et al., 2003a; Moll et al., 2003b; Katalinic et al., 2004; Lightfoot, 2004; Niemitz and Feinberg, 2004; Owen and Segars, 2009) to congenital anomalies such as septal heart defects and cleft lip or palate (Wennerholm et al., 2000; Anthony et al., 2002; Orstavik, 2003; Katalinic et al., 2004; Rimm et al., 2004; Bonduelle et al., 2005; Hansen et al., 2005; Kallen et al., 2005; Karpman et al., 2005; Olson et al., 2005; Schieve et al., 2005; Sutcliffe and Derom, 2006; Bertelsmann, 2008; Reefhuis et al., 2009; Poret et al., 2010; Williams et al., 2010), as well as neurological problems that may result in an intellectual lag (Katalinic et al., 2004; Bonduelle et al., 2005; Lidegaard et al., 2005). Unfortunately, the aetiology of these unwanted effects has proven difficult to determine, in part due to the patient-, clinic- and MD-specific practices that are associated with the use of these reproductive techniques.
The extent to which assisted reproduction treatment-induced defects can be traced back to an altered pattern of embryonic gene expression is unknown. However, aberrant gene expression within the blastocyst, the first differentiated stage of development that occurs after any manipulation of the gametes, could increase the incidence of, or predisposition to, the multitude of defects that have now been associated with the use of techniques such as intracytoplasmic sperm injection (ICSI). The objective of this study was to identify the genes and the biological pathways that they regulate, which differ in blastocysts generated by ICSI versus IVF. Because chemical activation of the oocyte is now included in some ICSI protocols, aiming to mimic the events within the oocyte that are induced by the penetrating spermatozoon, the experimental design was expanded to include ICSI with chemical activation (ICSI-A) as an independent procedure in itself. The hypothesis was that gene expression would differ in blastocysts derived by ICSI versus IVF and that these differences would be negated by the inclusion of the chemical activation procedure to the ICSI protocol. In effect, artificial activation of the oocyte would overcome some of the differences in gene expression induced by the ICSI procedure. Determining how these methodologies affect gene expression in the blastocyst should stimulate research that will translate to advances in the diagnosis, treatment and/or management of patients and offspring adversely affected by these technologies, as well in the refinement of these practices that benefit so many. To facilitate this, the data generated is provided in a manner as complete as possible, relying heavily upon the use of supplementary tables (available online only) that will allow the identification of specific gene targets for further investigation by others.
Materials and methods
Animals
All mice were purchased from Samtaco (IcrTacSam; Samtaco, Seoul, Korea), housed under a 12:12 h light/dark cycle in a temperature- and humidity-controlled room and provided with food and water ad libitum. The protocol for the use of these animals was approved by the Institutional Animal Care and Use Committee of CHA University, Seoul, Korea.
Sperm collection and preparation
Epididymal spermatozoa were obtained from male BDF1 mice at 8–10 weeks of age. Spermatozoa used for IVF were collected from the cauda epididymis in 200 μl drops of Quinn's Advantage Medium with Hepes (SAGE In-Vitro Fertilization; Pasadena, CA, USA) and capacitated by incubation for 1.5 h at 37°C under 5% CO2 in air. Spermatozoa used for ICSI and ICSI-A were squeezed from the cauda epididymis and placed in the bottom of 1.5-ml tubes containing 500 μl of the same Quinn's Advantage medium with Hepes. Spermatozoa were then allowed to ‘swim up’ for 5 min at room temperature before being collected for the intracytoplasmic injections.
Oocyte collection
Six-week old female B6D2F1 mice were treated with 5 IU pregnant mare's serum gonadotrophin (PMSG; Sigma-Aldrich, St Louis, MO, USA) to induce follicular development and 48 h later with 5 IU human chorionic gonadotrophin (Sigma-Aldrich) to induce ovulation. At 12 to 15 h after human chorionic gonadotrophin, ovulated cumulus–oocyte–complexes were retrieved from the ampullary region of each oviduct and placed in Quinn's Advantage Medium with Hepes. Cumulus–oocyte–complexes to be used for IVF were then transferred into fertilization drops of the same medium. Cumulus–oocyte–complexes to be used for ICSI and ICSI-A were first incubated in Quinn's medium with the addition of 0.1% bovine testicular hyaluronidase (Tokyo Kasei Kogyo, Tokyo, Japan), effectively dispersing cumulus cells. The cumulus-free oocytes were then washed in fresh Quinn's Advantage Medium with Hepes and used immediately for ICSI.
IVF
Cumulus-intact oocytes in 200 μl drops of Quinn's Advantage Fertilization medium were incubated with capacitated spermatozoa at a final concentration of 100 spermatozoa/μl medium. Gametes were co-incubated for 6 h at 37°C under 5% CO2 in air. After the 6 h co-incubation, oocytes were washed several times with fresh Quinn's Advantage Medium with Hepes. This protocol is based on the well-established methodology of Hogan et al. (1994).
Intracytoplasmic sperm injection
A small volume of the sperm suspension was mixed with Quinn's Advantage Medium with Hepes containing 12% polyvinyl pyrrolidone. Injections were performed immediately thereafter using a micromanipulator with a Piezo-electric actuator (PMM Controller, model PMAS-CT150; Prima Tech, Tsukuba, Japan). The head of each spermatozoa was separated from the tail by applying pulses to the head-tail junction by means of the Piezo-driven pipette. Only the head of one spermatozoon was injected into the cytoplasm of each metaphase II-stage oocyte and the oocytes were injected in groups of 10–15 (on a particular day), overall taking less than 10 min per group. A photographic time-course of the procedure is provided as Figure 1. Spermatozoa-injected oocytes were then transferred into KSOM medium and cultured at 37°C, 5% CO2 in air. To activate the oocytes after ICSI (ICSI-A treatment), injected oocytes were placed in Ca2+-free CZB medium containing 10 mmol/l SrCl2 at 60 min after the injection and cultured for 1 h. ICSI-A oocytes were then returned to culture in KSOM medium at 37°C, 5% CO2 in air. This protocol is based on the original work of Kimura and Yanagimachi (1995).
Figure 1.
