Abstract
The retinoblastoma gene, Rb, was originally identified as the tumor suppressor gene mutated in a rare childhood cancer called retinoblastoma (reviewed in [1]. Subsequent studies showed that Rb functions in a pathway that is often functionally inactivated in a large majority of human cancers. Interestingly, recent studies showed that in certain types of cancers, Rb function is actually required for cancer development. The intimate link between the Rb pathway and cancer development suggests that the status of Rb activity can potentially be used to develop targeted therapy. However, a prerequisite will be to understand the role of Rb and its interaction with other signaling pathways in cancer development. In this review, we will discuss the roles of Rb in proliferation, apoptosis and differentiation by reviewing the recent findings in both mammalian systems and different model organisms. In addition, we will discuss strategies that can be employed that specifically target cancer cells based on the status of the Rb pathway.
Rb AND E2F FAMILY PROTEINS
Mammalian Rb Family Proteins
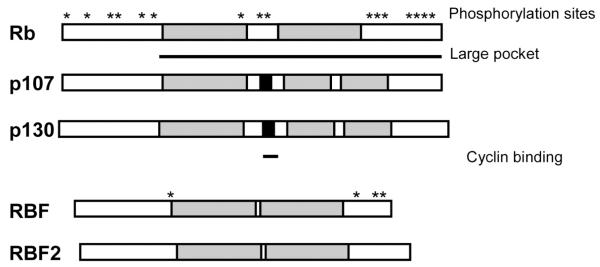
In addition to Rb, the mammalian system has two other Rb related proteins, p107 and p130 (Fig. (1)). These three proteins are also referred to as the “pocket proteins” because their main sequence similarity resides in a domain, the pocket domain, which mediates interactions with viral oncoproteins as well as cellular proteins to exert the biological functions of this family. A non-conserved spacer region separates the conserved pocket domain into two parts. Interestingly, the spacer region of p107 and p130 but not Rb contain binding sites for cyclin/cdk complexes [2, 3].
Fig. (1). The mammalian and Drosophila Rb family proteins.
The Rb family of proteins in mammals consists of Rb, p107 and p130, and in Drosophila contains RBF and RBF2. The pocket domain, which is shaded in grey, is conserved and is responsible for most protein-protein interactions. A cyclin/cdk interaction motif (shaded in black) is conserved in the spacer region of p107 and p130 but absent in Rb protein. The activity of Rb proteins is controlled by phosphorylation at numerous phosphorylation sites (indicated for Rb and RBF by *). The phosphorylation sites in other Rb family members have not been precisely mapped.
Phosphorylation plays a key role in regulating the activities of the Rb protein. The Rb protein contains numerous phosphorylation sites that are phosphorylated by cyclinD/cdk4, cyclinE/cdk2, and cyclinA/cdk2 kinases during cell cycle progression [4-7]. Generally speaking, hypophosphorylated Rb is active in the inhibition of cell proliferation and tumor suppression while the hyperphosphorylated Rb is inactive. In addition to regulation by phosphorylation, the Rb family proteins have differing expression patterns depending on the stage of the cell cycle and the type of tissue. For example, p130 is most abundant in quiescent, differentiated cells and in early G1 [8]. p107 expression increases in mid to late G1 while Rb expression is prominent in both proliferating and non-proliferating cells [8].
The biological functions of Rb include tumor suppression, regulation of the cell cycle, differentiation, and apoptosis. These functions of Rb are mediated by its interaction with a large number of cellular proteins. Over 100 proteins have been reported to interact with the Rb protein [9], and most, if not all, of these interactions also involve the pocket domain. The best studied binding partners of Rb are the E2F transcription factors.
Mammalian E2F Transcription Factors
In the mammalian system there have been eight E2F transcription factors identified so far. E2F1 through 6 contain a DNA binding domain and dimerization domain and require DP proteins to form a heterodimer for DNA binding [10-12]. There are at least three types of E2F transcription factors. Activating E2Fs, E2F1, E2F2, and E2F3, are the most well known, and serve to promote progression into S phase when Rb is inactivated by actively promoting the transcription of cell cycle genes. Repressive E2F proteins, E2F4 and E2F5, act to repress the transcription of E2F target genes in a complex with the Rb family members. The third category of E2F proteins, E2F6, E2F7, and E2F8, can function to repress E2F target gene expression but they function independently of Rb binding. Interestingly, a recent study showed that E2F7 and E2F8 form homodimers and heterodimers with each other and function redundantly in repressing E2F1 expression during S and G2 phases [13].
Drosophila RB and E2F Proteins
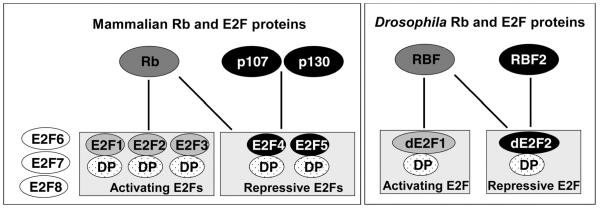
There are two E2F (dE2F1 and dE2F2), one DP (dDP), and two Rb family (RBF and RBF2) genes in the Drosophila genome [14-18]. The two Drosophila E2F proteins behave like the first two subgroups of the mammalian E2F proteins: dE2F1 mainly functions as a transcriptional activator [19, 20], comparable to the mammalian activating E2Fs proteins, while dE2F2 primarily mediates active repression, similar to the mammalian repressive E2F proteins E2F4 and 5 [20]. Furthermore, similar to the mammalian Rb protein that can bind to both the activating and the repressive E2F proteins, RBF can bind to both dE2F1 and dE2F2 proteins in Drosophila [20]. In contrast, RBF2 can only bind dE2F2 [16] analogous to the mammalian p107/p130 proteins that bind preferentially to the repressive E2F proteins (see Fig. (2)). Thus the Rb-E2F pathway is well conserved and is much simpler in Drosophila than in the mammalian systems.
Fig. (2). Similarities between the Rb and E2F proteins in mammals and Drosophila.
In mammals the E2F family is composed of eight E2F proteins, E2F1 through 8 and three DP proteins, DP1, DP2, and DP4. In Drosophila it consists of two E2Fs, dE2F1, dE2F2, and one DP protein, dDP. The eight mammalian E2F proteins can be divided into three groups: activating E2Fs (E2F1, E2F2, and E2F3), repressive E2Fs (E2F4 and E2F5), and Rb independent E2Fs (E2F6, E2F7, and E2F8). The mammalian activating E2Fs interact only with Rb. Similarly, the Drosophila activating E2F, dE2F1, interacts only with RBF. The mammalian repressive E2Fs interact with p107 and p130. In addition, E2F4 can also interact with Rb protein. Similarly, the Drosophila repressive E2F, dE2F2, interacts with both RBF and RBF2.
