Abstract
Mass spectrometric imaging (MSI) is a powerful analytical technique that provides two- and three-dimensional spatial maps of multiple compounds in a single experiment. This technique has been routinely applied to protein, peptide, and lipid molecules with much less research reporting small molecule distributions, especially pharmaceutical drugs. This review’s main focus is to provide readers with an up-to-date description of the substrates and compounds that have been analyzed for drug and metabolite composition using MSI technology. Additionally, ionization techniques, sample preparation, and instrumentation developments are discussed.
Keywords: Drug, Pharmaceuticals, Metabolite, MALDI, SIMS, NIMS, DESI, LAESI, Mass spectrometry, Imaging mass spectrometry (IMS), Mass spectrometric imaging (MSI)
1. Introduction
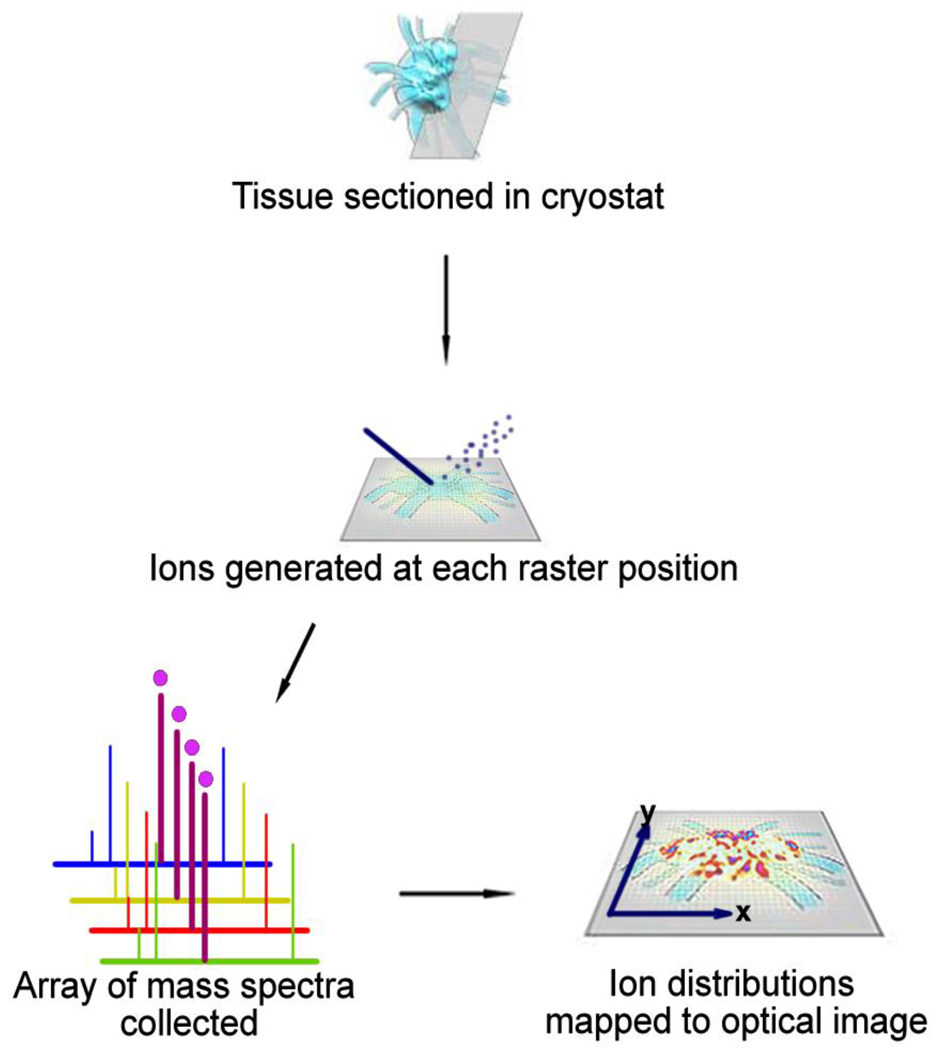
The development of imaging technologies to identify and quantitate molecular species in biological tissue has become essential in pharmaceutical discovery and development. One well-established technique, autoradiography (ARG), uses photographic methods to image the spatial distribution and relationship of radioisotopes inside or on a specimen [1]. ARG is generally divided into two techniques, macroautoradiography, which includes whole-body autoradiography (WBA), and microautoradiography (MARG) [2]. WBA is routinely used for assessing pharmacokinetic and distribution properties of drug candidates in small animals but is unable to distinguish between parent compound and potential metabolite distributions. This disadvantage necessitates coupling WBA with liquid chromatography mass spectrometry (LCMS) of tissue extracts to obtain specific compound and metabolite distributions [2]. An emerging technology, mass spectrometric imaging (MSI), combines the spatial resolution of WBA with the molecular specificity of mass spectrometry. MSI for drugs and metabolites typically involves rastering a laser, or other ionization source, across thin sections of dosed animal tissue by moving the sample stage in predefined x-y coordinates in order to generate thousands of position-dependent mass spectra. Collected mass spectra are then assembled into a data set, and distributions according to specific mass to charge ratios (m/z) can be extracted to display the spatial distribution of an analyte throughout a tissue section. The ability of MSI to perform parallel analyses of multiple molecules in complex samples without labeling gives it a distinct advantage over preexisting methods for label-free and simultaneous detection of drugs and metabolites. Figure 1 shows a general scheme for MSI.
Figure 1.
General scheme of mass spectrometry imaging technology. Acquisition occurs by collecting mass spectra for each pixel and processing this array into representative 2D images of specific m/z.
This review’s main focus is to provide readers with an up-to-date description of the substrates and compounds that have been analyzed for drug and metabolite composition using MSI technology. Table 1 summarizes the above emphasis. Recent reviews on small molecule MSI have highlighted the complementary nature of MSI to autoradiography [2] and described the technology developments in the field [3]. In this review, ionization techniques, sample preparation, and instrumentation developments in the field are described in detail.
Table 1.