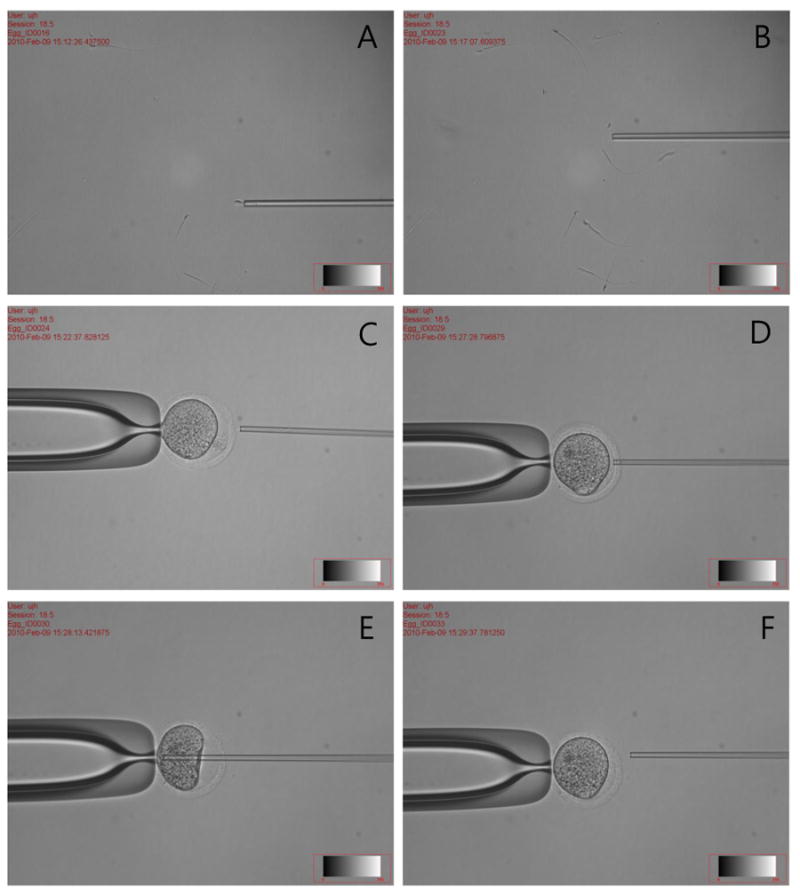
Representative images taken during intracytoplasmic sperm injection. (A) Loading of an epididymal spermatozoa. (B) Separation of head and tail. (C) The metaphase II-staged oocyte ready for injection. (D) Penetration of the zona pellucida. (E) Injection of the spermatozoa into the oocyte. (F) The injected oocyte.
Embryo culture
Following ICSI, ICSI-A and IVF, oocytes were placed in 50 μl drops of KSOM medium and incubated at 37°C, 5% CO2 in air. The culture drops were contained in plastic culture dishes and overlaid with mineral oil. Cultured embryos were then evaluated for developmental progress after 24 and 96 h. Embryos were generated by the hands of a single experienced embryologist, with the rates of fertilization and development shown in Table 1. A comparison of cell numbers (total, inner cell mass and trophectoderm) for blastocysts generated by ICSI, ICSI-A and IVF is presented as Table 2.
Table 1.
Development of embryos obtained by IVF, intracytoplasmic sperm injection (ICSI) and ICSI with artificial oocyte activation (ICSI-A).
| Technique | Fertilized oocytes cultured | 2-cell embryos | Blastocysts |
|---|---|---|---|
| IVF | 108 | 101 (93.52) | 79 (78.22) |
| ICSI | 130 | 111 (85.38) | 57 (51.35) |
| ICSI-A | 80 | 74 (92.50) | 40 (54.05) |
Data are the sum results from four independent experiments. Values are n or n (%).
Table 2.
Comparison of cell numbers in blastocysts derived by IVF, intracytoplasmic sperm injection (ICSI) and ICSI with chemical activation (ICSI-A).
| Technique (no. of blastocysts) | Mean cells/blastocyst (n) | Cells in trophectoderm | Cells in inner cell mass | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| IVF (n = 10) | 39.7 | 25.9 ± 1.2a | 65.3 ± 2.1b | 13.8 ± 1.0b | 34.7 ± 2.1b |
| ICSI (n = 10) | 39.4 | 29.2 ± 1.4a | 74.2 ± 1.7a | 10.2 ± 0.9a | 25.8 ± 1.7a |
| ICSI-A (n = 9) | 38.7 | 28.3 ± 3.5a | 72.6 ± 2.7a | 10.3 ± 1.1a | 28.0 ± 2.7a |
Values are mean ± SD.
Values in columns with different superscripts differ (P < 0.05).
Sample processing and microarray hybridization
Embryos that developed to the blastocyst stage in vitro were collected for the microarray analysis, chosen because at that stage of early development, blastocysts have differentiated to have an inner cell mass and trophectoderm and have a high chance of continuing in their development. Blastocysts were not collected from naturally bred mice in vivo as embryo-oviductal interactions that affect embryonic gene expression would confound the results. Ten blastocysts were pooled for each treatment group (ICSI, ICSI-A and IVF) and three independent replicates for each treatment procedure were collected. Pooling was required in order to obtain sufficient RNA for the analysis and also to reduce the overall chance of generating errors. Total RNA was extracted using Trizol and purified using an RNeasy kit (Qiagen, Valencia, CA, USA). Total RNA was then amplified and labelled using the Nugen WT Ovation One-Direct RNA Amplification and Nugen FL-Ovation cDNA Biotin Module V2 labelling kits, respectively (NuGen Technologies, San Carlos, CA, USA). Microarray hybridization was then performed using Affymetrix Mouse 430–2.0 whole genome arrays (Affymetrix, Santa Clara, CA, USA) by the Microarray Core Facility at the University of Kentucky, as previously described (Jo et al., 2004; Jeoung et al., 2010). Three independent microarray chips were hybridized for each treatment procedure.
Data analysis
Data from the microarray hybridization were sorted for each analysis, excluding probe sets that consistently exhibited absent or marginal detection calls (i.e. at least two of the three detection calls for both treatment groups in a particular analysis were absent or marginal). The resultant datasets were then processed using Pathways Studio 7.1 software (Ariadne Genomics, Rockville, MD, USA) to: (i) identify probe sets that differed among two treatment groups; and (ii) sort the data into biological pathways regulated by treatment. We have successfully used this software to analyse microarray datasets in the past (Jeoung et al., 2010). For the analysis to identify probe sets that differed among two treatment groups, differential expression was defined as those transcripts that exhibited a difference in expression of 2-fold or greater and were statistically different (P < 0.01), as determined by the software package that utilizes a statistical algorithm with a one-sided Mann–Whitney U-test to determine P-values indicating significance. To sort the data into biological processes regulated by treatment, the statistical algorithm was relaxed (P < 0.05) and a Gene Set Enrichment performed by the software package. The comparisons performed were ICSI versus IVF, ICSI-A versus IVF and ICSI-A versus ICSI, with only biological processes that exhibited at least five overlapping entities (genes) presented in the results.
Results
Gene expression I: ICSI versus IVF
Analysis of the microarray dataset identified 236 probes reflecting 197 known genes that differed in blastocysts generated by ICSI versus IVF (P < 0.01). For each of these genes, its probe set and Entrez gene identification number, fold-change in level of expression and P-value comparing the two treatment techniques is listed in Supplementary Table 1 (available online only). The dataset was also processed to reveal biological pathways that differed when blastocysts were generated by these two techniques (Table 3). Consistent with some of the developmental anomalies that have been associated with the use of assisted reproduction treatment (Wennerholm et al., 2000; Anthony et al., 2002; Orstavik, 2003; Katalinic et al., 2004; Rimm et al., 2004; Bonduelle et al., 2005; Hansen et al., 2005; Kallen et al., 2005; Karpman et al., 2005; Olson et al., 2005; Schieve et al., 2005; Sutcliffe and Derom, 2006; Bertelsmann, 2008; Reefhuis et al., 2009; Poret et al., 2010; Williams et al., 2010), the analysis identified development as a primary biological process affected by technique (ICSI versus IVF). Inclusive to this were several genes regulating structural and organ-specific processes, as well as those that were classified into more general cellular categories. Several metabolic pathways were identified to differ by procedure as well as a variety of response processes, signalling and transport mechanisms. As the purpose of this study is to provide the background genetic information required to stimulate further research, the results are presented without overt explanation or likely bias to any one physiological mechanism. The complete list of individual genes that were grouped into each biological function is also presented as an expanded version of Table 3 (Supplementary Table 2, available online only).