Transcriptional Targets of E2f Proteins
Several microarray experiments have demonstrated that E2F transcription factors are involved in many cellular processes. In addition to regulating transcription of genes involved in promoting the G1-S transition, E2F proteins were found to be involved in the transcription of other cell cycle genes including those involved in DNA repair, mitosis as well as the spindle checkpoint. There were several other categories of genes that were regulated by E2F transcription including genes involved in apoptosis and the DNA damage checkpoint [21-23]. Another category of genes regulated by E2F are genes involved in the Ras pathway, which may indicate that E2F is involved in some developmental processes or that E2F regulates the cells response to mitogens or growth factors which are regulated by the Ras pathway [24, 25].
The relative simplicity of the Drosophila Rb and E2F proteins has made it an ideal system for the genome wide identification of Rb/E2F targets. Specifically, RNAi has been used to deplete the activating E2F (dE2F1), the inhibitory E2F (dE2F2), dDP, RBF or RBF2 from Drosophila cell lines [26] or specific tissues [27]. Although it is possible that secondary changes in gene expression could have occurred, the screen did detect actual changes in expression, suggesting that the E2F/Rb interactions were rate limiting under these conditions. A surprising result of this study was that very few genes showed both a decrease in expression in the absence of dE2F1 and an increase in expression in the absence of dE2F2, suggesting that most genes are more influenced either by repression or by activation rather than by both activities equally [26]. Most of the genes found to be regulated largely by dE2F1 were involved in processes similar to those found in the ChIP on CHIP screens, including the cell cycle, DNA replication and repair, mitosis, chromosome segregation, and checkpoints. However, the genes regulated primarily by dE2F2 were not involved in DNA repair or S-phase processes. While the functions of many of these genes are unknown, a surprising number appear to be unrelated to the cell cycle and instead are involved in development, including male and female specific genes. The importance of dE2F2 repression was confirmed by finding that expression of many of these genes was deregulated in de2f2 null flies.
Further examination of repression of these differentiation genes has highlighted the complexity of Rb/E2F control of transcription and the importance of interactions with other factors. In comparing the requirement for dE2F2 and RBF2 to repress expression in Drosophila cell lines, ovaries, and embryos, microarray data showed that different genes were deregulated in each tissue with surprisingly little overlap, suggesting cell-type specific requirements for expression [27]. Comparison of dE2F2 and RBF2 binding to the promoters of these genes showed that in some cases the differences in expression correlated with dE2F2/RBF2 occupancy of the promoter, suggesting that cell type specific factors determine dE2F2/RBF2 distribution. In contrast, at other promoters dE2F2 and RBF2 were always present, suggesting that the functional relevance of promoter occupancy also varies with cell type. These findings emphasize the need to further understand the mechanisms determining Rb and E2F functions in specific contexts.
ROLE OF Rb FAMILY PROTEINS IN CELL PROLIFERATION
It is well established that Rb is a key inhibitor of entry into S-phase of the cell cycle, thereby regulating cell proliferation. The Rb family of proteins plays a role in regulating other stages of the cell cycle including G1 progression, S-phase entry and even exit from mitosis. The role of the Rb family of proteins can be both E2F dependent and independent. In addition, recent studies suggest that Rb also plays a role in transmitting the signals from both DNA damage and spindle checkpoint machinery to cell cycle machinery to prevent proliferation in the presence of damaged DNA. Interestingly, recent reports showed that Rb may be required for proliferation rather than acting as an inhibitor or proliferation in certain cell types [28]. Further understanding the different roles of Rb in cell proliferation, differentiation, and apoptosis could help us develop rational therapies either to prevent proliferation of cancer cells or to induce the apoptosis of cancer cells.
Regulation of G1/S
The Rb family proteins have differing levels of expression throughout G1 progression [29]. In early G1, p130 and p107 are expressed at high levels and are found in association with the repressive E2Fs. Due to being weak repressors and not having a nuclear localization signal, the repressive E2Fs require the Rb family proteins in order to repress gene expression [30]. In addition, Rb protein is also expressed in early G1 and associates with the activating E2F proteins. The Rb family proteins are hypophosphorylated in early G1 and associate with the E2F proteins to prevent expression of cell cycle progression genes.
The Rb family proteins recruit chromatin remodeling factors to regulate the expression of genes required for S-phase entry [31]. Some of the chromatin remodeling factors that are recruited are histone deacetylases (HDACs) and the SWI/SNF chromatin remodeling complex. As high levels of cyclin E drive the G1-S transition, cyclin E transcription is repressed in early G1. Rb protein is required for the recruitment of the HDACs in cyclin E transcriptional initiation sites [32]. Rb can also bind to components of chromatin remodeling complexes to affect gene expression. Rb was found in a complex with Brg1 and Brm which are ATP-dependent helicases that are involved in chromatin remodeling [33]. Rb can recruit Brg to specific promoters to inhibit E2F mediated transcription and to inhibit cell cycle progression. Interestingly, Rb recruits Brg to cyclin A but not cyclin E promoters. Since cyclin A drives S-phase progression and peaks in late S-phase, it is possible that this may be one mechanism by which the ordered expression of the cyclins is regulated [34]. Another way Rb may regulate E2F mediated transcription is via methylation of chromatin which can inhibit transcription. Rb was found in a complex with DNA methyltransferase 1 (DNMT1) and the repressive effects of Rb were enhanced by DNMT1 [35].
As cells progress to the middle of the G1 stage, cyclin D/cdk4 or cdk6 promote the hyperphosphorylation of the Rb family proteins resulting in dissociation of the Rb family proteins from E2F transcription factors allowing progression through G1 and eventually S-phase entry [6, 36]. In one current model the repressor E2Fs dissociate from the E2F target genes allowing gene expression and the activator E2Fs remain associated with the target genes and recruit histone acetyltransferases (HATs) to promote transcription of the cell cycle genes [37, 38]. In addition, activator E2Fs may occupy the promoter regions of all E2F target genes including those previously occupied by the repressor E2Fs.
Rb also has a role in regulating S-phase entry independent of transcriptional regulation via E2F proteins. Rb was found to associate with several proteins involved in the initiation of DNA replication including the DNA replication licensing factor MCM7, the replication factor RFC and the single stranded DNA binding protein, purα, suggesting that Rb plays a role in regulating the initiation of replication [39, 40]. CyclinD/Cdk4 was found to associate with MCM7 and this complex was catalytically active; however, CyclinD/Cdk4 did not phosphorylate MCM7. Instead it catalyzed the dissociation of Rb from the MCM7 complex in preparation for DNA replication [41].
Senescence is the non-reversible exit from the cell cycle. Rb seems to be particularly important in the regulation of senescence. Under conditions of stress p16 is upregulated leading to upregulation of Rb function. This activation leads to chromatin reorganization with repressive heterochromatin forming at loci containing E2F targets especially those regulating progression of the cell cycle, thus leading to exit from the cell cycle. The heterochromatin formation is not reversible, so p16 and Rb protein activation does not need to be retained [42]. Promoting cellular senescence could be one way to prevent proliferation of cancer cells.