Summary of drugs and metabolites studied using MSI
| Primary Analyte | Analyte Metabolites Studied?a |
Tissue Type | Ionization Method |
Detection Mode | Instrument | Analyte Dosingb | Reference |
|---|---|---|---|---|---|---|---|
| AQ4N | + | tumour tissue (human) | MALDI | MS | AB Qstar Pulsar i | 100 mg/kg | [23] |
| artesunic acid | − | artesunate tablets | DESI | MS | LCQ DECA XP+ quadrupole ion-trap | - | [58] |
| ATP (primary metabolites) | + | brain (rat) | MALDI | MS and MS/MS | AB 4800 TOF/TOF | - | [42] |
| + | tumour tissue (human) | MALDI | MS | AB Qstar Pulsar i | - | [23] | |
| biflavonoids | + | plant (A. thaliana and Hypericum) | LDI | MS and MS/MS | Bruker Daltonics Autoflex III MALDI TOF/TOF | - | [61] |
| BMS-X-P (proprietary drug candidate) | + | liver, heart, lung, spleen (rat) | MALDI | MS/MS | Finnigan LTQ linear ion trap | 30 to 1000 mg/kg | [45] |
| chlorisondamine | − | brain (rat) | MALDI | MS and MS/MS | AB 4700 TOF/TOF | 20 mg/kg | [49] |
| cocaine | − | brain (rat) | MALDI | MS and MS/MS | AB 4700 TOF/TOF | 20 mg/kg | [49] |
| clozapine | + | brain (rat) | NIMS | MS | Bruker Daltonics Autoflex III MALDI TOF/TOF and AB Sciex 4700 TOF/TOF | 3 mg/kg | [13] |
| + | brain (rat) | MALDI | SRM | AB Qstar Pulsar i | 5 mg/kg | [24] | |
| + | brain, lung, kidney, testis (rat) | DESI | MS/MS | Thermo-Fisher Scientific LTQ | 50 mg/kg | [52] | |
| − | kidney (rat) | MALDI | MS and MS/MS | Bruker Daltonics Ultraflex III TOF/TOF | 50 mg/kg | [30,46] | |
| epicuticular was metabolites | + | plant (arabidopsis) | LDI (Ag colloid) | MS | Thermo Finnigan LTQ LIT with vMALDI source | - | [60] |
| erlotinib | + | liver, spleen, muscle (rat) | MALDI | SRM | AB Qstar XL | 5 mg/kg | [47] |
| flavonoids | + | plant (A. thaliana and Hypericum) | LDI | MS and MS/MS | Bruker Daltonics Autoflex III MALDI TOF/TOF | - | [61] |
| glucosinolates | + | plant (A. thaliana) | MALDI | MS and MS/MS | Waters MALDI micro MX MALDI TOF | - | [63] |
| ketoconazole | − | porcine skin | MALDI | MS | AB Qstar Pulsar i | - | [44] |
| multiple active pharmaceutical components | − | pharmaceutical tablets | MALDI | MS | AB Qstar Pulsar i hybrid quadrupole TOF | various | [59] |
| mystery compound | + | whole body (rat) | MALDI | MS | AB 4700 TOF/TOF | 0.5 mg/kg | [48] |
| naphthodianthrones | + | plant (A. thaliana and Hypericum) | LDI | MS and MS/MS | Bruker Daltonics Autoflex III MALDI TOF/TOF | - | [61] |
| nelfinavir | − | cells (Mono Mac 6) | MALDI | MS | Bruker Daltonics Ultraflex III and Apex IV Q 9.4 T | 0.1 to 10 µM | [29] |
| nicosulfuron | + | plant (sunflower) | MALDI | MS | AB Qstar Pulsar i quadrupole TOF with oMALDI 2 orthogonal MALDI source | 20, 40 ppm and 1.25 mg/mL | [62] |
| olanzapine | + | whole body (rat) | MALDI | MRM | AB Qstar XL | 8 mg/kg | [25] |
| oxaliplatin | + | kidney (rat) | MALDI | MS | AB 4800 Plus TOF/TOF | 5 mg/mL | [43] |
| paclitaxel | − | liver, cancer tissue (rat) | MALDI | MS/MS | home built instrument: Finnigan ITS-40 ion trap with MALDI ion source | 10 to 50 mg/kg | [40] |
| paracetamol | − | multilayer drug beads | SIMS | MS | Physical Electronics TRIFT II | - | [53] |
| prednisolone | − | multilayer drug beads | SIMS | MS | Physical Electronics TRIFT II | - | [53] |
| primary metabolites | + | plant (peace lily and zebra plant) | LAESI | MS and MS/MS | Waters Q-TOF Premier | - | [20] |
| propranolol | − | whole body (mice) | DESI | SRM | AB Sciex 4000 QTRAP | 7.5 mg/kg | [19] |
| rapamycin | − | poly(lactic-co-glycolic acid) | SIMS | MS | Ion-TOF | 5–50% (w/w) | [54] |
| saquinavir | − | cells (Mono Mac 6) | MALDI | MS | Bruker Daltonics Ultraflex III and Apex IV Q 9.4 T | 0.1 to 10 µM | [29] |
| SCH 226374 | − | brain (mouse) | MALDI | SRM | AB Qstar Pulsar i | 80 mg/kg | [4] |
| sirolimus | − | poly(lactic-co-glycolic acid) | SIMS | MS | Ion-TOF IV and TRIFT V nanoTOF | 0–50% (w/w) | [54,56] |
| SSR180711 | − | kidney (rat) | MALDI | MS and MS/MS | Bruker Daltonics Ultraflex III TOF/TOF | 30 mg/kg | [30,46] |
| theophylline | − | multilayer drug beads | SIMS | MS | Physical Electronics TRIFT II | - | [53] |
| tiotropium | − | lung (rat) | MALDI | MS/MS | Bruker Daltonics UltraFlex II | 1 mg/kg | [27] |
| − | lung (rat) | MALDI | MS and MS/MS | Thermo LTQ Orbitrap XL | - | [28] | |
| vinblastine | − | whole body (rat) | MALDI | IM-MS/MS | Waters Synapt HDMS | 6 mg/kg | [26] |
analtyes were ("+") or were not ("−") analyzed in the work.
("-") analyte dosing was not specified or analyte was endogenous
2. Ionization Techniques
Matrix-assisted laser desorption/ionization (MALDI) requires matrix application on tissue to desorb and ionize compounds with a laser and is particularly useful for protein, peptide, and oligo-nucleotide detection. Matrix and instrumentation developments have expanded MALDI’s mass range so that this method can image small-molecule pharmaceuticals, metabolites, and lipids [3–5]. Unfortunately, MALDI MSI has relatively poor spatial resolution (~20–100 µm), making it incapable of imaging at the cellular level [6]. Furthermore, the sensitivity and spatial resolution of MALDI depend on matrix selection and application. One MALDI variant, scanning microprobe MALDI (SMALDI), has been shown to be capable of imaging biological tissue at ~5 µm [7]. A detailed description of this technique is given in the instrumentation developments section. Another technique that provides greater spatial resolution than MALDI is secondary ion mass spectrometry (SIMS). SIMS is a matrix-free approach that directs an ion beam at a tissue surface to eject “secondary” ions that are drawn into a mass analyzer, typically a time-of-flight (TOF) analyzer [8]. Aun+ and Bin+ (n=1–7) cluster liquid metal ion guns (LMIGs) allow for high resolution imaging of small molecules in tissue but cause significant subsurface damage to tissue upon primary ion impact, limiting the useful spatial resolution to about 1 µm [9]. For this reason, dual beam analyses in SIMS can be performed. In this analysis, a well-focused LMIG probes the tissue’s surface chemistry between polyatomic etch cycles to obtain resolutions below 1 µm [10]. Although SIMS has unmatched spatial resolution and is matrix-free, its hard ionization results in extensive fragmentation, and commercial SIMS instruments lack tandem MS capabilities important for metabolite identification [11–12]. Another matrix-free approach, nanostructure initiator mass spectrometry (NIMS), utilizes “initiator” molecules trapped in nanostructured surfaces to release/ionize intact molecules adsorbed from the surface upon laser irradiation. This method has imaged xenobiotics and metabolites in tissue at a spatial resolution of 15–20 µm, and appears to be ideally suited to small molecule ionization [13–16].
In the above MSI ionization techniques, tissues need to be introduced into high vacuum regions and subjected to laser or ion beam irradiation. To mitigate these conditions, several ambient ionization techniques have been developed and applied to MSI of drugs and metabolites. One such technique, desorption electrospray ionization (DESI), channels charged droplets and ions of solvent from an electrospray source onto a surface, yielding gaseous ions that enter the mass spectrometer via an elongated capillary source [17]. DESI MSI is useful for fast screening purposes, but unfortunately suffers from poor spatial resolution (~180–200 µm), although values of ~40 µm have been reported on non-biological tissues [18–19]. Another ambient ionization technique, laser ablation electrospray ionization (LAESI), is designed for biological samples containing water [20]. Using this mode, a mid-IR laser excites –OH vibrations in a sample’s water molecules, and phase explosion causes a rapid microscale ablation, ejecting a mixture of molecules, clusters, and particulate matter from the surface. Ejected biomolecules coalesce with charged droplets formed from an electrospray source, and a portion of them are converted into gas-phase ions for introduction into a mass analyzer [21]. Similar to DESI, LAESI also suffers from relatively poor resolution (300–400 µm) [20].
3. Sample preparation
3.1. MALDI sample preparation
The most critical step of a mass spectrometric imaging experiment is sample preparation. This is especially true for MALDI based approaches where a matrix is deposited onto the tissue surface to extract analyte molecules and facilitate the desorption/ionization process. The selection of matrix and matrix deposition technique can determine the success or failure of an experiment. Here, we will discuss the general workflow for an imaging experiment as well as the types of matrices and application techniques that have been successful in drug and metabolite analysis.
Most drug and metabolite MSI experiments begin with tissue collection from dosed animals. After harvesting the tissue, it is collected and immediately snap frozen and placed in a −80 °C freezer until sectioning. A cryostat is used to slice the tissue into thin sections (3 µm to 50 µm). These thin tissue sections are then transferred to a conductive stainless steel plate or indium-tin oxide (ITO)-coated glass slide, freeze-dried, and stored at −80 °C until matrix application. Matrix has been applied to drug dosed tissue using wet-spray deposition (pneumatic air sprayer [4, 22–26], a commercial matrix applicator [27–28]), sublimation [29], dry-coating [30], or use of a homemade nanospotter [28] and allowed to dry prior to mass spectrometric analysis. The matrix-coated tissue is then subjected to MSI by rastering a laser across the area of interest in predefined x-y coordinates by moving the sample stage. Mass spectra are collected from each sampling point and reassembled into a 2D image using post acquisition software.