Table 3.
Functional classification of genes that differed in blastocysts derived by intracytoplasmic sperm injection (ICSI) versus IVF (P < 0.05).
| Type | Name | Entities | P-value | |
|---|---|---|---|---|
| Development | Cellular | Multicellular organismal development | 550 | 0.00062 |
| Cell differentiation | 332 | 0.00160 | ||
| Ureteric bud branching | 17 | 0.00264 | ||
| Cell development | 11 | 0.00627 | ||
| Metanephros development | 10 | 0.00767 | ||
| Morphogenesis of a branching structure | 5 | 0.00911 | ||
| Growth | Growth | 18 | 0.00091 | |
| Neural | Nervous system development | 219 | 0.00419 | |
| Organ | Organ morphogenesis | 95 | 0.00008 | |
| Pancreas development | 12 | 0.00142 | ||
| Sex differentiation | 11 | 0.00427 | ||
| Endocrine pancreas development | 9 | 0.00898 | ||
| Inner ear morphogenesis | 30 | 0.00905 | ||
| Forebrain development | 44 | 0.00979 | ||
| Sex determination | 9 | 0.00993 | ||
| Structural | Anatomical structure development | 9 | 0.00047 | |
| Embryonic gut development | 7 | 0.00061 | ||
| Proximal–distal pattern formation | 14 | 0.00119 | ||
| Other | Pattern specification process | 27 | 0.00294 | |
| Metabolism | Lipid catabolic process | 57 | 0.00018 | |
| Xenobiotic metabolic process | 14 | 0.00059 | ||
| Lipid glycosylation | 6 | 0.00155 | ||
| Cholesterol metabolic process | 42 | 0.00581 | ||
| Taurine metabolic process | 5 | 0.00958 | ||
| Response | Cellular | Negative regulation of chondrocyte differentiation | 5 | 0.00442 |
| Positive regulation of ossification | 10 | 0.00781 | ||
| Negative regulation of erythrocyte differentiation | 5 | 0.00854 | ||
| Positive regulation of cell proliferation | 209 | 0.00065 | ||
| Negative regulation of angiogenesis | 17 | 0.00089 | ||
| Positive regulation of vasodilation | 10 | 0.00223 | ||
| Chemotaxis | 57 | 0.00309 | ||
| Proteoglycan biosynthetic process | 6 | 0.00315 | ||
| Positive regulation of survival gene product expression | 7 | 0.00521 | ||
| Neurotransmitter secretion | 21 | 0.00580 | ||
| Positive regulation of mitosis | 11 | 0.00942 | ||
| Response to glucocorticoid stimulus | 58 | 0.01000 | ||
| Immune | Inflammatory response | 111 | 0.00060 | |
| Immune response | 202 | 0.00263 | ||
| Response to wounding | 31 | 0.00630 | ||
| Positive regulation of T cell-mediated cytotoxicity | 8 | 0.00637 | ||
| Mechanical | Response to mechanical stimulus | 30 | 0.00023 | |
| Sperm motility | 12 | 0.00102 | ||
| Ciliary or flagellar motility | 5 | 0.00812 | ||
| Positive regulation of smooth muscle contraction | 8 | 0.00868 | ||
| Other | Visual perception | 93 | 0.00095 | |
| Response to stimulus | 80 | 0.00139 | ||
| Digestion | 18 | 0.00204 | ||
| Synaptic transmission | 93 | 0.00014 | ||
| Response to external stimulus | 5 | 0.00568 | ||
| Signalling | G-protein coupled receptor protein signalling pathway | 210 | 0.00000 | |
| Signal transduction | 789 | 0.00022 | ||
| Cell–cell signalling | 104 | 0.00069 | ||
| Inositol phosphate-mediated signalling | 5 | 0.00399 | ||
| Wnt receptor signalling pathway, calcium modulating pathway | 5 | 0.00455 | ||
| Cytokine-mediated signalling pathway | 41 | 0.00590 | ||
| Elevation of cytosolic calcium ion concentration | 38 | 0.00867 | ||
| Transport | Iron ion transport | 22 | 0.00460 | |
| Ion transport | 281 | 0.00902 | ||
| Miscellaneous | Feeding behaviour | 11 | 0.00016 |
Gene expression II: ICSI-A versus IVF
A total of 146 probe sets representing 132 known genes were found to differ in blastocysts generated by ICSI-A versus IVF (P < 0.01). Again, the individual probes and associated information are listed in full (Supplementary Table 3, available online only). The biological process classification for differentially expressed genes in blastocysts derived by ICSI-A versus IVF (P < 0.05) is presented in Table 4 and expanded to include gene listings in Supplementary Table 4 (available online only). Interestingly, while differences in developmental processes were still found to differ in blastocysts generated by ICSI-A versus IVF, the number of classes identified was approximately halved. A large number of response-type pathways were identified that ranged from the specific (e.g. the regulation of ossification) to the general (e.g. immune response) that should be evaluated in the future in concert with the specific probes listed in the supplementary tables.
Table 4.
Functional classification of genes that differed in blastocysts derived by ICSI with chemical activation (ICSI-A) versus IVF (P < 0.05). The number of entities and level of statistical significance are indicated for each biological function.