Regulation of Mitosis and Cell Cycle Checkpoints
Rb also regulates the cell cycle beyond the G1-S transition. Work done in Drosophila showed that larvae mutant for the Rb homologe, Rbf, exhibited extensive defects in chromatin condensation in mitosis suggesting that Rb plays a role in mitosis. Rbf was found to interact with dCAP-D3 a component of the condensing II complex and was required for the accumulation of dCAP-D3 on chromatin [43]. These findings were found to be conserved in a human tumor cell line. Expression of Rb in an Rb deficient cell line promoted the association of hCAP-D3 with chromatin [43]. These findings provide one explanation for why inactivation of Rb will lead to genomic instability.
One recent study suggests that Rb plays a role in exiting mitosis and the initiation of G1 stage of the cell cycle. Rb may help regulate the targets of the anaphase promoting complex/cyclosome (APC/C) via a direct physical link between the Rb protein and the APC/C specificity factor, Cdh1. Cdh1 directs the destruction of the inhibitors of mitotic exit and G1 by ubiquitination via the APC/C and degradation via the proteosome. In this study Binne and colleagues found that Rb was required to direct the destruction of Skp2 the f-box protein of the Skp1cullin F-box protein, SCF, which is an E3 ubiquitin ligase. The targeted degradation of Skp2 led to the stabilization of p27(kip1), a CDK inhibitor, thereby promoting a G1 arrest [44].
Rb has also been shown to be involved in mediating a checkpoint induced response. One study showed that the spindle checkpoint kinase Mad2 is a direct transcriptional target of E2F1 and is overexpressed in Rb deficient cells. Overexpression of Mad2 can lead to chromosomal instability and tumorigenesis, contributing to cancer [45]. Rb may also play a role in the DNA damage checkpoint. Inoue and Taya showed that Rb was phosphorylated in an ATM, Chk1/2 dependent fashion in response to DNA damage on S612. Phosphorylation of S612 in response to DNA damage promotes the association of Rb with E2F and inhibits E2F mediated transcription, halting the cell cycle [46].
Although a role for Rb in inhibiting proliferation is well established, a recent study suggests that Rb is required for cell proliferation in certain cells. Work done by Morris and others showed that E2F1 can inhibit beta-catenin/T cell factor (TCF) dependent transcription of c-Myc and other targets that are important for cellular proliferation and survival. They also showed that E2F1 can upregulate the expression of proteins that result in beta-catenin degradation. Since the Wnt signaling pathway promotes cell proliferation and survival in some cell types, inhibition of E2F1 by Rb is required for the proliferation of these cells. E2F1 is also downregulated by CDK8, a colorectal oncogene [28]. This study supports the finding that Rb is often overexpressed or amplified in colorectal cancers [47, 48] unlike other cancer types in which Rb is often inactivated.
ROLE OF Rb FAMILY PROTEINS IN APOPTOSIS
In addition to regulation of the cell cycle, Rb also regulates other functions in cells and organisms including apoptosis. Rb may regulate apoptosis directly by controlling the expression of apoptosis regulators or indirectly by regulating cell cycle progression since cycling cells are more likely to undergo apoptosis than G1 arrested cells. Induction of apoptosis in cancer cells is one of the most promising areas of research in cancer therapy. Therefore identifying targets that would specifically induce apoptosis in Rb deficient cells could be a good strategy for the development of new cancer therapies.
Rb and Apoptosis in Mammalian Systems
Apoptosis may occur via a death receptor-dependent (extrinsic) or independent (intrinsic or mitochondrial) mechanism [49]. The mitochondrial pathway of cell death is mediated by the Bcl-2 family of proteins and caspases. The upstream activators of apoptosis are responsible for receiving and transmitting cell death signals, leading to the expression and/or activation of BH3 domain containing proteins. These proteins can signal to other Bcl-2 family proteins that can insert into the mitochondrial outer membrane and this leads to release of cytochrome-C (Cyt-C) [50]. Cyt-C binds to APAF-1 which leads to activation of the activating caspases including caspase 9 [51]. The activating caspases turn on the effector caspases to induce cell death. Apoptosis is further regulated by the inhibitors of apoptosis proteins (IAPs) which bind to and inhibit caspase function (reviewed in [52]. The inhibition of caspase activation by IAP proteins is counteracted by another protein released from mitochondria during apoptosis, Smac/Diablo [53, 54].
The Rb pathway can regulate apoptosis via transcriptional regulation of pro-apoptotic factors. E2F1 overexpression induces apoptosis via transcriptional activation of pro-apoptotic genes including Arf, p73, APAF-1, Smac/Diablo and Omi HTRA2 [55, 56]. Most BH3 only proteins are induced by E2F as well as some of the initiator and effector caspases [57]. E2F can also regulate apoptosis via stabilization of p53 protein by upregulating the levels of Arf and pin [56]. The regulation of proapoptotic target genes expression by Rb/E2F proteins can be further modulated by other regulators and signaling pathways. For example, GABP was found to bind directly to E2F1 and specifically inhibit E2F1 dependent apoptosis [58]. Another example is that DNA damage induced phosphorylation of E2F1 by the ATM/Chk2 pathway which results in activation of E2F1 transcriptional activity and promotes apoptosis [59-61]. In addition, DNA damage signals can lead to acetylation of Rb protein, which disrupt its binding to E2F1 and activates the pro-apoptotic transcriptional activity of E2F1 [62, 63].
Rb and Apoptosis in Drosophila
The general mechanisms of inducing apoptosis are conserved in Drosophila and mammalian systems. Activation of caspases induces cell death in both systems and both systems use an apoptosome complex. However, in Drosophila cyt-C does not seem to be necessary for apoptosis although it is still released from the mitochondria [64]. Reaper, Hid and Grim, which are inhibitors of the IAPs similar to Smac/Diablo, are the key regulators of apoptosis in Drosophila [65]. Reaper, Hid and Grim localize to the mitochondria and this is essential for their full cell death effect [66-68]. Apoptosis induction in Drosophila also involves disruption of the mitochondria similar to the mammalian system [64].
The role of the Rb (Rbf) pathway in apoptosis is also conserved in Drosophila. Overexpression of dE2F1 is pro-apoptotic in most developing cells and results in transcription of dArk/Apaf1, and Reaper [69, 70]. Rbf is able to suppress apoptosis in an E2F dependent manner by limiting the expression of Hid [71, 72]. The effects of the Rb pathway on apoptosis are dependent on the developmental context of the cell. For example, in the larval eye disc, removal of Rbf results in increased apoptosis but only in cells along the morphogenetic furrow [19]. The conservation of the apoptotic pathways along with the ease of carrying out genetics screens makes Drosophila a good system for identifying potential new drug targets that would result in the induction of apoptosis in the absence of Rbf.