There are several matrices that have been used for MSI. The most commonly used matrices in drug and metabolite imaging experiments are 2,5-dihydroxybenzoic acid (DHB), α-cyano-4-hydroxycinnamic acid (CHCA), and sinapinic acid. The selection of a matrix for targeting a small molecule is often empirical and necessitates the need to try several matrices to find the one that provides the greatest selectivity for an analyte without introducing interfering matrix ions. Recently a more rational approach to matrix selection has been developed for metabolomic studies which employs an “ionless matrix” (1,8-bis(dimethylamino)naphthalene (DMAN)) devoid of interfering ions [31]. This matrix is most suitable for negative-ion mode analyses whereas the previous mentioned matrices are suitable for positive-ion mode analyses. In any case, it is suggested that a researcher investigates several matrices before performing large scale imaging studies.
3.2. SIMS sample preparation
One advantage of SIMS compared to MALDI is that only drying is necessary after tissue sectioning and attachment. Surface modification approaches like metal-assisted-SIMS (MetA-SIMS) and matrix-enhanced SIMS (ME-SIMS) improve secondary ion yields of larger molecules, but have limited applicability to drug and metabolite imaging [32–33]. Briefly, MetA-SIMS involves depositing a thin layer (~1 nm) of metal (e.g., gold or silver) on the sample surface by a sputter coater. ME-SIMS typically uses electrospray deposition for matrix application to ensure that crystal size will not hinder resolution. As mentioned above, primary ion beam choice is critical in SIMS experiments. High-energy atomic primary ions like Ga+ and In+ LMIGs analyze tissue sample surfaces with excellent lateral resolutions in the nanometer range but inflict severe subsurface chemical damage [34]. Consequently, ion impacts must be limited, decreasing the number of ions collected per unit area and negatively influencing spatial resolution. The minimal yield of molecular ions above m/z 300 compounds the sensitivity limitations from these LMIGs. To increase secondary ion yield, Aun+ and Bin+ (n=1–7) cluster LMIGs coupled with TOF SIMS instruments are used, but as mentioned above, subsurface chemical damage still occurs [9]. Molecular primary ions like SF5+ and C60+ deliver substantial increases in secondary ion yields, generating minimal subsurface damage and allowing for depth profiling [35–36]. Unfortunately, these molecular ion beams are harder to focus than liquid metal beams. For this reason, experiments concentrating on molecular depth profiling and maximum resolution use a C60 source to etch away surface monolayers while a LMIG source acquires images with high lateral resolution [37].
3.3. NIMS sample preparation
Sample preparation for NIMS is similar to MALDI except the tissue needs to be sliced less than 5 µm thick and no matrix is applied [13]. Briefly, after collecting and snap-freezing the tissue it is sliced to 3 to 5 µm sections and thaw-mounted onto a NIMS plate. The tissue is then allowed to dry for ~ 30 min before mass spectrometry imaging is performed.
3.4. DESI sample preparation
DESI is a mass spectrometry technique that can be used for imaging biomaterials at ambient pressures. The sample preparation method is again similar to MALDI except that matrix does not need to be applied to the surface thereby mitigating the chances of analyte delocalization prior to analysis [38]. Briefly, after collecting and snap-freezing the tissue it is sliced and placed on glass slides. The tissues are then stored in a dry atmosphere until DESI analysis.
3.5. LAESI sample preparation
Sample preparation for LAESI is similar to DESI. However, the sample should not be dried and there is no need to section plant samples prior to analysis [21, 39].
4. Drug and metabolite imaging in animal tissues
The majority of drug/metabolite MSI studies have been conducted using the MALDI strategy, but more recently matrix free strategies, including SIMS, NIMS, DESI, and LAESI, have been investigated because of their simpler sample preparation steps and lack of a matrix. Here we will discuss the experiments that utilized MALDI, SIMS, NIMS, DESI, and LAESI for drug and metabolite imaging in biological tissues. This review does its best at only presenting reports of pharmaceutical drugs and metabolites, but interesting imaging studies of primary metabolites (AMP, ADP, ATP, etc) and of plant metabolites are also briefly described. A summary of the different drugs and metabolites that have been subjected to MSI analysis is listed in Table 1.
4.1. MALDI applications
One of the earliest examples of MALDI MS profiling of drugs in tissue was presented by Troendle et al. in 1999. The authors constructed a custom laser microprobe quadrupole ion trap mass spectrometer equipped with a MALDI ion source to investigate MALDI’s ability to detect the pharmaceutical compound paclitaxel (MW 853) in intact tissue [40]. The novel ion trap instrument was constructed to minimize the spectral noise in the low mass region generated from matrix ions in order to allow confident identification of low mass analytes in MS and MSn scans [41]. Experimentally, a 2.0 mg liver section was incubated for 60 minutes in a methanol solution containing 100 ng of the anticancer drug. In addition, human tumor tissue that had been transplanted into a mouse which was intravenously dosed with 10 to 50 mg/kg paclitaxel was analyzed by MALDI as well. Unambiguous identification of paclitaxel was achieved by comparing collisionally activated dissociation (CAD) mass spectra of the analyte cations in the two tissue specimens with the CAD mass spectra of the standard drug. This work demonstrated the potential of mass spectrometry to detect drugs in biological tissues, but it took another four years before the first MALDI MSI paper of these analytes was published.
In 2003 the Caprioli group reported the first images of pharmaceutical compounds in animal tissue using MALDI MSI [4]. In these experiments spatial distributions of two antitumor drugs (SCH 226374 and “Compound A”) in mouse brain were elucidated using a quadrupole time-of-flight (QTOF) instrument operated in selected reaction monitoring (SRM) mode. SRM mode was selected to provide the greatest sensitivity for tissue analysis. This work demonstrated the proof-of-principle that MALDI MSI could be used for biological tissues to monitor drug localization post dosing.
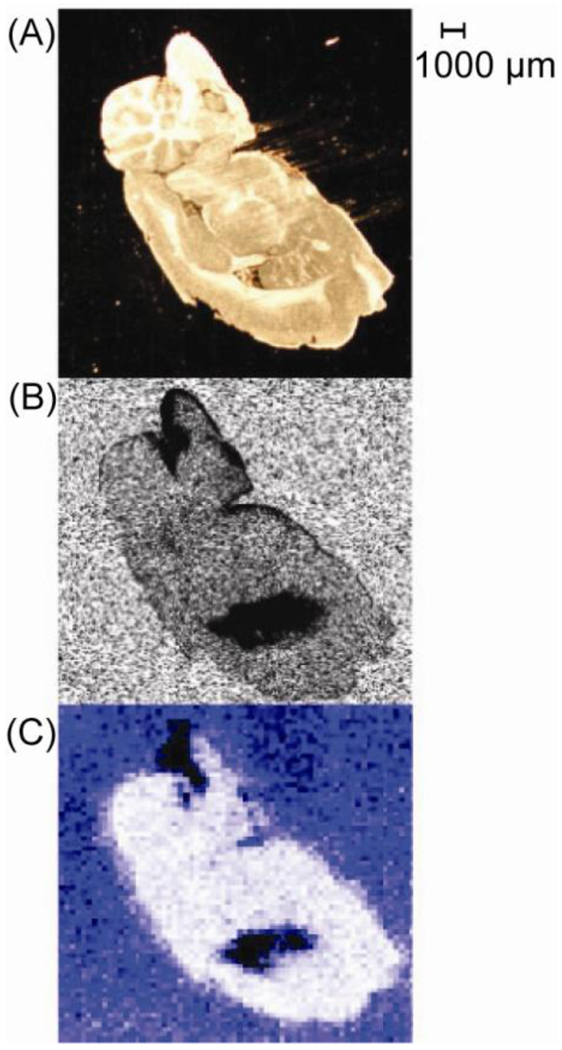
Hsieh et al. (2006) built upon Reyzer’s advancement by addressing two important questions: 1) The effect matrix deposition has on analyte delocalization and 2) How well MALDI MSI data compares to autoradiography images of the same tissue [24]. The first question was addressed by using a microsyringe to place clozapine and norclozapine rich droplets onto rat brain to form the two letters S and P. Matrix was then applied with an airbrush followed by mass spectrometric imaging. The results clearly demonstrated that when matrix is applied with care to tissue the redistribution of drugs is minimal. In addressing the second question, the group found that both autoradiography and MALDI MSI indicated that clozapine is distributed throughout the brain with the highest concentrations being located in the lateral ventricle (Figure 2). The results presented in this paper indicated that MALDI MSI provides similar results compared to the more expensive autoradiography technique without sacrificing spatial resolution.
Figure 2.