| Type | Name | Entities | P-value | |
|---|---|---|---|---|
| Development | Cellular | Ureteric bud branching | 16 | 0.00469 |
| Neural | Peripheral nervous system development | 14 | 0.00398 | |
| Neural crest cell development | 5 | 0.00798 | ||
| Organ | Organ morphogenesis | 98 | 0.00007 | |
| Thyroid gland development | 5 | 0.00147 | ||
| Inner ear morphogenesis | 29 | 0.00197 | ||
| Middle ear morphogenesis | 6 | 0.00324 | ||
| Embryonic gut development | 5 | 0.00792 | ||
| Pattern specification process | 28 | 0.00890 | ||
| Structural | Cartilage development | 28 | 0.00659 | |
| Embryonic skeletal system morphogenesis | 26 | 0.00720 | ||
| Metabolism | Lipid glycosylation | 7 | 0.00147 | |
| Retinol metabolic process | 10 | 0.00297 | ||
| Response | Cellular | Glial cell differentiation | 10 | 0.00072 |
| Elevation of cytosolic calcium ion concentration | 40 | 0.00005 | ||
| Cell adhesion | 265 | 0.00036 | ||
| Chemotaxis | 59 | 0.00217 | ||
| Regulation of ossification | 7 | 0.00378 | ||
| Positive regulation of cellular protein metabolic process | 5 | 0.00605 | ||
| Positive regulation of mitosis | 12 | 0.00678 | ||
| NAD biosynthetic process | 6 | 0.00710 | ||
| Cell–cell adhesion | 52 | 0.00812 | ||
| Cellular calcium ion homeostasis | 36 | 0.00877 | ||
| Cholesterol biosynthetic process | 26 | 0.00889 | ||
| Protein amino acid N-linked glycosylation | 18 | 0.00963 | ||
| Immune | Inflammatory response | 112 | 0.00000 | |
| Immune response | 203 | 0.00002 | ||
| Defence response to bacterium | 26 | 0.00055 | ||
| Antigen processing and presentation | 18 | 0.00128 | ||
| Complement activation, alternative pathway | 5 | 0.00259 | ||
| Complement activation, classical pathway | 15 | 0.00723 | ||
| Mechanical | Response to mechanical stimulus | 33 | 0.00154 | |
| Vasoconstriction | 5 | 0.00188 | ||
| Neural | Axon guidance | 59 | 0.00678 | |
| Synaptic transmission | 98 | 0.00005 | ||
| Other | Detection of chemical stimulus involved in sensory perception of smell | 5 | 0.00647 | |
| Visual perception | 93 | 0.00001 | ||
| Response to stimulus | 84 | 0.00006 | ||
| Digestion | 17 | 0.00629 | ||
| Acute-phase response | 15 | 0.00840 | ||
| Signalling | G-protein coupled receptor protein signalling pathway | 216 | 0.00000 | |
| Signal transduction | 805 | 0.00008 | ||
| Cell surface receptor linked signal transduction | 115 | 0.00014 | ||
| G-protein signalling, coupled to cyclic nucleotide second messenger | 14 | 0.00026 | ||
| Cell–cell signalling | 103 | 0.00115 | ||
| Inositol phosphate-mediated signalling | 5 | 0.00651 | ||
| G-protein signalling, coupled to cAMP nucleotide second messenger | 10 | 0.00783 | ||
| Transport | Ion transport | 286 | 0.00006 | |
| Calcium ion transport | 68 | 0.00078 | ||
| Iron ion transport | 22 | 0.00197 | ||
| Miscellaneous | Feeding behaviour | 11 | 0.00024 | |
| Axonal fasciculation | 7 | 0.00188 | ||
| Memory | 21 | 0.00749 |
Gene expression III: ICSI or ICSI-A versus IVF
From the 197 genes that were found to differ when blastocysts were derived by ICSI versus IVF, only 18 probe sets representing 17 genes were common to those identified when ICSI-A and IVF were compared. These 17 genes, their average intensities, fold changes and individual P-values are identified in Table 5.
Table 5.
Identification of the 17 genes that differed when blastocysts were derived by IVF versus intracytoplasmic sperm injection (ICSI) and IVF versus ICSI with chemical activation (ICSI-A).
| Name | IVF versus ICSI | IVF versus ICSI-A | Probe set ID | Intensity average | ||||
|---|---|---|---|---|---|---|---|---|
| Fold change | P-value | Fold change | P-value | IVF | ICSI | ICSI-A | ||
| Unknown | −3.5516 | 0.0020 | −2.334 | 0.0035 | 1439599_at | 598.6 | 172.9 | 259.6 |
| ABHD14B | −2.2566 | 0.0087 | −2.0973 | 0.0055 | 1451326_at | 1313.4 | 582.5 | 619.9 |
| C10orf32 | 2.3728 | 0.0013 | 2.4465 | 0.0013 | 1419299_at | 137.2 | 327.6 | 337.9 |
| CA12 | −6.8079 | 0.0026 | −3.9913 | 0.0059 | 1428485_at | 186.6 | 29.0 | 48.9 |
| CUBN | −2.1933 | 0.0041 | −2.0282 | 0.0018 | 1452270_s_at | 1921.2 | 886.1 | 949.9 |
| DDA1 | −12.5618 | 0.0100 | −12.0678 | 0.0019 | 1429039_s_at | 239.0 | 22.1 | 19.6 |
| EPAS1 | 3.2595 | 0.0005 | 2.5005 | 0.0011 | 1449888_at | 173.2 | 567.6 | 434.7 |
| FNTB | 4.7982 | 0.0061 | 4.1234 | 0.0057 | 1459043_at | 82.3 | 381.3 | 321.4 |
| GRB10 | −5.5633 | 0.0004 | −4.8841 | 0.0005 | 1425457_a_at | 755.8 | 135.6 | 154.3 |
| GYLTL1B | −2.162 | 0.0065 | −2.2273 | 0.0027 | 1434007_at | 671.5 | 315.6 | 304.4 |
| HAS2 | −14.8595 | 0.0064 | −24.3663 | 0.0051 | 1449169_at | 272.3 | 20.7 | 13.0 |
| HMGA2 | 2.2213 | 0.0022 | 2.1215 | 0.0048 | 1450780_s_at | 1068.8 | 2346.3 | 2250.7 |
| IL6 | −3.569 | 0.0017 | −4.072 | 0.0054 | 1450297_at | 1257.0 | 355.5 | 321.3 |
| LARGE | −2.0839 | 0.0075 | −4.2598 | 0.0027 | 1417435_at | 1017.6 | 478.2 | 240.3 |
| NR3C1 | 2.8113 | 0.0007 | 2.1535 | 0.0007 | 1460303_at | 414.5 | 1174.6 | 895.2 |
| RAB17 | 2.6639 | 0.0005 | 2.1843 | 0.0010 | 1422178_a_at | 702.5 | 1868.9 | 1531.4 |
| Sco2 | −5.5218 | 0.0026 | −3.4909 | 0.0046 | 1432181_s_at | 450.7 | 81.4 | 126.5 |
| TBX20 | 3.6076 | 0.0000 | 3.5279 | 0.0004 | 1453351_at | 686.4 | 2477.4 | 2451.8 |
Gene expression IV: ICSI versus ICSI-A
The analysis identified 74 probe sets representing 65 known genes that differed between the ICSI and ICSI-A treatment groups (P < 0.01). Each of these 65 genes, its identity, fold-change in level of expression and P-value comparing the two treatment techniques is listed in Supplementary Table 5 (available online only). The method of ICSI (i.e. with or without artificial activation of the oocytes) used to generate blastocysts was also found to affect developmental, metabolic and response processes in the biological pathway analysis (P < 0.05; Table 6) with this information expanded to include gene information in Supplementary Table 6 (available online only).
Table 6.
Functional classification of genes that differed in blastocysts derived by intracytoplasmic sperm injection (ICSI) versus ICSI with chemical activation (ICSI-A) (P < 0.05).
| Type | Name | Entities | P-value | |
|---|---|---|---|---|
| Development | Cellular | Somitogenesis | 25 | 0.00686 |
| Neural | Peripheral nervous system development | 12 | 0.00474 | |
| Central nervous system development | 72 | 0.00824 | ||
| Hindbrain development | 28 | 0.00687 | ||
| Structural | Embryonic forelimb morphogenesis | 6 | 0.00478 | |
| Negative regulation of striated muscle development | 10 | 0.00266 | ||
| Anatomical structure development | 8 | 0.00717 | ||
| Metabolism | Insulin secretion | 15 | 0.00180 | |
| Fucose metabolic process | 6 | 0.00192 | ||
| Response | Cellular | Receptor clustering | 10 | 0.00151 |
| Fertilization | 19 | 0.00266 | ||
| Protein localization | 49 | 0.00039 | ||
| Synaptic vesicle endocytosis | 7 | 0.00847 | ||
| Vesicle organization | 7 | 0.00901 | ||
| Immune | Cytokine production | 15 | 0.00822 | |
| Neural | Synaptonemal complex assembly | 8 | 0.00598 |
The number of entities and level of statistical significance are indicated for each biological function.