Moon and colleagues showed that in the absence of Rbf function the E2F dependent apoptosis was dependent on hid and reaper [73]. In support of this finding, Hid was identified in a screen to identify novel genes that regulated apoptosis in Rbf deficient cells [72]. In addition, our lab showed that hid was deregulated in rbf mutant larvae and that hid was repressed directly by Rbf/E2F proteins via association with an E2F binding site in the hid enhancer [72]. In one screen for suppressors of dE2F1 mediated apoptosis, the Dyson lab found AAC11 (mammalian homologue Api5) AAC11 drives cell death in an E2F dependent fashion but does not affect dE2F1 mediated transcription of cell cycle or pro-apoptotic genes suggesting that dE2F1 mediated apoptosis may be regulated at least to some extent in a non-transcriptional fashion [74]. In support of this finding, Bantam, a miRNA that regulates the translation of Hid, can modulate the survival of rbf mutant cells [72]. These results suggest that there are potential molecular drug targets that can allow us to induce the specific apoptosis of Rb mutant cells.
ROLE OF Rb FAMILY PROTEINS IN DIFFERENTIATION
Results from model organisms have shown that the Rb pathway is also involved in regulating the differentiation of cells in developing organisms. In developing cancer therapies, one way to stop tumor growth could be to reactivate differentiation pathways so that the tumor cells undergo terminal differentiation and stop proliferating. Understanding the role of Rb pathway in regulating differentiation will potentially give us novel targets for promoting terminal differentiation and blocking proliferation in cancer cells.
Mammalian
There is some evidence that the Rb pathway regulates differentiation in mammalian cells. The Majority of the evidence of Rb in differentiation may be related to its role in regulating cell cycle exit. Cells lacking Rb function cannot exit the cell cycle and may continue to proliferate when they should be terminally differentiated. For example, Rb null hematopoietic cells cannot fully differentiate and this may result in myloproliferative disorders which could lead to an increased number of precursor cells and more tumors [75]. Rb may also coordinate cell cycle exit with differentiation. In Rb null skin cells, the cells continue to divide despite having markers for being differentiated [76], and in the sensory hair cells of the ear, removal of Rb results in fully differentiated cells continuing to proliferate [77]. Work done by Guo and colleagues showed that Rb is required for the quiescence and differentiation of enterocytes in the intestine. Deletion of Rb protein in the small intestine enterocytes resulted in ectopic cell cycle reentry and while these cells had a higher rate of apoptosis, the remainder of the cycling cells did not completely differentiate which may lead to the continued proliferation of the undifferentiated cells [78]. Therefore, inactivation of Rb in the small intestine could lead to uncontrolled cell growth of undifferentiated cells. In conditional knockout mice where Rb deficient mice could live until birth there was abnormal development and impaired ossification of bones. The Rb deficient osteoblasts impaired cell cycle exit suggesting that Rb was required for linking cell differentiation and cell cycle exit [79]. These results suggest that Rb has an important role in maintaining or establishing cell cycle exit and the quiescent state of differentiated cells.
There is also evidence that Rb may play a more direct role in differentiation. In MEFs that have been activated to become adipocytes, removal of Rb blocks while removal of E2F4 promotes differentiation. Interestingly, E2F4 loss does not override the differentiation defect resulting from Rb loss even though it completely suppresses the proliferation defect, suggesting that Rb promotes adipogenesis independent of its cell cycle role [80]. Additionally, mutation of Rb in mouse lungs led to more neuroendocrine cell differentiation, which suggested that Rb can regulate differentiation in lung cells by specifically inhibiting neuroendocrine cell fate [81]. Rb has been shown to have protein-protein interactions with proteins other than the E2F family of proteins and many of these interactions are with proteins involved in differentiation. Rb can interact with the transcription factor involved in differentiation of muscle tissue, MyoD [82]. Rb may help promote differentiation of muscle tissue by coordinating with MyoD to induce transcription of muscle target genes. There is some evidence that shows that acetylation of Rb may help promote its role in muscle development, but not affect the role of Rb in cell cycle. The acetylation mutant is able to arrest the cell cycle, but is not able to cooperate with MyoD to regulate MyoD transcription [83]. In rat neural stem cells overexpression of Rb or p130 affected the lineage specification of differentiating cells, but did not inhibit cell proliferation or apoptosis [84]. These results suggest that the Rb family proteins can regulate differentiation independent of their role in the cell cycle. Consistent with this, cell cycle independent roles of Rb proteins have also been observed in C elegans and in Drosophila systems, suggesting that the role of Rb proteins in differentiation could also be conserved.
C. elegans
The C. elegans system is an excellent model system for studying the role of specific genes in a developmental context. C. elegans contains the homologues of the Rb pathway including Lin-35 (Rb), EFL-1 (E2F transcription factor), and DPL-1 (DP). Mutation of these genes has revealed a role for the Rb pathway in embryonic development and oocyte maturation, and Rb-E2F mediated transcription repression regulates the expression of developmental genes to the proper context. The Rb pathway was found to play a role in C. elegans vulval development. In C. elegans, epidermal growth factor (Lin-3) signaling is essential for appropriate vulval development. Lin-3 signals via RAS and MAPK signaling pathways to activate the transcription factor Lin-1 in the 22 cells that will form the vulva. Null mutants of Lin-1 result in multipule vulval type structures or Muv (reviewed in [85]. In a screen to find other components of this pathway that resulted in Muv phenotype, the Horvitz lab identified two classes of mutants, class A and B. Interestingly, one mutation in class A in combination with one mutation in class B resulted in a synergistic Muv phenotype, but multiple mutations in the same classes resulted in a wild type phenotype [86]. Lin-35, the Rb homologue in C. elegans, was identified as a class B gene suggesting that the Rb pathway plays an important role in regulating differentiation. The identification of the Rb mutation in one of the two classes of the synergistic Muv mutants indicates that the role of Rb in vulval development is at least partially redundant with other mechanisms.
Drosophila
Drosophila is a good model organism for studying the role of the Rb pathway in development. Similar to the role of Rb in differentiation in the mouse, deletion of rbf was shown to have mild differentiation defects on its own [19, 87]. This suggests that the role of Rbf in differentiation in flies may also involve partially redundant mechanisms as observed in C. elegans. Using a mosaic eye screen, our lab was able to identify genes that are required for the proper differentiation of rbf mutant cells. In particular, a gene called rhinoceros (rno) was identified to have a role in regulating eye cell differentiation in the absence of Rb. Tissue mutant for both rbf and rno displayed developmental defects in the larval, pupal and adult stages of development. Notch signaling is critical for the proper R8 determination. It was found that rbf and rno exhibited partially redundant regulation of the expression of Dl, the ligand of Notch signaling. In addition, this regulation was dependent on the dE2F1 transcription factor (Steele et al , submitted). Using this type of screening, it is possible that novel roles for Rb in regulating differentiation can be uncovered, and proteins that work in tandem with Rb to promote differentiation can be identified.