Distribution of clozapine in rat brain. (A) Optical image. (B) Autoradiography image. (C) MALDI MS/MS image. Adapted with permission from Ref. [24].
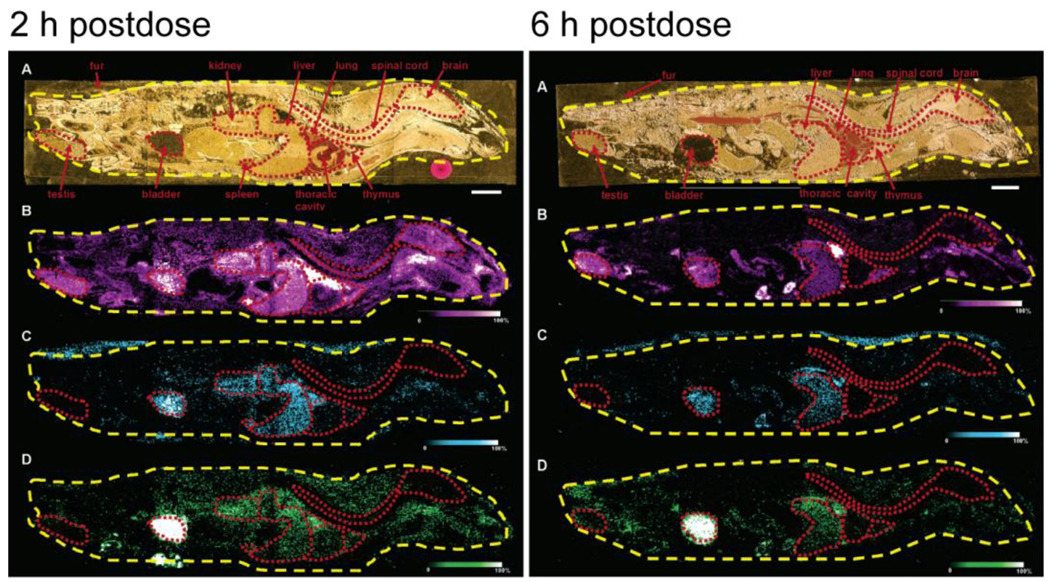
A limitation of autoradiography is that it is impossible to differentiate drug and metabolite distributions from one another. Khatib-Shahidi et al. reported that MALDI MSI has the ability to provide spatial information for both drugs and their metabolites separately (Figure 3) [24]. In this work the drug olanzapine was administered to 10-week old male rats and the dosed animals were sacrificed the day following administration. Whole-body tissue sections were collected, mounted onto MALDI target plates, and analyzed using an Applied Biosystems QSTAR XL mass spectrometer. Figure 3 shows that the drug olanzapine (B) and its metabolites N-desmethyl metabolite (C) and 2-hydroxymethyl metabolite (D) have different distributions 2 hr and 6 hr post dosing. The demonstration of simultaneous imaging of both drug and metabolite in the same tissue section showcases MALDI MSI’s advantage over conventional WBA.
Figure 3.
Detection of drug and metabolite distribution at 2 hr and 6 hr postdose in a whole rat sagittal tissue section by MALDI IMS. (A) Optical image of post OLZ dosed tissue section across four gold MALDI target plates. (B) Organs outlined in red. MS/MS ion image of OLZ (m/z 256). (C) MS/MS ion image of N-desmethyl metabolite (m/z 256). (D) MS/MS ion image of 2-hydroxymethyl metabolite (m/z 272). Bar, 1 cm. Adapted with permission from Ref. [25].
There are approximately another dozen original reports of MALDI MSI of drugs and metabolites in biological tissues in the literature [22–23, 26–30, 42–49] (Table 1). A general theme of all of these studies is the use of tandem mass spectrometry to verify analyte peak identification among the matrix and endogenous small molecule chemical noise in the low mass range. The use of high resolution mass spectrometers and novel gas phase ion separation techniques have been reported in more recent drug imaging studies to reduce the degree at which low mass chemical noise affects confident MS identification with great success [26, 28–29].
4.2. DESI and NIMS
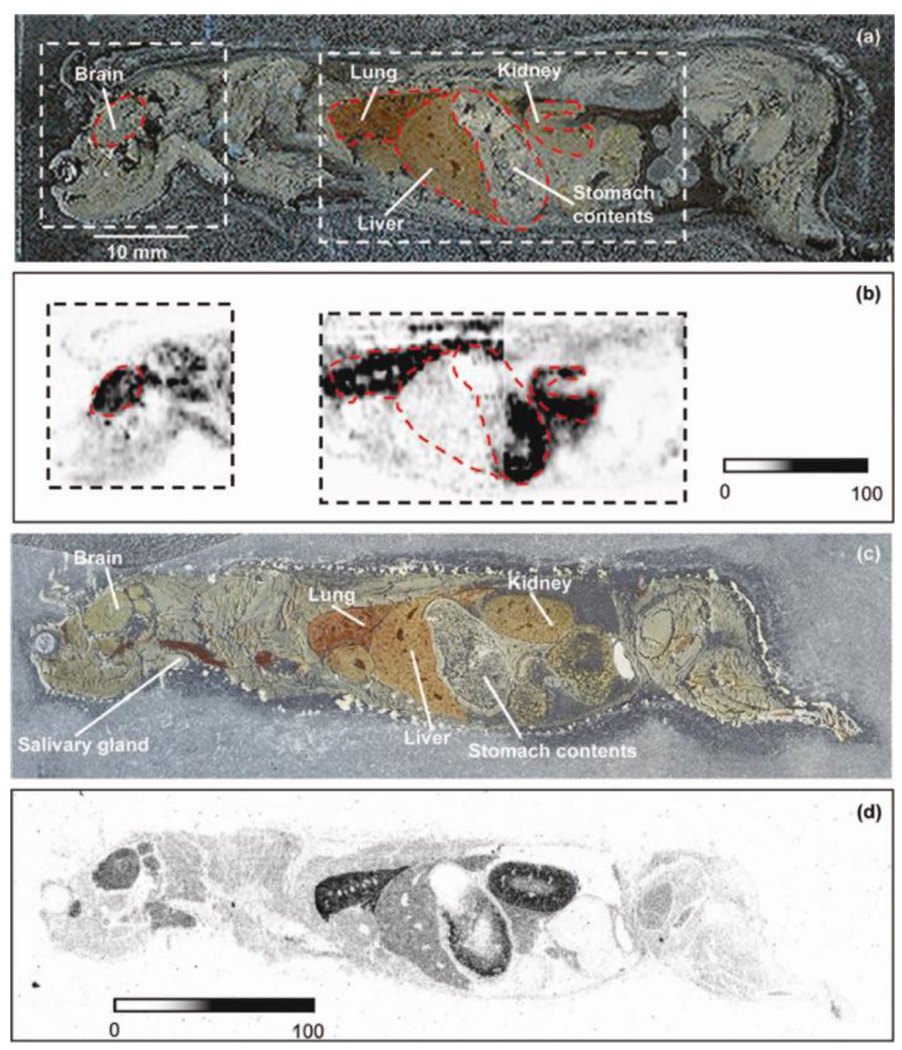
In addition to mass analyzer advancements, two alternative ionization methods have been developed and utilized in drug/metabolite MSI studies. These two methods are desorption electrospray ionization (DESI) [17] and nanostructure initiator mass spectrometry (NIMS) [50]. Both of these ionization methods do not require matrix, thereby eliminating the chemical noise that plagues low mass analysis in MALDI platforms. Two separate DESI studies have reported successful imaging of drugs propranolol and clozapine in mouse whole-body and rat tissue sections [38, 51–52]. These studies have shown that DESI imaging results are comparable to WBA results (Figure 4). An advantage of DESI compared to MALDI experiments is that a whole body mouse tissue can be imaged in roughly 79 minutes whereas it would take hours for a MALDI experiment to finish. This is due to the ionization efficiency of the method as well as the decreased imaging spatial resolution inherent to the method (0.25 to 1 mm). Recent advances in DESI operational parameters have facilitated increased spatial resolution down to 40 µm to be achieved, but these parameters have only been applied to nonbiological surfaces (e.g. paper and thin-layer chromatography plates) [19].
Figure 4.