Discussion
Recent data from the US Centres for Disease Control and Prevention indicate that ∼7.3 million US couples suffer from infertility and that ∼140,000 assisted reproduction cycles are performed per year (Wright et al., 2007; http://www.cdc.gov/ART/). The benefit from the development and use of these techniques is unquestionable; however assisted reproduction treatment and especially ICSI have become increasingly linked to a broad range of unwanted and often serious consequences to the mother and/or her offspring. The objective of this study was to determine, in the developing mouse embryo, genetic pathways affected by ICSI versus the less manipulative IVF. In addition to this, this study evaluated whether artificial activation of oocytes would overcome some of the differences in gene expression attributed specifically to the ICSI procedure. Mice were used to generate the embryos as they represent a population of fertile, genetically homogenous and healthy subjects. Determination of procedural-specific changes in embryonic gene expression should stimulate research that will translate into advances in clinical practice, both in the management of prior treatment-induced effects, as well as in the refinement of these techniques in the future. Immediate advances could include identifying an array of treatment-induced genes as genetic markers, signalling the need for planned intervention and management of certain diseases in the newborn, or increasing the breadth of preimplantation genetic diagnosis for the most serious consequences associated with these treatments for infertility. The first live births after IVF and ICSI were reported in 1978 (Steptoe and Edwards, 1978) and 1992 (Palermo et al., 1992), respectively. The field of assisted reproduction treatment has grown at an unprecedented pace, making this analysis of treatment-induced gene function a timely and necessary report.
The majority of treatment cycles performed in the USA utilize IVF and/or ICSI (>50% ICSI) with the other treatment types, gamete and zygote intra-Fallopian transfer (GIFT and ZIFT) accounting for <1% of all procedures (Wright et al., 2007; http://www.cdc.gov/ART/). However, the data from these reports also indicate that ICSI is proportionally increasing in its use and routinely prescribed over IVF, regardless of the aetiology of infertility; i.e. even when the cause of infertility would indicate success with IVF alone. This is extremely pertinent given the data presented herein; 236 probe sets accounting for 197 genes differed in blastocysts generated by ICSI versus IVF, suggesting procedural-driven changes in embryonic gene expression and, potentially, the unwanted consequences that have been associated with assisted reproduction treatment.
Of the birth defects related to the use of these techniques, the manifestation of aberrant growth and development appears a common consequence (Wennerholm et al., 2000; Anthony et al., 2002; Orstavik, 2003; Katalinic et al., 2004; Rimm et al., 2004; Bonduelle et al., 2005; Hansen et al., 2005; Kallen et al., 2005; Karpman et al., 2005; Olson et al., 2005; Schieve et al., 2005; Sutcliffe and Derom, 2006; Bertelsmann, 2008; Reefhuis et al., 2009; Poret et al., 2010; Williams et al., 2010). One of the most readily recognizable of these defects includes cleft lip and palate, condition(s) reflecting an asymmetry in development. A septal heart defect is less overtly recognizable, although a serious defect that is also related to symmetry and assisted reproduction treatment. Interestingly, this study identified the differential expression of 14 genes (ICSI versus IVF, Table 3 and Supplementary Table 2) that were categorized to regulate proximal–distal pattern formation (i.e. symmetry) in these early embryos. That being said, developmental problems associated with assisted reproduction treatment are not confined to those manifested from asymmetry. Several specific syndromes that include Beckwith–Wiedemann (Filippi and McKusick, 1970; Neelanjana and Sabaratnam, 2008) and Silver–Russell (Abu-Amero et al., 2008; Neelanjana and Sabaratnam, 2008), which are characterized by overgrowth and intrauterine growth retardation, respectively, are also well recognized (DeBaun et al., 2003; Gicquel et al., 2003; Gosden et al., 2003; Maher et al., 2003a; Maher et al., 2003b; Halliday et al., 2004; Katalinic et al., 2004; Lucifero et al., 2004; Niemitz et al., 2004; Niemitz and Feinberg, 2004; Lidegaard et al., 2005; Sutcliffe et al., 2006; Owen and Segars, 2009). In addition to this, aberrant development extends to the manifestation of neurological defects (Katalinic et al., 2004; Bonduelle et al., 2005; Lidegaard et al., 2005) and the present study identified procedural-induced changes in several genes regulating neural development when the expression profiles of blastocysts derived from IVF and ICSI were compared.
Similar to the wide scope of developmental disorders implicated with assisted reproduction treatment, a range of metabolic consequences have been described including neonatal hypoglycaemia (Beckwith–Wiedemann syndrome (Hussain et al., 2005; Kapoor et al., 2009; Palladino et al., 2009)) and childhood-onset obesity (Prader–Willi and MatUPD14 syndromes, (O'Neill et al., 2005; Bouchard, 2009; Butler et al., 2009)). Analysis of the present data indicated differences in the expression of genes regulating several metabolic pathways, including cholesterol and lipid metabolism/catabolism, in blastocysts derived from ICSI versus IVF. The concomitant change in genes regulating organ morphogenesis, as an example, pancreatic development, is consistent with a metabolic consequence to assisted reproduction treatment and suggests an underlying genetic cause for the development of certain metabolic diseases. Overall, the changes in gene expression that this study has identified between ICSI and IVF indicate that further investigation into these pathways is vital. Although the refinement of techniques to circumvent adverse consequences is obviously a primary goal for the future, the development of genetic markers indicating susceptibility to disease could have a dramatic impact on the diagnosis and management of several of the adverse consequences, including those of a metabolic origin, that are associated with assisted reproduction treatment.
A more recent modification to the procedure of ICSI is the inclusion of a chemical activation step (Dozortsev et al., 1995), reviewed by Nasr-Esfahani et al., (2010), which aims to mimic the events within the oocyte that occur during the normal fertilization process; i.e. under normal conditions, spermatozoon–oocyte fusion is followed by the incorporation of, and interaction between, a demembranated sperm nucleus and the cytoplasm of the oocyte. The nucleus of the spermatozoa is readily accessible to ooplasmic factors, with fusion of the gametes stimulating pivotal intracellular calcium oscillations. Modifications to the zona pellucida and release from meiotic arrest ensue. Artificial methods to activate the oocyte have now been developed, that improve the rate of fertilization (especially in cases of low fertility) and can be performed using chemical agents such as strontium chloride (Kumagai, 2006) and calcium ionophores (Murase et al., 2004; Borges et al., 2009) or by mechanical (Dirican et al., 2008) or electrical (Yanagida et al., 1999) stimulation. In the experiments described herein, strontium chloride was used. Although this technique has proven very effective (Suttner et al., 2000; Murase et al., 2004), the utilization of activation after ICSI (ICSI-A) appears to be clinic-specific with only a very limited number of clinics activating oocytes after microinjection.