THE STATUS OF THE Rb PATHWAY AS A GUIDE FOR CANCER THERAPIES
The general approaches employed in cancer therapies are to induce apoptosis of cancer cells or to inhibit their cell proliferation/induce differentiation. Successful cancer therapeutic drugs such as Gleevec that have relatively low side effects generally have the abilities to preferentially target the cancer cells while mostly sparing normal cells. The development of such successful cancer drugs require that distinct features of the cancer cells be targeted. As shown in Fig. (3), there are at least three different functional states of Rb in cancer cells. While a majority of cancers have an inactivated Rb pathway, either by mutation/deletion of Rb or by its functional inactivation (Fig. (3), category I and II), a small subset of cancers require functional Rb and may have overexpression or amplification of Rb (Fig. (3), category III). Depending on the status of the Rb pathway, different strategies can potentially be used to specifically target cancer cells (see Fig. (3)).
Fig. (3). The Rb pathway status in cancers and possible therapeutic approaches.
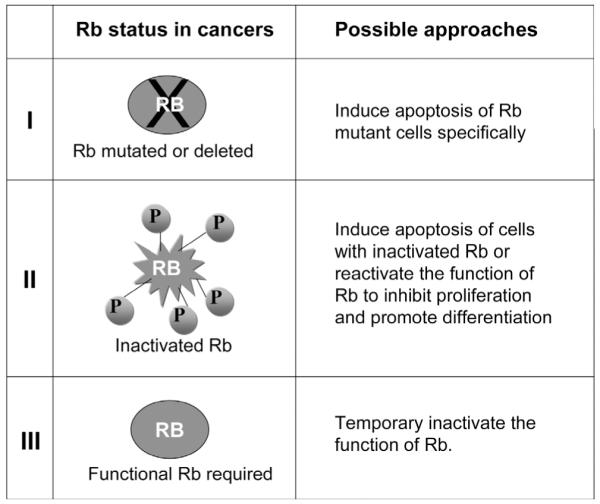
There are at least three different functional states of Rb in cancer cells. While a majority of cancers have an inactivated Rb pathway, either by mutation/deletion of Rb or by its functional inactivation (category I and II), a small subset of cancers require functional Rb (category III). Depending on the status of the Rb pathway, different strategies can be potentially used to specifically target cancer cells. See text for more detailed discussion.
Inducing apoptosis is a commonly used strategy in cancer therapies. Since the inactivated Rb pathway is a common feature that distinguishes cancer cells from normal cells for a majority of cancers, an approach that specifically kills Rb deficient cells can potentially be useful to treat large majority of cancers. Because the expression of a number of apoptosis regulators are controlled by E2F, Rb deficient cells are often somewhat sensitized to apoptosis and are more dependent on survival signals. However, currently we do not know how to specifically kill cancer cells with an inactivated Rb pathway and a lot of research will be required to identify these types of drug targets. It is likely that studies using model organisms to identify genes that regulate the apoptosis of Rb mutant cells could lead to such new drug targets. For example, identification of hid as a modifier of rbf null cells in Drosophila uncovered the role of the bantam miRNA in regulating the apoptosis of rbf cells. While removal of bantam alone was not sufficient to induce apoptosis, increased apoptosis was observed in bantam, rbf double mutant cells [72]. Although these studies were carried out in Drosophila, it is likely that conserved regulators of apoptosis of Rb deficient cells can be identified in such studies, which will potentially allow specific induction of apoptosis to cells that have deficient Rb pathway without significant effect to cells with normal Rb pathway activity.
Although a majority of human cancers have an inactivated Rb pathway, functional Rb is actually required in a subset of cancers to maintain the proliferation and prevent apoptosis of cancer cells (Fig. (3), category III). For such kind of cancers, a possible approach will be to develop small molecule inhibitors of Rb function. Temporary inhibition of the Rb function can potentially lead to the killing of cancer cells or sensitization of th ese cancer cells to other chemotherapeutic agents. However, the success of such an approach hinges on the possibility that these cancer cells will be more sensitive to the loss of Rb function than their normal cell counterparts in the body. The observation that Rb is often overexpressed or amplified in colorectal cancers [47, 48], a cancer type that requires Rb function, certainly suggest the possibility that colon cancer cells could be more sensitive than wild type cells to the loss of Rb function. Further research will be needed to determine the feasibility of this approach.
While most research on drug therapy for cancer focuses on inducing apoptosis, other viable options include inhibiting cell proliferation, inducing cellular senescence, or reactivation of differentiation pathways so that the cells undergo terminal differentiation and stop proliferation. For cancers in which Rb is not irreversibly inactivated, it may be possible to reactivate Rb function to halt tumorigenesis. Much more research needs to be done to fully understand the cell types and cellular context in which this would work. Understanding the role of the Rb pathway in regulating differentiation will potentially give us novel targets for promoting terminal differentiation and blocking proliferation in cancer cells.
In summary, it is possible that the Rb status in cancers can be used to develop new cancer therapeutic drugs that could lead to better treatments with potentially fewer side effects. However, much more work will be needed to understand the control of cell proliferation, apoptosis and differentiation in the presence or absence of Rb.
ACKNOWLEDGEMENTS
This work is supported by grants from National Institute of Health and the American Cancer Society. JS is supported by a postdoctoral training grant from the Committee on Cancer Biology (T32CA009594).