Detection of propranolol in whole mouse sagittal tissue by DESI-MS/MS. (a) Scanned optical image of a 40 µm thick sagittal whole-body tissue section of a mouse dosed intravenously with 7.5 mg/kg propranolol and euthanized 20 min after dose. (b) Distribution of propranolol in 20 mm × 20 mm and 38 mm × 20 mm areas measured by DESI-MS/MS (SRM: m/z 260 → 116) using 80/20 (v/v) methanol/water as DESI solvent at a flow rate of 5 µL/min. Surface scan rate was 0.1 mm/s, dwell time was 100 ms, and the images were created from 41 lanes with 500 µm spacing. Pixel size was 84 µm (h) × 500 µm (v), and experiment times were 150 and 285 min for the 20 mm × 20 mm and 38 mm × 20 mm areas, respectively. (c) Scanned optical image of a 40 µm thick sagittal whole-body tissue section of a mouse dosed intravenously with 7.5 mg/kg [3H]propranolol and euthanized 20 min after dose. (d) Autoradioluminograph of [3H]propranolol-related material in the tissue section presented in (c). Adapted with permission from Ref. [38].
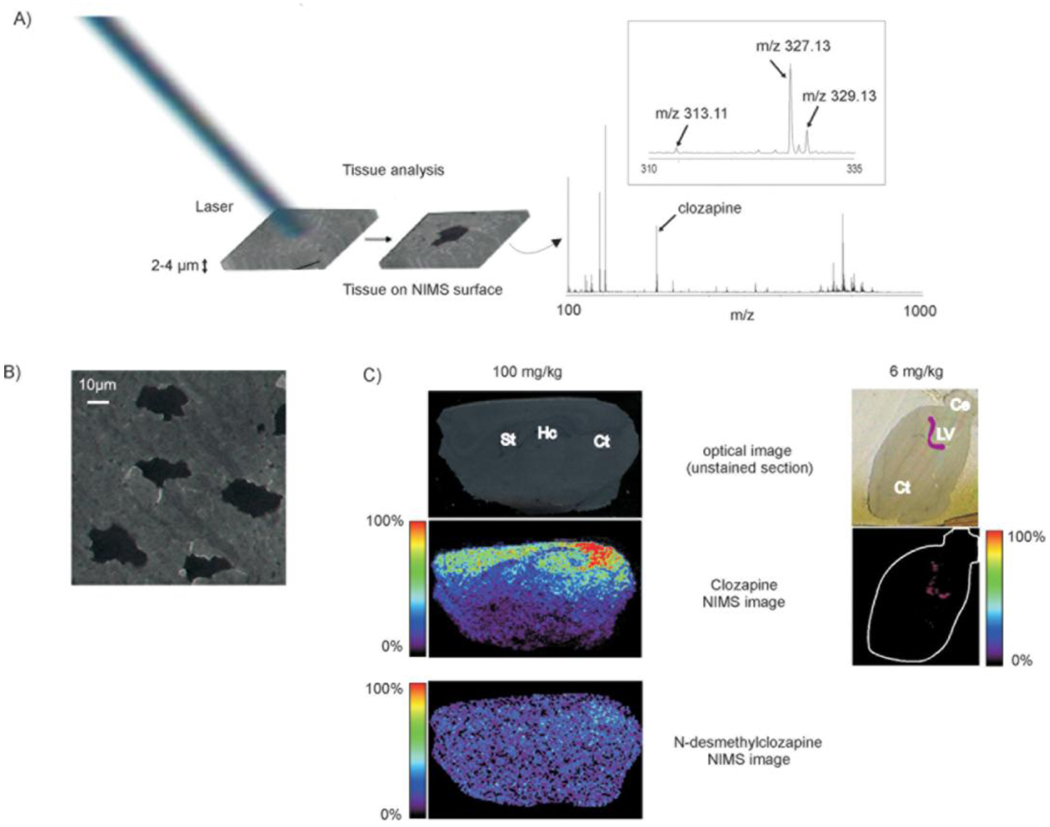
More recently NIMS has been a pplied to the imaging of clozapine and its metabolites in dosed rat brains using TOF/TOF mass analyzers [13]. Figure 5 shows the imaging results of 100 mg/kg and 6 mg/kg clozapine dosed rats. The indication that clozapine is concentrated in the lateral ventricle agrees with earlier MALDI MSI and autoradiography experiments showing the same distribution [24]. The inset of Figure 5 shows the minimal interferences present in the low mass region when using this ionization method. NIMS MSI is compatible with both ion beam and laser sources available on commercial SIMS and MALDI instruments. In addition, fewer laser shots are required per spot as compared to MALDI methods. This is because all the analyte is removed from the NIMS surface after a few shots whereas the matrix used in MALDI experiments is ablated slowly and is rich in analyte. A drawback to NIMS imaging is that it requires very thin tissue slices (3 to 5 µm) that are difficult to collect serially using a manual cryostat. If serial sections are needed for a study then an automatic slicing cryostat is recommended.
Figure 5.
The concept of tissue imaging with laser-NIMS: (A) The tissue slice (2–4 µm thick sections) is placed directly on the NIMS surface and is subjected to laser irradiation (~0.1 J/cm2 pulse)) resulting in desorption/ionization of endogenous metabolites and xenobiotics. The full-MS mode mass spectrum shows the presence of intact clozapine (m/z 327.13) and N-desmethylclozapine (m/z 313.11) in the brain tissue. Inset shows the isotopic distribution of clozapine characterized by the chloride atom. (B) Nitrogen laser beam irradiation produces 15–20 µm diameter “etched” areas (black area). The photograph shows the sagittal unstained section of a mouse brain after MS acquisition on NIMS chip (scale bar: 10 µm). (C) Top, unstained sections of brain slices (sagittal) before NIMS analysis. Hc, hippocampus; St, striatum; Ce, cerebellum; LV, lateral ventricle; Ct, cortex. Middle, NIMS clozapine images (dose: 100 mg/kg rat and 6 mg/kg mouse). Bottom, NIMS N-desmethylclozapine image (dose: 100 mg/kg). For easy visualization, the purple mark in the unstained section (6 mg/kg) indicates clozapine localization by NIMS. In the NIMS image, the edge of the tissue has been also highlighted in white. Adapted with permission from Ref. [13].
5. Tablet and powder analysis
One important application of MSI in pharmaceutical analysis is imaging of drug release systems like stents, tablets, and powders. In 2000, Belu et al. analyzed three multilayer drug beads that serve as controlled-release drug delivery systems with TOF SIMS [53]. The integrity of each layer was evaluated by imaging specific ion species for the drug (theophylline, paracetamol, prednisolone), excipient, and coating materials at submicrometer resolution. Each bead had a unique distribution of ingredients, and imaging of the active drug showed their distribution ranged from micrometer-sized particles in one bead cross section to almost homogeneous in another bead cross section [53]. Furthermore, TOF SIMS data showed the ingredients of the bead did not exactly match the manufacturer’s specifications. TOF SIMS has also analyzed drug eluting stent coatings [54–56]. One group employed cluster SIMS with a SF5+ polyatomic primary ion sputter source and Bi3+ analysis source [54]. Depth profiles indicated that in stent coatings of various compositions, the drug sirolimus tended to diffuse to the surface, forming a diffusion profile. The diffusion profile consisted of a drug-enriched surface region, a drug-depletion region, and a constant bulk composition region [54]. Another study obtained chemical images of the surfaces and interiors of rapamycin in poly(lactic-co-glycolic acid) (PLGA) coatings [55] using surface sensitive electron microscopy for chemical analysis (ESCA), TOF SIMS, and Raman microscopy. As rapamycin concentration was increased from 5 % (wt.) to 50 % (wt.) in the rapamycin/PLGA coating, increased segregation within the bulk of the coating was apparent [55]. However, all formulations except for the cap-coated stent contained a significant enrichment of rapamycin at the surface. The 3D spatial distribution of pharmaceutical molecules in a coronary stent coating was analyzed in order to visualize drug distribution according to elution time of the stent [56]. The TOF SIMS instrument analyzed the 3D localization of sirolimus in PLGA matrix as a function of elution time with an Au+ LMIG coupled to a C60+ ion beam. The examined stent contained large portions of the surface and subsurface channels composed primarily of sirolimus, followed by a drug-depleted region, and then relatively homogeneous sirolimus dispersion in the polymer matrix. In order to determine pharmaceutical elution from the stent, 3D chemical distributions were analyzed with TOF-SIMS by characterizing stents at 0 h, 1 h, and 1 day. Elution was found to occur on the drug-enriched surface region much quicker compared to the gradual elution in the subsurface regions, revealing that much of the drug (~55%) had eluted in the first day [56].