It is very interesting that when compared with IVF, the inclusion of this chemical activation step to the ICSI protocol brought the sample clustering of the probe sets closer together. i.e. ICSI-A clustered closer to IVF than to ICSI alone, suggesting that this artificial activation step effectively mimics, at the genetic level, a proportion of the events initiated by sperm penetration. When gene expression in blastocysts derived by ICSI-A and IVF were analysed independently, 146 probe sets were identified to differ, 90 fewer than the 236 probes identified to differ in the ICSI versus IVF analysis. In itself, this is a striking reduction in differential gene expression and further illustrative of the alignment of IVF and ICSI-A observed in the clustering analysis. When gene expression in blastocysts derived by ICSI was compared directly to those generated with the optional activation step (ICSI-A), 74 probe sets reflecting the differential expression of 65 known genes was revealed. The classification of development was highlighted by the biological pathway grouping and differences in the expression of genes regulating structural and neural developmental pathways uncovered, consistent with known treatment-induced dysfunction.
Overall, the expression of genes in blastocysts was affected by the procedure used to generate the embryo and classification of differentially expressed genes into biological pathways revealed consistency to known treatment-induced adverse consequences. Obviously, further investigation is needed before these findings can be translated into clinical advances and the reader must remain cognizant that these results were generated using blastocysts of a murine origin, and not those obtained from an assisted reproduction clinic. However, with the use of ICSI now appearing to be dominant over IVF, regardless of the aetiology of infertility, and ICSI-A not yet standardized into the operating procedure of the majority of assisted reproduction clinics, deadlines for teasing out procedure-specific consequences cannot be delayed.
The field of assisted reproduction treatment has grown exponentially over the last 30 years, providing an unprecedented opportunity for infertile couples to conceive a child. This genetic analysis was performed to stimulate research in this field with the overall goal of advancing understanding (and performance) of these most valuable treatment options for infertility. It is hoped that future investigation into the genes and pathways uncovered in this report will prove fruitful and that advances made will be timely.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health (grant P20 RR15592, CK and PB; and K12 DA014040, PB) and the Priority Research Centres Program funded by the Ministry of Education, Science and Technology, Republic of Korea (grant 2009-0093821, DRL).
Footnotes
The authors report no financial or commercial conflicts of interest.
References
- Abu-Amero S, Monk D, Frost J, Preece M, Stanier P, Moore GE. The genetic aetiology of Silver-Russell syndrome. J Med Genet. 2008;45:193–199. doi: 10.1136/jmg.2007.053017. [DOI] [PubMed] [Google Scholar]
- Anthony S, Buitendijk SE, Dorrepaal CA, Lindner K, Braat DD, den Ouden AL. Congenital malformations in 4224 children conceived after IVF. Hum Reprod. 2002;17:2089–2095. doi: 10.1093/humrep/17.8.2089. [DOI] [PubMed] [Google Scholar]
- Arav A, Gavish Z, Elami A, Natan Y, Revel A, Silber S, Gosden RG, Patrizio P. Ovarian function 6 years after cryopreservation and transplantation of whole sheep ovaries. Reprod Biomed Online. 2010;20:48–52. doi: 10.1016/j.rbmo.2009.10.019. [DOI] [PubMed] [Google Scholar]
- Aytoz A, De Catte L, Camus M, Bonduelle M, Van Assche E, Liebaers I, Van Steirteghem A, Devroey P. Obstetric outcome after prenatal diagnosis in pregnancies obtained after intracytoplasmic sperm injection. Hum Reprod. 1998;13:2958–2961. doi: 10.1093/humrep/13.10.2958. [DOI] [PubMed] [Google Scholar]
- Bertelsmann H, de Carvalho G. The risk of malformation following assisted reproduction. Dtsch Arztebl Int. 2008;1:105. doi: 10.3238/arztebl.2008.0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenfeld Z, Dirnfeld M, Abramovici H, Amit A, Bronshtein M, Brandes JM. Spontaneous fetal reduction in multiple gestations assessed by transvaginal ultrasound. Br J Obstet Gynaecol. 1992;99:333–337. doi: 10.1111/j.1471-0528.1992.tb13734.x. [DOI] [PubMed] [Google Scholar]
- Bonduelle M, Wennerholm UB, Loft A, Tarlatzis BC, Peters C, Henriet S, Mau C, Victorin-Cederquist A, Van Steirteghem A, Balaska A, Emberson JR, Sutcliffe AG. A multi-centre cohort study of the physical health of 5-year-old children conceived after intracytoplasmic sperm injection, in vitro fertilization and natural conception. Hum Reprod. 2005;20:413–419. doi: 10.1093/humrep/deh592. [DOI] [PubMed] [Google Scholar]
- Borges E, Jr, de Almeida Ferreira Braga DP, de Sousa Bonetti TC, Iaconelli A, Jr, Franco JG., Jr Artificial oocyte activation with calcium ionophore A23187 in intracytoplasmic sperm injection cycles using surgically retrieved spermatozoa. Fertil Steril. 2009;92:131–136. doi: 10.1016/j.fertnstert.2008.04.046. [DOI] [PubMed] [Google Scholar]
- Bouchard C. Childhood obesity: are genetic differences involved? Am J Clin Nutr. 2009;89:1494S–1501S. doi: 10.3945/ajcn.2009.27113C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckett WM, Chian RC, Dean NL, Sylvestre C, Holzer HE, Tan SL. Pregnancy loss in pregnancies conceived after in vitro oocyte maturation, conventional in vitro fertilization, and intracytoplasmic sperm injection. Fertil Steril. 2008;90:546–550. doi: 10.1016/j.fertnstert.2007.06.107. [DOI] [PubMed] [Google Scholar]
- Butler JV, Whittington JE, Holland AJ, McAllister CJ, Goldstone AP. The transition between the phenotypes of Prader-Willi syndrome during infancy and early childhood. Dev Med Child Neurol. 2009 doi: 10.1111/j.1469-8749.2009.03530.x. [DOI] [PubMed] [Google Scholar]
- DeBaun MR, Niemitz EL, Feinberg AP. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am J Hum Genet. 2003;72:156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devroey P, Van Steirteghem A. A review of ten years experience of ICSI. Hum Reprod Update. 2004;10:19–28. doi: 10.1093/humupd/dmh004. [DOI] [PubMed] [Google Scholar]