REFERENCES
- [1].Knudsen ES, Knudsen KE. Retinoblastoma tumor suppressor: where cancer meets the cell cycle. Exp Biol Med (Maywood) 2006;231(7):1271–81. doi: 10.1177/153537020623100713. [DOI] [PubMed] [Google Scholar]
- [2].Zhu L, Harlow E, Dynlacht BD. p107 uses a p21CIP1-related domain to bind cyclin/cdk2 and regulate interactions with E2F. Genes Dev. 1995;9(14):1740–52. doi: 10.1101/gad.9.14.1740. [DOI] [PubMed] [Google Scholar]
- [3].Dynlacht BD, Flores O, Lees JA, Harlow E. Differential regulation of E2F transactivation by cyclin/cdk2 complexes. Genes Dev. 1994;8(15):1772–86. doi: 10.1101/gad.8.15.1772. [DOI] [PubMed] [Google Scholar]
- [4].Hinds PW, Mittnacht S, Dulic V, Arnold A, Reed SI, Weinberg RA. Regulation of retinoblastoma protein functions by ectopic expression of human cyclins. Cell. 1992;70(6):993–1006. doi: 10.1016/0092-8674(92)90249-c. [DOI] [PubMed] [Google Scholar]
- [5].Ewen ME, Sluss HK, Sherr CJ, Matsushime H, Kato J, Livingston DM. Functional interactions of the retinoblastoma protein with mammalian D-type cyclins. Cell. 1993;73(3):487–97. doi: 10.1016/0092-8674(93)90136-e. [DOI] [PubMed] [Google Scholar]
- [6].Kato J, Matsushime H, Hiebert SW, Ewen ME, Sherr CJ. Direct binding of cyclin D to the retinoblastoma gene product (pRb) and pRb phosphorylation by the cyclin D-dependent kinase CDK4. Genes Dev. 1993;7(3):331–42. doi: 10.1101/gad.7.3.331. [DOI] [PubMed] [Google Scholar]
- [7].Resnitzky D, Hengst L, Reed SI. Cyclin A-associated kinase activity is rate limiting for entrance into S phase and is negatively regulated in G1 by p27Kip1. Mol Cell Biol. 1995;15(8):4347–52. doi: 10.1128/mcb.15.8.4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Claudio PP, Zamparelli A, Garcia FU, Claudio L, Ammirati G, Farina A, et al. Expression of cell-cycle-regulated proteins pRb2/p130, p107, p27(kip1), p53, mdm-2, and Ki-67 (MIB-1) in prostatic gland adenocarcinoma. Clin Cancer Res. 2002;8(6):1808–15. [PubMed] [Google Scholar]
- [9].Morris EJ, Dyson NJ. Retinoblastoma protein partners. Adv Cancer Res. 2001;82:1–54. doi: 10.1016/s0065-230x(01)82001-7. [DOI] [PubMed] [Google Scholar]
- [10].Dyson N. The regulation of E2F by pRB-family proteins. Genes Dev. 1998;12(15):2245–62. doi: 10.1101/gad.12.15.2245. [DOI] [PubMed] [Google Scholar]
- [11].Attwooll C, Denchi E Lazzerini, Helin K. The E2F family: specific functions and overlapping interests. Embo J. 2004;23(24):4709–16. doi: 10.1038/sj.emboj.7600481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Du W, Pogoriler J. Retinoblastoma family genes. Oncogene. 2006;25(38):5190–200. doi: 10.1038/sj.onc.1209651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Li J, Ran C, Li E, Gordon F, Comstock G, Siddiqui H, et al. Synergistic function of E2F7 and E2F8 is essential for cell survival and embryonic development. Dev Cell. 2008;14(1):62–75. doi: 10.1016/j.devcel.2007.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Dynlacht BD, Brook A, Dembski M, Yenush L, Dyson N. DNA-binding and trans-activation properties of Drosophila E2F and DP proteins. Proc Natl Acad Sci USA. 1994;91(14):6359–63. doi: 10.1073/pnas.91.14.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Du W, Vidal M, Xie JE, Dyson N. RBF, a novel RB-related gene that regulates E2F activity and interacts with cyclin E in Drosophila. Genes Dev. 1996;10(10):1206–18. doi: 10.1101/gad.10.10.1206. [DOI] [PubMed] [Google Scholar]
- [16].Stevaux O, Dimova D, Frolov MV, Taylor-Harding B, Morris E, Dyson N. Distinct mechanisms of E2F regulation by Drosophila RBF1 and RBF2. Embo J. 2002;21(18):4927–37. doi: 10.1093/emboj/cdf501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Sawado T, Yamaguchi M, Nishimoto Y, Ohno K, Sakaguchi K, Matsukage A. dE2F2, a novel E2F-family transcription factor in Drosophila melanogaster. Biochem Biophys Res Commun. 1998;251(2):409–15. doi: 10.1006/bbrc.1998.9407. [DOI] [PubMed] [Google Scholar]
- [18].Ohtani K, Nevins JR. Functional properties of a Drosophila homolog of the E2F1 gene. Mol Cell Biol. 1994;14(3):1603–12. doi: 10.1128/mcb.14.3.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Du W. Suppression of the rbf null mutants by a de2f1 allele that lacks transactivation domain. Development. 2000;127(2):367–79. doi: 10.1242/dev.127.2.367. [DOI] [PubMed] [Google Scholar]
- [20].Frolov MV, Huen DS, Stevaux O, Dimova D, Balczarek-Strang K, Elsdon M, et al. Functional antagonism between E2F family members. Genes Dev. 2001;15(16):2146–60. doi: 10.1101/gad.903901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, et al. E2F integrates cell cycle progression with DNA repair, replication, and G(2)/M checkpoints. Genes Dev. 2002;16(2):245–56. doi: 10.1101/gad.949802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Weinmann AS, Yan PS, Oberley MJ, Huang TH, Farnham PJ. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 2002;16(2):235–44. doi: 10.1101/gad.943102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Muller H, Bracken AP, Vernell R, Moroni MC, Christians F, Grassilli E, et al. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 2001;15(3):267–85. doi: 10.1101/gad.864201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Young AP, Nagarajan R, Longmore GD. Mechanisms of transcriptional regulation by Rb-E2F segregate by biological pathway. Oncogene. 2003;22(46):7209–17. doi: 10.1038/sj.onc.1206804. [DOI] [PubMed] [Google Scholar]
- [25].Korotayev K, Chaussepied M, Ginsberg D. ERK activation is regulated by E2F1 and is essential for E2F1-induced S phase entry. Cell Signal. 2008;20(6):1221–6. doi: 10.1016/j.cellsig.2008.02.012. [DOI] [PubMed] [Google Scholar]
- [26].Dimova DK, Stevaux O, Frolov MV, Dyson NJ. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 2003;17(18):2308–20. doi: 10.1101/gad.1116703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Stevaux O, Dimova DK, Ji JY, Moon NS, Frolov MV, Dyson NJ. Retinoblastoma family 2 is required in vivo for the tissue-specific repression of dE2F2 target genes. Cell Cycle. 2005;4(9):1272–80. doi: 10.4161/cc.4.9.1982. [DOI] [PubMed] [Google Scholar]
- [28].Morris EJ, Ji JY, Yang F, Di Stefano L, Herr A, Moon NS, et al. E2F1 represses beta-catenin transcription and is antagonized by both pRB and CDK8. Nature. 2008;455(7212):552–6. doi: 10.1038/nature07310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Classon M, Dyson N. p107 and p130: versatile proteins with interesting pockets. Exp Cell Res. 2001;264(1):135–47. doi: 10.1006/excr.2000.5135. [DOI] [PubMed] [Google Scholar]