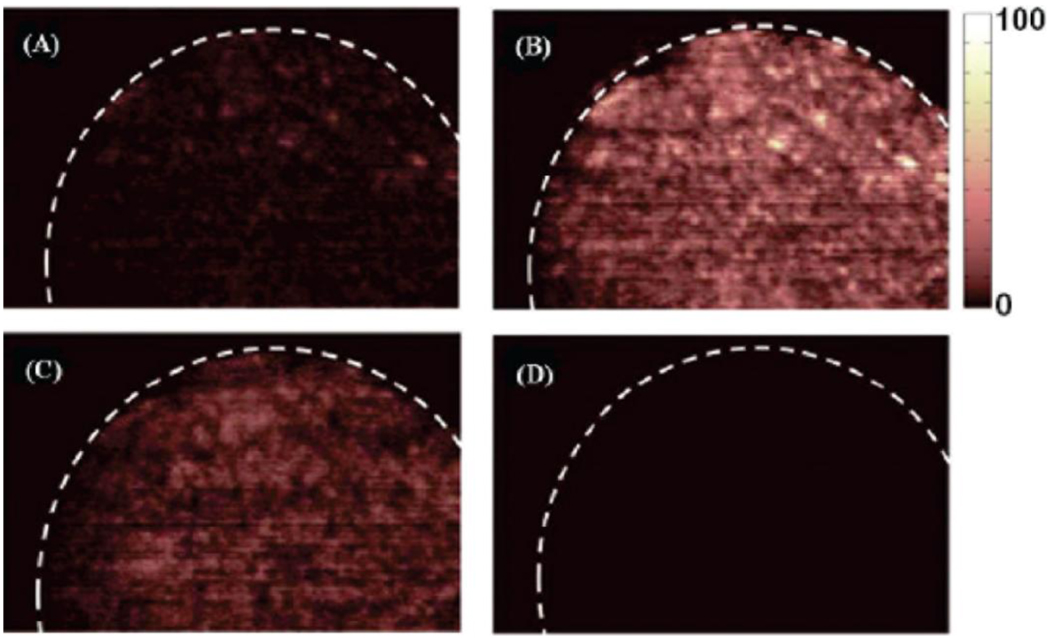
DESI MSI research has imaged drug tablets and powders with limited sample preparation. The Hopfgartner lab performed simple and rapid DESI-MS analysis of drug tablets and powders without sample preparation [57]. A home-made DESI source coupled to a triple-quadrupole linear-ion trap (QqQLIT) mass spectrometer enabled analysis of twenty-one commercial drugs and illicit Ecstasy tablets and powders. Although imaging was not performed in these experiments, the authors found that DESI identified different ions at different locations on the tablet surfaces and hypothesized that DESI MSI could be useful [57]. One DESI-MSI study of pharmaceuticals combined two-dimensional diffusion-ordered nuclear magnetic resonance spectroscopy (2D DOSY 1H NMR) with direct analysis in real-time mass spectrometry (DART MS) and DESI [58]. The study’s aim was to assess the complementarity of these methods for pharmaceutical forensics of counterfeit medicines like fake artesunate-based antimalarial drugs. One group used DESI MSI to test their hypothesis that chemical species were distributed heterogeneously in counterfeit tablets. DESI MS imaging results showed that artesunatic acid was homogeneously distributed on the tablet surface. Surprisingly, they also showed that in fake tablets, acetaminophen was distributed homogeneously in the sample, as shown in Figure 6. However, artesunate was not detected in the fake tablets (Figure 6) [58].
Figure 6.
DESI MS images of a counterfeit artesunate sample: (A) sodiated acetaminophen (m/z 174.1), (B) sodiated acetaminophen dimer (m/z 325.1), (C) sodiated lactose (m/z 365.3), and (D) sodiated artesunic acid monomer (m/z 407.2). Adapted with permission from Ref. [58].
A MALDI MSI study also directly analyzed a range of pharmaceutical tablets to assess the homogeneity of the active drug compound [59]. Commercially available and prescription tablets including aspirin, paracetamol, sildenafil citrate (Viagra®) and a batch of tablets in development were analyzed. In these experiments, tablet sections ~1mm thick were crafted with a tablet cutter to fracture the tablet and expose the surface. Tablet surfaces were then coated with a matrix (α-CHCA 25 mg/mL in EtOH with 0.1% TFA) by airspray before MALDI analysis. Although results indicated that MALDI MSI is an effective tool to analyze spatial distributions of active pharmaceutical components in tablets, tablet shape and texture controls the effectiveness and feasibility of this approach [59]. Furthermore, an edge effect was observed that suggests position of the laser relative to the tablet section is critical in the success of a tablet imaging experiment.
6. Metabolite imaging in plant tissue or bacteria colonies
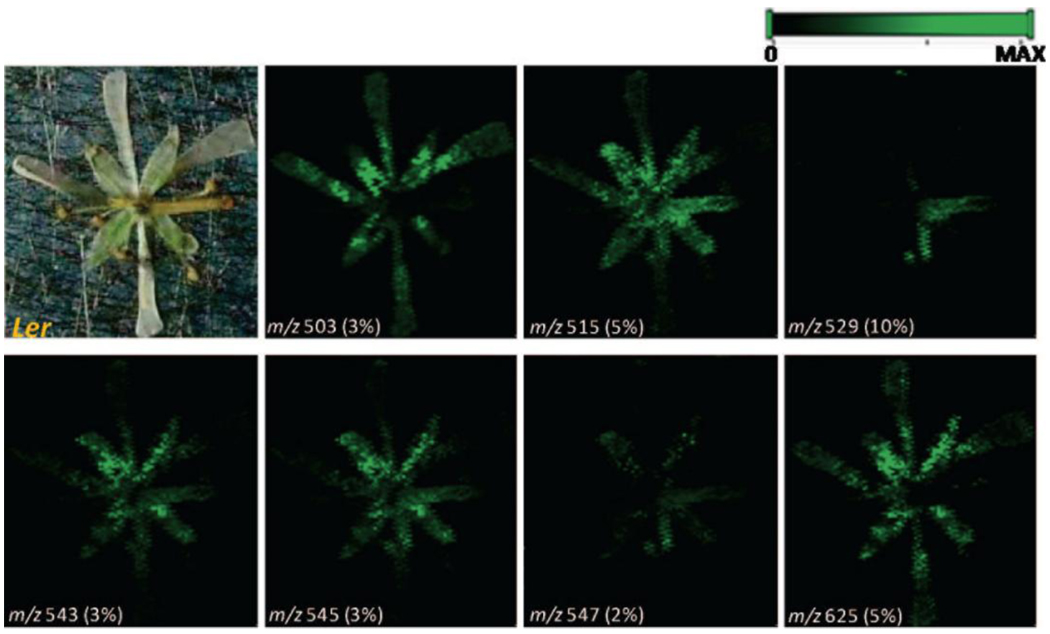
MSI of metabolites and pesticides in plants is another burgeoning imaging application. The Vertes group demonstrated LAESI 3D imaging MS of metabolites in the leaf tissues of Peace lily (Spathiphyllum lynise) and Zebra plant (Aphelandra squarrosa) [39]. LAESI’s ability to depth profile provided ambient molecular imaging at lateral and depth resolutions of ~300 µm and 30–40 µm. MS/MS studies assisted in structural determination of the metabolites, and results showed that 3D distributions of metabolites exhibited tissue-specific accumulation patterns correlating with their biochemical roles in defense and photosynthesis [39]. A novel LDI MSI approach employed colloidal silver as a matrix to profile and image epicuticular wax metabolites on Arabidopsis thaliana leaves and flowers [60]. A controllable spraying device reproducibly deposited colloidal silver solution onto the plant’s surface, generating homogeneous and constant surface coverage. Silver dimer ion intensity was then used as a reference to normalize mass spectra. This method profiled surface metabolites like long-chain fatty acids, alcohols, alkanes, and ketones by detecting them as silver adduct ions from different flower organs (carpels, petals, and sepals) at a spatial resolution of 100 µm [60] as shown by Figure 7. Another LDI MSI publication studied UV-absorbing secondary metabolites in plant tissues of the genera Arabidopsis and Hypericum [61]. Hypericin and pseudohypericin, two naphthodianthrones, were imaged using the Ultraflex III® at 10 µm lateral resolution in the secretory cavities of H. reflexum leaves, and hypericin, pseudohypericin, quercetin, and rutin were also detected. Laser microdissection (LMD) assisted with the difficulties in performing LDI MSI experiments on three-dimensional bulky stamens and styli [61]. Hypericins and biflavanoid signals were also detected on the stigma, resulting in a study to determine if other phenolic compounds could be desorbed from plant material using LDI. Ion images with 10 µm resolution and cell-like structures were obtained with the Ultraflex III®. These ion images contained signals for many putative flavanoids and their glycosides [61]. A MALDI MSI study by Anderson et al. imaged the distribution of the pesticide nicosulfuron (2-[[(4,6-dimethoxypyrimidin-2-yl)aminocarbonyl]aminosulfonyl]-N,N-dimethyl-3-pyridinecarboxamide) in sunflower tissue after root and foliar uptake [62]. Sunflower plants introduced to nicosulfuron were sectioned horizontally at 10, 30, 50, and 140 mm from the root bundle region and coated in CHCA matrix prior to MALDI MSI. Foliar absorption of nicosulfuron was also imaged by applying nicosulfuron to leaf veins and sectioning tissue horizontally at varying distances from the root bundle region after 24 or 48 hours. Nicosulfuron and a nicosulfuron phase 1 metabolite distributions were imaged according to time period and distance from the root region [62]. Another MALDI MSI study aimed to determine the spatial distribution of plant defenses in an Arabidopsis thaliana rosette leaf by imaging three major glucosinolates, 4-methylsulfinylbutylglucosinolate, indol-3-ylmethylglucosinolate, and 8-methylsulfinyloctylglucosinolate [63]. MALDI-TOF images revealed that the glucosinolates were more abundant in the leaf’s midvein and periphery than the inner lamina, correlating with the avoidance of these regions by Helicoverpa armigera (the cotton bollworm) when feeding [63].