- Dirican EK, Isik A, Vicdan K, Sozen E, Suludere Z. Clinical pregnancies and livebirths achieved by intracytoplasmic injection of round headed acrosomeless spermatozoa with and without oocyte activation in familial globozoospermia: case report. Asian J Androl. 2008;10:332–336. doi: 10.1111/j.1745-7262.2008.00248.x. [DOI] [PubMed] [Google Scholar]
- Dozortsev D, Rybouchkin A, De Sutter P, Qian C, Dhont M. Human oocyte activation following intracytoplasmic injection: the role of the sperm cell. Hum Reprod. 1995;10:403–407. doi: 10.1093/oxfordjournals.humrep.a135952. [DOI] [PubMed] [Google Scholar]
- Filippi G, McKusick VA. The Beckwith-Wiedmann syndrome. Medicine (Baltimore) 1970;49:279–298. doi: 10.1097/00005792-197007000-00002. [DOI] [PubMed] [Google Scholar]
- Gicquel C, Gaston V, Mandelbaum J, Siffroi JP, Flahault A, Le Bouc Y. In vitro fertilization may increase the risk of Beckwith-Wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am J Hum Genet. 2003;72:1338–1341. doi: 10.1086/374824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosden R, Trasler J, Lucifero D, Faddy M. Rare congenital disorders, imprinted genes, and assisted reproductive technology. Lancet. 2003;361:1975–1977. doi: 10.1016/S0140-6736(03)13592-1. [DOI] [PubMed] [Google Scholar]
- Halliday J, Oke K, Breheny S, Algar E, D JA. Beckwith-Wiedemann syndrome and IVF: a case-control study. Am J Hum Genet. 2004;75:526–528. doi: 10.1086/423902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Bower C, Milne E, de Klerk N, Kurinczuk JJ. Assisted reproductive technologies and the risk of birth defects--a systematic review. Hum Reprod. 2005;20:328–338. doi: 10.1093/humrep/deh593. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the mouse embryo: A laboratory manual. 2. Cold Spring Harbor Laboratory; 1994. pp. 146–150. [Google Scholar]
- Hussain K, Cosgrove KE, Shepherd RM, Luharia A, Smith VV, Kassem S, Gregory JW, Sivaprasadarao A, Christesen HT, Jacobsen BB, Brusgaard K, Glaser B, Maher EA, Lindley KJ, Hindmarsh P, Dattani M, Dunne MJ. Hyperinsulinemic hypoglycemia in Beckwith-Wiedemann syndrome due to defects in the function of pancreatic beta-cell adenosine triphosphate-sensitive potassium channels. J Clin Endocrinol Metab. 2005;90:4376–4382. doi: 10.1210/jc.2005-0158. [DOI] [PubMed] [Google Scholar]
- Jeoung M, Lee S, Hawng HK, Cheon YP, Jeong YK, Gye MC, Iglarz M, Ko C, Bridges PJ. Identification of a novel role for endothelins within the oviduct. Endocrinology. 2010;151:2858–2867. doi: 10.1210/en.2009-1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jo M, Gieske MC, Payne CE, Wheeler-Price SE, Gieske JB, Ignatius IV, Curry TE, Jr, Ko C. Development and application of a rat ovarian gene expression database. Endocrinology. 2004;145:5384–5396. doi: 10.1210/en.2004-0407. [DOI] [PubMed] [Google Scholar]
- Kallen B, Finnstrom O, Nygren KG, Olausson PO. In vitro fertilization (IVF) in Sweden: infant outcome after different IVF fertilization methods. Fertil Steril. 2005;84:611–617. doi: 10.1016/j.fertnstert.2005.02.038. [DOI] [PubMed] [Google Scholar]
- Kapoor RR, James C, Hussain K. Hyperinsulinism in developmental syndromes. Endocr Dev. 2009;14:95–113. doi: 10.1159/000207480. [DOI] [PubMed] [Google Scholar]
- Karpman E, Williams DH, Lipshultz LI. IVF and ICSI in male infertility: update on outcomes, risks, and costs. ScientificWorldJournal. 2005;5:922–932. doi: 10.1100/tsw.2005.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katalinic A, Rosch C, Ludwig M. Pregnancy course and outcome after intracytoplasmic sperm injection: a controlled, prospective cohort study. Fertil Steril. 2004;81:1604–1616. doi: 10.1016/j.fertnstert.2003.10.053. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Yanagimachi R. Intracytoplasmic sperm injection in the mouse. Biol Reprod. 1995;52:709–720. doi: 10.1095/biolreprod52.4.709. [DOI] [PubMed] [Google Scholar]
- Kumagai ST, A Three successful pregnancies and deliveries after oocyte. Fertil Steril. 2006;86:S156–S157. [Google Scholar]
- Lidegaard O, Pinborg A, Andersen AN. Imprinting diseases and IVF: Danish National IVF cohort study. Hum Reprod. 2005;20:950–954. doi: 10.1093/humrep/deh714. [DOI] [PubMed] [Google Scholar]
- Lightfoot T, Bunch K, Ansell P, Murphy M. Ovulation induction, assisted conception and childhood cancer. European Journal of Cancer. 2004;41:715–724. doi: 10.1016/j.ejca.2004.07.032. [DOI] [PubMed] [Google Scholar]
- Lucifero D, Mann MR, Bartolomei MS, Trasler JM. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum Mol Genet. 2004;13:839–849. doi: 10.1093/hmg/ddh104. [DOI] [PubMed] [Google Scholar]
- Maher ER, Afnan M, Barratt CL. Epigenetic risks related to assisted reproductive technologies: epigenetics, imprinting, ART and icebergs? Hum Reprod. 2003a;18:2508–2511. doi: 10.1093/humrep/deg486. [DOI] [PubMed] [Google Scholar]
- Maher ER, Brueton LA, Bowdin SC, Luharia A, Cooper W, Cole TR, Macdonald F, Sampson JR, Barratt CL, Reik W, Hawkins MM. Beckwith-Wiedemann syndrome and assisted reproduction technology (ART) J Med Genet. 2003b;40:62–64. doi: 10.1136/jmg.40.1.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moll AC, Imhof SM, Cruysberg JR, Schouten-van Meeteren AY, Boers M, van Leeuwen FE. Incidence of retinoblastoma in children born after in-vitro fertilisation. Lancet. 2003a;361:309–310. doi: 10.1016/S0140-6736(03)12332-X. [DOI] [PubMed] [Google Scholar]
- Moll AC, Imhof SM, Schouten-van Meeteren AY, van Leeuwen FE. In-vitro fertilisation and retinoblastoma. Lancet. 2003b;361:1392. doi: 10.1016/S0140-6736(03)13065-6. [DOI] [PubMed] [Google Scholar]
- Murase Y, Araki Y, Mizuno S, Kawaguchi C, Naito M, Yoshizawa M. Pregnancy following chemical activation of oocytes in a couple with repeated failure of fertilization using ICSI: case report. Hum Reprod. 2004;19:1604–1607. doi: 10.1093/humrep/deh294. [DOI] [PubMed] [Google Scholar]
- Nasr-Esfahani MH, Deemeh MR, Tavalaee M. Artificial oocyte activation and intracytoplasmic sperm injection. Fertil Steril. 2010;94:520–526. doi: 10.1016/j.fertnstert.2009.03.061. [DOI] [PubMed] [Google Scholar]
- Neelanjana M, Sabaratnam A. Malignant conditions in children born after assisted reproductive technology. Obstet Gynecol Surv. 2008;63:669–676. doi: 10.1097/OGX.0b013e318181a9f0. [DOI] [PubMed] [Google Scholar]