- [30].Dimova DK, Dyson NJ. The E2F transcriptional network: old acquaintances with new faces. Oncogene. 2005;24(17):2810–26. doi: 10.1038/sj.onc.1208612. [DOI] [PubMed] [Google Scholar]
- [31].Frolov MV, Dyson NJ. Molecular mechanisms of E2F-dependent activation and pRB-mediated repression. J Cell Sci. 2004;117(Pt 11):2173–81. doi: 10.1242/jcs.01227. [DOI] [PubMed] [Google Scholar]
- [32].Morrison AJ, Sardet C, Herrera RE. Retinoblastoma protein transcriptional repression through histone deacetylation of a single nucleosome. Mol Cell Biol. 2002;22(3):856–65. doi: 10.1128/MCB.22.3.856-865.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Dahiya A, Wong S, Gonzalo S, Gavin M, Dean DC. Linking the Rb and polycomb pathways. Mol Cell. 2001;8(3):557–69. doi: 10.1016/s1097-2765(01)00346-x. [DOI] [PubMed] [Google Scholar]
- [34].Shanahan F, Seghezzi W, Parry D, Mahony D, Lees E. Cyclin E associates with BAF155 and BRG1, components of the mammalian SWI-SNF complex, and alters the ability of BRG1 to induce growth arrest. Mol Cell Biol. 1999;19(2):1460–9. doi: 10.1128/mcb.19.2.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat Genet. 2000;25(3):338–42. doi: 10.1038/77124. [DOI] [PubMed] [Google Scholar]
- [36].Sherr CJ. Cancer cell cycles. Science. 1996;274(5293):1672–7. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- [37].Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14(7):804–16. [PMC free article] [PubMed] [Google Scholar]
- [38].Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, et al. E2F-dependent histone acetylation and recruitment of the Tip60 acetyltransferase complex to chromatin in late G1. Mol Cell Biol. 2004;24(10):4546–56. doi: 10.1128/MCB.24.10.4546-4556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Pennaneach V, Salles-Passador I, Munshi A, Brickner H, Regazzoni K, Dick F, et al. The large subunit of replication factor C promotes cell survival after DNA damage in an LxCxE motif- and Rb-dependent manner. Mol Cell. 2001;7(4):715–27. doi: 10.1016/s1097-2765(01)00217-9. [DOI] [PubMed] [Google Scholar]
- [40].Sterner JM, Dew-Knight S, Musahl C, Kornbluth S, Horowitz JM. Negative regulation of DNA replication by the retinoblastoma protein is mediated by its association with MCM7. Mol Cell Biol. 1998;18(5):2748–57. doi: 10.1128/mcb.18.5.2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gladden AB, Woolery R, Aggarwal P, Wasik MA, Diehl JA. Expression of constitutively nuclear cyclin D1 in murine lymphocytes induces B-cell lymphoma. Oncogene. 2006;25(7):998–1007. doi: 10.1038/sj.onc.1209147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Takahashi A, Ohtani N, Yamakoshi K, Iida S, Tahara H, Nakayama K, et al. Mitogenic signalling and the p16INK4a-Rb pathway cooperate to enforce irreversible cellular senescence. Nat Cell Biol. 2006;8(11):1291–7. doi: 10.1038/ncb1491. [DOI] [PubMed] [Google Scholar]
- [43].Longworth MS, Herr A, Ji JY, Dyson NJ. RBF1 promotes chromatin condensation through a conserved interaction with the Condensin II protein dCAP-D3. Genes Dev. 2008;22(8):1011–24. doi: 10.1101/gad.1631508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Binne UK, Classon MK, Dick FA, Wei W, Rape M, Kaelin WG, Jr., et al. Retinoblastoma protein and anaphase-promoting complex physically interact and functionally cooperate during cell-cycle exit. Nat Cell Biol. 2007;9(2):225–32. doi: 10.1038/ncb1532. [DOI] [PubMed] [Google Scholar]
- [45].Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, et al. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 2004;430(7001):797–802. doi: 10.1038/nature02820. [DOI] [PubMed] [Google Scholar]
- [46].Inoue Y, Kitagawa M, Taya Y. Phosphorylation of pRB at Ser612 by Chk1/2 leads to a complex between pRB and E2F-1 after DNA damage. Embo J. 2007;26(8):2083–93. doi: 10.1038/sj.emboj.7601652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Firestein R, Bass AJ, Kim SY, Dunn IF, Silver SJ, Guney I, et al. CDK8 is a colorectal cancer oncogene that regulates beta-catenin activity. Nature. 2008;455(7212):547–51. doi: 10.1038/nature07179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Gope R, Christensen MA, Thorson A, Lynch HT, Smyrk T, Hodgson C, et al. Increased expression of the retinoblastoma gene in human colorectal carcinomas relative to normal colonic mucosa. J Natl Cancer Inst. 1990;82(4):310–4. doi: 10.1093/jnci/82.4.310. [DOI] [PubMed] [Google Scholar]
- [49].Fulda S, Debatin KM. Extrinsic versus intrinsic apoptosis pathways in anticancer chemotherapy. Oncogene. 2006;25(34):4798–811. doi: 10.1038/sj.onc.1209608. [DOI] [PubMed] [Google Scholar]
- [50].Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116(2):205–19. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- [51].Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an a poptotic protease cascade. Cell. 1997;91(4):479–89. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- [52].Salvesen GS, Duckett CS. IAP proteins: blocking the road to death’s door. Nat Rev Mol Cell Biol. 2002;3(6):401–10. doi: 10.1038/nrm830. [DOI] [PubMed] [Google Scholar]
- [53].Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102(1):33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- [54].Wu G, Chai J, Suber TL, Wu JW, Du C, Wang X, et al. Structural basis of IAP recognition by Smac/DIABLO. Nature. 2000;408(6815):1008–12. doi: 10.1038/35050012. [DOI] [PubMed] [Google Scholar]
- [55].Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29(8):409–17. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- [56].Iaquinta PJ, Lees JA. Life and death decisions by the E2F transcription factors. Curr Opin Cell Biol. 2007;19(6):649–57. doi: 10.1016/j.ceb.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 2002;4(11):859–64. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- [58].Hauck L, Kaba RG, Lipp M, Dietz R, von Harsdorf R. Regulation of E2F1-dependent gene transcription and apoptosis by the ETS-related transcription factor GABPgamma1. Mol Cell Biol. 2002;22(7):2147–58. doi: 10.1128/MCB.22.7.2147-2158.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15(14):1833–44. [PMC free article] [PubMed] [Google Scholar]
- [60].Urist M, Tanaka T, Poyurovsky MV, Prives C. p73 induction after DNA damage is regulated by checkpoint kinases Chk1 and Chk2. Genes Dev. 2004;18(24):3041–54. doi: 10.1101/gad.1221004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Stevens C, Smith L, La Thangue NB. Chk2 activates E2F-1 in response to DNA damage. Nat Cell Biol. 2003;5(5):401–9. doi: 10.1038/ncb974. [DOI] [PubMed] [Google Scholar]