Figure 7.
Chemically selective images of Arabidopsis wild-type (Ler) whole flowers. Major ions detected at m/z 529 correspond to [C29 ketone + 107Ag]+. Ions detected at m/z 515 and 543 are mainly silver adduct ions of C29 and C31 alkanes. Ions at m/z 547 are from C30 alcohol, and the image for the peak at m/z 545 corresponds to the overlapped image of C31 alkane and C30 alcohol as [C31 alkane + 109Ag]+ and [C30 alcohol + 107Ag]+. Images for silver adduct ions of C26 fatty acids (at m/z 503) and an unknown compound (at m/z 625) are also shown. Adapted with permission from Ref. [60].
One important and novel application of MALDI-MSI enabled visualization of the spatial and temporal production of metabolites from a single bacterial species as well as observation of multiple microbial signals involved in an interspecies interaction [64]. Application of MSI to an intact bacterial colony was performed by layering a thin film of agar media on top of a MALDI target plate and inoculating with B. subtilis. Following incubation before MSI analysis, a digital image was acquired to record bacterial growth. This MSI method allowed characterization of natural products from B. subtilis and S. coelicolor. Furthermore, data revealed that chemical interactions between bacteria involve multiple signals functioning simultaneously to control the results of interspecies encounters [64].
7. Instrumentation developments
The need to improve MSI performance has led to recent instrumentation developments that are applicable to conducting drug and metabolite imaging. Bruker Daltonics has developed a proprietary kHz smartbeam-II™ MALDI laser integrated with a novel FlashDetector™ on their MALDI TOF/TOF mass spectrometer, ultrafleXtreme™ [65]. This 1 kHz laser allows for focus diameters down to 10 µm for high resolution imaging and a resolving power of more than 40000. Another advantage is the 1kHz data acquisition for TOF and TOF/TOF operation ensures high-throughput sampling and the self-cleaning Perpetual ion source ensures robust operation of the instrument [65]. Similarly, the Karger group has built a MALDI TOF instrument with a 2-kHz laser and ~140 µm spot diameter that would also be capable of high-throughput imaging experiments [66]. A continuous laser rastering technique has been developed by Applied Biosystems that rasters the laser continuously in rows across the sample surface instead of using the conventional but time-consuming “stop-and-go” method of spectra acquisition [67]. Lastly, the Caprioli group developed a MALDI TOF instrument operating in continuous scan mode with a 5 kHz laser to promote significant acquisition time improvements over conventional MALDI TOF systems [68].
The current spatial resolution limits of traditional MALDI sources hinder cellular imaging possibilities with this technology. However, one technique, scanning microprobe MALDI (SMALDI) combines laser microprobe mass spectrometry (LMMS) and MALDI-MSI to enable high spatial resolution imaging of biomolecular substances [69]. Focusing of a laser beam occurs through a prefocusing step outside of the vacuum and another focusing step in the vacuum above the sample stage. Both prefocusing and final focusing steps utilize suprasil quartz lenses to achieve sub-micrometer focusing of the laser beam [69]. SMALDI has been used to investigate cultured A-498 cells of human renal carcinoma and mouse urinary bladder at cellular resolution [7, 70]. SMALDI’s application to biological substances is quite recent, and optimization of matrix preparation procedures must be investigated in order to ensure this technique’s future applicability.
Mass resolving power deficiencies in TOF MS limit its usefulness for MALDI MSI, especially for small molecular weight compounds like drugs and metabolites, due to possible presence of interfering molecular species with similar molecular masses or overlapped isotopic clusters. One alternative, MALDI FT-ICR, has been applied to record accurate mass measurements that provide molecular specificity for ion images of drugs and metabolites in tissue on the basis of elemental composition [71]. Accurate mass fragment ions generated in an external quadrupole-collision cell allowed structural confirmation of targeted compounds. Unfortunately, the low throughput of MALDI FT-ICR hinders its application to imaging studies. In the study above, images contained 1000–1700 pixels and the typical imaging speed was ~0.067 pixels/sec whereas TOF mass analyzers are capable of 0.50–0.83 pixels/sec [71]. High mass resolution and accuracy imaging can also be accomplished with Orbitrap mass spectrometers. One study performed experiments on an LTQ Orbitrap mass spectrometer with a DESI ion source (Thermo Scientific) and used a 30 k mass resolution setting while a 100 k setting with lock masses enabled exact mass measurements for molecular formulae calculations [72]. Lipids in sectioned rat brain were imaged at a rate of 0.94 pixels/sec (230 × 230 µm) but accurate mass data was not always sufficient to unambiguously identify unknown peaks because of isomeric lipid species and unresolved peaks. Our group also performed high mass resolution and accuracy imaging of several neuropeptide families in the lobster Homarus americanus brain using a MALDI LTQ Orbitrap mass spectrometer [73]. In this study, MALDI Orbitrap once again demonstrated its superior throughput by acquiring data at a rate of ~0.15 pixels/sec in addition to excellent mass resolving power and accuracy. Ion mobility based MSI is another technique addressing the low mass resolving power of TOF MS. In ion mobility-mass spectrometry (IM-MS) imaging applications, analyte is ionized by MALDI and directed into the IM drift tube. The drift tube contains a neutral background drift gas that collides with ions traversing the drift cell [74]. Ion separation is due to differences in collisional cross-section (apparent surface area), creating a gas-phase separation dimension orthogonal to the MS dimension useful for identification of isobaric analytes extracted from tissue [74–75]. One IMMS imaging experiment differentiated between an anticancer drug, vinblastine and an endogenous isobaric lipid [26].
SIMS instrumentation has primarily developed through advances in cluster-beam source technology to improve sensitivity, mass range, and lateral resolution for biological imaging applications. Although cluster SIMS experiments are now routine for characterization of complex organic samples, commercial TOF SIMS instruments are not equipped with MS/MS capabilities required for analysis. The Winograd group developed a C60 SIMS hybrid quadrupole orthogonal TOF MS capable of MS/MS [76]. This instrument has mapped the spatial distribution of gramicidin S under a copper grid with ~25–30 µm resolution and is sufficiently sensitive to conduct MS/MS experiments during imaging. The hybrid instrument is also advantageous in that it utilizes a continuous (dc) primary ion beam rather than a pulsed beam to increase incident ion beam current by as much as four orders of magnitude, improving S/N ratio by several orders of magnitude [76]. The Vickerman group also developed an instrument that overcomes the long analysis times and inability to maintain both high spatial and mass resolution inherent in TOF-SIMS [77]. This instrument removes the pulsed primary ion beam and employs a continuous stream of secondary ions from a surface with a primary ion beam in dc mode. Reports showed this hybrid imaging instrument has the mass detection and range of a TOF analyzer and the ability to perform nanoscale imaging without sacrifice of mass resolution. Furthermore, the C60 ion beam delivers an ultimate spot size of 200 nm while still maintaining a mass accuracy of 5 ppm [77]. The instrument’s MS/MS capabilities enable further structural elucidation for drugs and metabolites, and it has provided high mass resolution subcellular imaging in 2D and 3D.