- Niemitz EL, DeBaun MR, Fallon J, Murakami K, Kugoh H, Oshimura M, Feinberg AP. Microdeletion of LIT1 in familial Beckwith-Wiedemann syndrome. Am J Hum Genet. 2004;75:844–849. doi: 10.1086/425343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemitz EL, Feinberg AP. Epigenetics and assisted reproductive technology: a call for investigation. Am J Hum Genet. 2004;74:599–609. doi: 10.1086/382897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill MA, Farooqi IS, Wevrick R. Evaluation of Prader-Willi Syndrome gene MAGEL2 in severe childhood-onset obesity. Obes Res. 2005;13:1841–1842. doi: 10.1038/oby.2005.224. [DOI] [PubMed] [Google Scholar]
- Odone-Filho V, Cristofani LM, Bonassa EA, Braga PE, Eluf-Neto J. In vitro fertilization and childhood cancer. J Pediatr Hematol Oncol. 2002;24:421–422. doi: 10.1097/00043426-200206000-00023. [DOI] [PubMed] [Google Scholar]
- Olson CK, Keppler-Noreuil KM, Romitti PA, Budelier WT, Ryan G, Sparks AE, Van Voorhis BJ. In vitro fertilization is associated with an increase in major birth defects. Fertil Steril. 2005;84:1308–1315. doi: 10.1016/j.fertnstert.2005.03.086. [DOI] [PubMed] [Google Scholar]
- Orstavik KH. Intracytoplasmic sperm injection and congenital syndromes because of imprinting defects. Tidsskr Nor Laegeforen. 2003;123:177. [PubMed] [Google Scholar]
- Osmanagaoglu K, Kolibianakis E, Tournaye H, Camus M, Van Steirteghem A, Devroey P. Cumulative live birth rates after transfer of cryopreserved ICSI embryos. Reprod Biomed Online. 2004;8:344–348. doi: 10.1016/s1472-6483(10)60915-7. [DOI] [PubMed] [Google Scholar]
- Owen CM, Segars JH., Jr Imprinting disorders and assisted reproductive technology. Semin Reprod Med. 2009;27:417–428. doi: 10.1055/s-0029-1237430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo G, Joris H, Devroey P, Van Steirteghem AC. Pregnancies after intracytoplasmic injection of single spermatozoon into an oocyte. Lancet. 1992;340:17–18. doi: 10.1016/0140-6736(92)92425-f. [DOI] [PubMed] [Google Scholar]
- Palladino AA, Bennett MJ, Stanley CA. Hyperinsulinism in infancy and childhood: when an insulin level is not always enough. Ann Biol Clin (Paris) 2009;67:245–254. doi: 10.1684/abc.2009.0330. [DOI] [PubMed] [Google Scholar]
- Pinborg A, Lidegaard O, la Cour Freiesleben N, Andersen AN. Consequences of vanishing twins in IVF/ICSI pregnancies. Hum Reprod. 2005;20:2821–2829. doi: 10.1093/humrep/dei142. [DOI] [PubMed] [Google Scholar]
- Poret H, Blanchard M, Lemseffer M, Royere D, Guerif F. Conjoined twins after intracytoplasmic sperm injection and transfer of a single day 2 embryo: case report. Fertil Steril. 2010;93:268, e267–269. doi: 10.1016/j.fertnstert.2009.08.054. [DOI] [PubMed] [Google Scholar]
- Reefhuis J, Honein MA, Schieve LA, Correa A, Hobbs CA, Rasmussen SA. Assisted reproductive technology and major structural birth defects in the United States. Hum Reprod. 2009;24:360–366. doi: 10.1093/humrep/den387. [DOI] [PubMed] [Google Scholar]
- Rimm AA, Katayama AC, Diaz M, Katayama KP. A meta-analysis of controlled studies comparing major malformation rates in IVF and ICSI infants with naturally conceived children. J Assist Reprod Genet. 2004;21:437–443. doi: 10.1007/s10815-004-8760-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieve LA, Rasmussen SA, Reefhuis J. Risk of birth defects among children conceived with assisted reproductive technology: providing an epidemiologic context to the data. Fertil Steril. 2005;84:1320–1324. doi: 10.1016/j.fertnstert.2005.04.066. discussion 1327. [DOI] [PubMed] [Google Scholar]
- Steptoe PC, Edwards RG. Birth after the reimplantation of a human embryo. Lancet. 1978;2:366. doi: 10.1016/s0140-6736(78)92957-4. [DOI] [PubMed] [Google Scholar]
- Sutcliffe AG, Derom C. Follow-up of twins: health, behaviour, speech, language outcomes and implications for parents. Early Hum Dev. 2006;82:379–386. doi: 10.1016/j.earlhumdev.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Sutcliffe AG, Peters CJ, Bowdin S, Temple K, Reardon W, Wilson L, Clayton-Smith J, Brueton LA, Bannister W, Maher ER. Assisted reproductive therapies and imprinting disorders--a preliminary British survey. Hum Reprod. 2006;21:1009–1011. doi: 10.1093/humrep/dei405. [DOI] [PubMed] [Google Scholar]
- Suttner R, Zakhartchenko V, Stojkovic P, Muller S, Alberio R, Medjugorac I, Brem G, Wolf E, Stojkovic M. Intracytoplasmic sperm injection in bovine: effects of oocyte activation, sperm pretreatment and injection technique. Theriogenology. 2000;54:935–948. doi: 10.1016/S0093-691X(00)00403-9. [DOI] [PubMed] [Google Scholar]
- Toren A, Sharon N, Mandel M, Neumann Y, Kenet G, Kaplinsky C, Dor J, Rechavi G. Two embryonal cancers after in vitro fertilization. Cancer. 1995;76:2372–2374. doi: 10.1002/1097-0142(19951201)76:11<2372::aid-cncr2820761128>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Unger S, Hoopmann M, Bald R, Foth D, Nawroth F. Monozygotic triplets and monozygotic twins after ICSI and transfer of two blastocysts: case report. Hum Reprod. 2004;19:110–113. doi: 10.1093/humrep/deh039. [DOI] [PubMed] [Google Scholar]
- Wennerholm UB, Bergh C, Hamberger L, Lundin K, Nilsson L, Wikland M, Kallen B. Incidence of congenital malformations in children born after ICSI. Hum Reprod. 2000;15:944–948. doi: 10.1093/humrep/15.4.944. [DOI] [PubMed] [Google Scholar]
- White L, Giri N, Vowels MR, Lancaster PA. Neuroectodermal tumours in children born after assisted conception. Lancet. 1990;336:1577. doi: 10.1016/0140-6736(90)93350-x. [DOI] [PubMed] [Google Scholar]
- Williams C, Sutcliffe A, Sebire NJ. Congenital malformations after assisted reproduction: risks and implications for prenatal diagnosis and fetal medicine. Ultrasound Obstet Gynecol. 2010;35:255–259. doi: 10.1002/uog.7589. [DOI] [PubMed] [Google Scholar]
- Woldringh G. Intracytoplamsmic sperm injection: a review of risks and complications. British Journal of Urology International. 2005;96:749–753. doi: 10.1111/j.1464-410X.2005.05708.x. [DOI] [PubMed] [Google Scholar]
- Wright VC, Chang J, Jeng G, Chen M, Macaluso M. Assisted reproductive technology surveillance–United States, 2004. MMWR Surveill Summ. 2007;56:1–22. [PubMed] [Google Scholar]
- Yanagida K, Katayose H, Yazawa H, Kimura Y, Sato A, Yanagimachi H, Yanagimachi R. Successful fertilization and pregnancy following ICSI and electrical oocyte activation. Hum Reprod. 1999;14:1307–1311. doi: 10.1093/humrep/14.5.1307. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