- [62].Dick FA, Dyson N. pRB contains an E2F1-specific binding domain that allows E2F1-induced apoptosis to be regulated separately from other E2F activities. Mol Cell. 2003;12(3):639–49. doi: 10.1016/s1097-2765(03)00344-7. [DOI] [PubMed] [Google Scholar]
- [63].Markham D, Munro S, Soloway J, O’Connor DP, La Thangue NB. DNA-damage-responsive acetylation of pRb regulates binding to E2F-1. EMBO Rep. 2006;7(2):192–8. doi: 10.1038/sj.embor.7400591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Abdelwahid E, Yokokura T, Krieser RJ, Balasundaram S, Fowle WH, White K. Mitochondrial disruption in Drosophila apoptosis. Dev Cell. 2007;12(5):793–806. doi: 10.1016/j.devcel.2007.04.004. [DOI] [PubMed] [Google Scholar]
- [65].Bangs P, White K. Regulation and execution of apoptosis during Drosophila development. Dev Dyn. 2000;218(1):68–79. doi: 10.1002/(SICI)1097-0177(200005)218:1<68::AID-DVDY6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- [66].Haining WN, Carboy-Newcomb C, Wei CL, Steller H. The proapoptotic function of Drosophila Hid is conserved in mammalian cells. Proc Natl Acad Sci U S A. 1999;96(9):4936–41. doi: 10.1073/pnas.96.9.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Claveria C, Caminero E, Martinez AC, Campuzano S, Torres M. GH3, a novel proapoptotic domain in Drosophila Grim, promotes a mitochondrial death pathway. Embo J. 2002;21(13):3327–36. doi: 10.1093/emboj/cdf354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Olson MR, Holley CL, Gan EC, Colon-Ramos DA, Kaplan B, Kornbluth S. A GH3-like domain in reaper is required for mitochondrial localization and induction of IAP degradation. J Biol Chem. 2003;278(45):44758–68. doi: 10.1074/jbc.M308055200. [DOI] [PubMed] [Google Scholar]
- [69].Zhou L, Steller H. Distinct pathways mediate UV-induced apoptosis in Drosophila embryos. Dev Cell. 2003;4(4):599–605. doi: 10.1016/s1534-5807(03)00085-6. [DOI] [PubMed] [Google Scholar]
- [70].Asano M, Nevins JR, Wharton RP. Ectopic E2F expression induces S phase and apoptosis in Drosophila imaginal discs. Genes Dev. 1996;10(11):1422–32. doi: 10.1101/gad.10.11.1422. [DOI] [PubMed] [Google Scholar]
- [71].Moon NS, Frolov MV, Kwon EJ, Di Stefano L, Dimova DK, Morris EJ, et al. Drosophila E2F1 has context-specific pro- and antiapoptotic properties during development. Dev Cell. 2005;9(4):463–75. doi: 10.1016/j.devcel.2005.08.015. [DOI] [PubMed] [Google Scholar]
- [72].Tanaka-Matakatsu M, Xu J, Cheng L, Du W. Regulation of apoptosis of rbf mutant cells during Drosophila development. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Moon NS, Di Stefano L, Dyson N. A gradient of epidermal growth factor receptor signaling determines the sensitivity of rbf1 mutant cells to E2F-dependent apoptosis. Mol Cell Biol. 2006;26(20):7601–15. doi: 10.1128/MCB.00836-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Morris EJ, Michaud WA, Ji JY, Moon NS, Rocco JW, Dyson NJ. Functional identification of Api5 as a suppressor of E2F-dependent apoptosis in vivo. PLoS Genet. 2006;2(11):e196. doi: 10.1371/journal.pgen.0020196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Spike BT, Dirlam A, Dibling BC, Marvin J, Williams BO, Jacks T, et al. The Rb tumor suppressor is required for stress erythropoiesis. Embo J. 2004;23(21):4319–29. doi: 10.1038/sj.emboj.7600432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Ruiz S, Santos M, Segrelles C, Leis H, Jorcano JL, Berns A, et al. Unique and overlapping functions of pRb and p107 in the control of proliferation and differentiation in epidermis. Development. 2004;131(11):2737–48. doi: 10.1242/dev.01148. [DOI] [PubMed] [Google Scholar]
- [77].Sage C, Huang M, Karimi K, Gutierrez G, Vollrath MA, Zhang DS, et al. Proliferation of functional hair cells in vivo in the absence of the retinoblastoma protein. Science. 2005;307(5712):1114–8. doi: 10.1126/science.1106642. [DOI] [PubMed] [Google Scholar]
- [78].Guo J, Longshore S, Nair R, Warner BW. pRb, but not p107 or p130 is required for maintenance of enterocyte quiescence and differentiation in small intestine. J Biol Chem. 2008 doi: 10.1074/jbc.M806133200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Berman SD, Yuan TL, Miller ES, Lee EY, Caron A, Lees JA. The retinoblastoma protein tumor suppressor is important for appropriate osteoblast differentiation and bone development. Mol Cancer Res. 2008;6(9):1440–51. doi: 10.1158/1541-7786.MCR-08-0176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Landsberg RL, Sero JE, Danielian PS, Yuan TL, Lee EY, Lees JA. The role of E2F4 in adipogenesis is independent of its cell cycle regulatory activity. Proc Natl Acad Sci USA. 2003;100(5):2456–61. doi: 10.1073/pnas.0138064100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Wikenheiser-Brokamp KA. Rb family proteins differentially regulate distinct cell lineages during epithelial development. Development. 2004;131(17):4299–310. doi: 10.1242/dev.01232. [DOI] [PubMed] [Google Scholar]
- [82].Gu W, Schneider JW, Condorelli G, Kaushal S, Mahdavi V, Nadal-Ginard B. Interaction of myogenic factors and the retinoblastoma protein mediates muscle cell commitment and differentiation. Cell. 1993;72(3):309–24. doi: 10.1016/0092-8674(93)90110-c. [DOI] [PubMed] [Google Scholar]
- [83].Nguyen DX, Baglia LA, Huang SM, Baker CM, McCance DJ. Acetylation regulates the differentiation-specific functions of the retinoblastoma protein. Embo J. 2004;23(7):1609–18. doi: 10.1038/sj.emboj.7600176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Jori FP, Galderisi U, Napolitano MA, Cipollaro M, Cascino A, Giordano A, et al. RB and RB2/P130 genes cooperate with extrinsic signals to promote differentiation of rat neural stem cells. Mol Cell Neurosci. 2007;34(3):299–309. doi: 10.1016/j.mcn.2006.11.009. [DOI] [PubMed] [Google Scholar]
- [85].Fay DS, Han M. The synthetic multivulval genes of C. elegans: functional redundancy, Ras-antagonism, and cell fate determination. Genesis. 2000;26(4):279–84. doi: 10.1002/(sici)1526-968x(200004)26:4<279::aid-gene100>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- [86].Ferguson EL, Horvitz HR. Identification and characterization of 22 genes that affect the vulval cell lineages of the nematode Caenorhabditis elegans. Genetics. 1985;110(1):17–72. doi: 10.1093/genetics/110.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Du W, Dyson N. The role of RBF in the introduction of G1 regulation during Drosophila embryogenesis. Embo J. 1999;18(4):916–25. doi: 10.1093/emboj/18.4.916. [DOI] [PMC free article] [PubMed] [Google Scholar]