8. Conclusions-outlook
Recent advancements in the field of drug and metabolite MSI have shown that this technique has great potential in pharmaceutical discovery and development. Multiple ionization techniques ensure a variety of options for imaging drugs and metabolites in multiple tissue types at different spatial resolutions. Although sample preparation for MALDI MSI is somewhat involved, matrix-free ionization methods like NIMS and ambient m ethods like DESI and LAESI simplify sample preparation greatly while remaining suitable for small molecule imaging. MALDI, DESI, and NIMS have been employed to image drugs and metabolites in animal tissues while SIMS has been heavily utilized for pharmaceutical detection in drug release systems. DESI has also been utilized for imaging of counterfeit tablets, and LAESI has shown promise in metabolite imaging in plant tissue.
Instrumentation developments are improving almost all areas of MSI. High repetition rate lasers, continuous laser rastering, and self-cleaning sources have increased the throughput and robustness of laser ionization sources. Furthermore, tandem mass analyzers and high mass resolving power instruments have improved the confidence of MSI measurements and structural characterization. Nonetheless, there are still other areas in this field that need to be developed. Sample preparation, assay sensitivity, and the quantitative ability of MSI must be further investigated. In addition, there are no reports of using MSI for drug/protein binding or drug conjugate studies. The current mass spectrometry technology for elucidating protein-drug interactions is drug affinity responsive target stability (DARTS) assays [78–79]. This method uses a bottom-up approach to compare a biological sample treated with a drug to a biological sample not treated with the drug to identify molecular targets.
When compared with the fields of protein and peptide MSI, the field of drug and metabolite MSI is still in its infancy but rapidly maturing. Analysis of these small molecules has been difficult due to the great amount of chemical interferences in the low mass region, but recent advances in mass analyzers’ mass resolution as well as ion transmission optics have increased the confidence and sensitivity of these types of experiments. Continued advancement in sample preparation tec hniques as well as instrument and software development will enable MSI of drugs and metabolites to become routine.
Acknowledgement
Preparation of this manuscript was supported in part by National Science Foundation (CHE-0957784), National Institutes of Health through grant 1R01DK071801. L.L. acknowledges an Alfred P. Sloan Research Fellowship, a Vilas Associate Award, and an H. I. Romnes Faculty Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hahn EJ. Autoradiography: a review of basic principles. Am Lab. 1983;15:64–71. [Google Scholar]
- 2.Solon E, Schweitzer A, Stoeckli M, Prideaux B. Autoradiography, MALDI-MS, and SIMS-MS imaging in pharmaceutical discovery and development. AAPS Journal. 2010;12:11–26. doi: 10.1208/s12248-009-9158-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Svatos A. Mass spectrometric imaging of small molecules. Trends Biotechnol. 2010;28:425–434. doi: 10.1016/j.tibtech.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Reyzer ML, Hsieh Y, Ng K, Korfmacher WA, Caprioli RM. Direct analysis of drug candidates in tissue by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 2003;38:1081–1092. doi: 10.1002/jms.525. [DOI] [PubMed] [Google Scholar]
- 5.Cohen LH, Gusev AI. Small molecule analysis by MALDI mass spectrometry. Anal Bioanal Chem. 2002;373:571–586. doi: 10.1007/s00216-002-1321-z. [DOI] [PubMed] [Google Scholar]
- 6.Todd PJ, Schaaff TG, Chaurand P, Caprioli RM. Organic ion imaging of biological tissue with secondary ion mass spectrometry and matrix-assisted laser desorption/ionization. J Mass Spectrom. 2001;36:355–369. doi: 10.1002/jms.153. [DOI] [PubMed] [Google Scholar]
- 7.Rompp A, Guenther S, Schober Y, Schulz O, Takats Z, Kummer W, et al. Histology by mass spectrometry: label-free tissue characterization obtained from high-accuracy bioanalytical imaging. Angew Chem Int Ed Engl. 2010;49:3834–3838. doi: 10.1002/anie.200905559. [DOI] [PubMed] [Google Scholar]
- 8.Fletcher JS, Lockyer NP, Vickerman JC. Developments in molecular SIMS depth profiling and 3D imaging of biological systems using polyatomic primary ions. Mass Spectrom Rev. 2010 doi: 10.1002/mas.20275. [DOI] [PubMed] [Google Scholar]
- 9.Jones E, Lockyer N, Vickerman J. Mass spectral analysis and imaging of tissue by ToF-SIMS - The role of buckminsterfullerene, C-60(+), primary ions. International Journal of Mass Spectrometry. 2007;260:146–157. [Google Scholar]
- 10.Breitenstein D, Rommel CE, Mollers R, Wegener J, Hagenhoff B. The chemical composition of animal cells and their intracellular compartments reconstructed from 3D mass spectrometry. Angew Chem Int Ed Engl. 2007;46:5332–5335. doi: 10.1002/anie.200604468. [DOI] [PubMed] [Google Scholar]
- 11.Chehade F, de Labriolle-Vaylet C, Moins N, Moreau MF, Papon J, Labarre P, et al. Secondary ion mass spectrometry as a tool for investigating radiopharmaceutical distribution at the cellular level: the example of I-BZA and (14)C-I-BZA. J Nucl Med. 2005;46:1701–1706. [PubMed] [Google Scholar]
- 12.Clerc J, Fourre C, Fragu P. SIMS microscopy: methodology, problems and perspectives in mapping drugs and nuclear medicine compounds. Cell Biol Int. 1997;21:619–633. doi: 10.1006/cbir.1998.0205. [DOI] [PubMed] [Google Scholar]
- 13.Yanes O, Woo H-K, Northen TR, Oppenheimer SR, Shriver L, Apon J, et al. Nanostructure Initiator Mass Spectrometry: Tissue Imaging and Direct Biofluid Analysis. Analytical Chemistry. 2009;81:2969–2975. doi: 10.1021/ac802576q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, et al. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–U3. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 15.Patti GJ, Woo HK, Yanes O, Shriver L, Thomas D, Uritboonthai W, et al. Detection of Carbohydrates and Steroids by Cation-Enhanced Nanostructure-Initiator Mass Spectrometry (NIMS) for Biofluid Analysis and Tissue Imaging. Analytical Chemistry. 2010;82:121–128. doi: 10.1021/ac9014353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reindl W, Northen TR. Rapid Screening of Fatty Acids Using Nanostructure-Initiator Mass Spectrometry. Analytical Chemistry. 2010;82:3751–3755. doi: 10.1021/ac100159y. [DOI] [PubMed] [Google Scholar]
- 17.Takáts Z, Wiseman JM, Gologan B, Cooks RG. Mass spectrometry sampling under ambient conditions with desorption electrospray ionization. Science. 2004;306:471–473. doi: 10.1126/science.1104404. [DOI] [PubMed] [Google Scholar]
- 18.Ifa D, Wu C, Ouyang Z, Cooks R. Desorption electrospray ionization and other ambient ionization methods: current progress and preview. Analyst. 2010;135:669–681. doi: 10.1039/b925257f. [DOI] [PubMed] [Google Scholar]
- 19.Kertesz V, Van Berkel GJ. Improved imaging resolution in desorption electrospray ionization mass spectrometry. Rapid Commun Mass Spectrom. 2008;22:2639–2644. doi: 10.1002/rcm.3662. [DOI] [PubMed] [Google Scholar]
- 20.Nemes P, Vertes A. Laser ablation electrospray ionization for atmospheric pressure, in vivo, and imaging mass spectrometry. Anal Chem. 2007;79:8098–8106. doi: 10.1021/ac071181r. [DOI] [PubMed] [Google Scholar]
- 21.Nemes P, Woods AS, Vertes A. Simultaneous imaging of small metabolites and lipids in rat brain tissues at atmospheric pressure by laser ablation electrospray ionization mass spectrometry. Anal Chem. 2010;82:982–988. doi: 10.1021/ac902245p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Acquadro E, Cabella C, Ghiani S, Miragoli L, Bucci E, Corpillo D. Matrix-Assisted laser Desorption Ionization Imaging Mass Spectrometry Detection of a Magnetic Resonance Imaging Contrast Agent in Mouse liver. Analytical Chemistry. 2009;81:2779–2784. doi: 10.1021/ac900038y. [DOI] [PubMed] [Google Scholar]
- 23.Atkinson SJ, Loadman PM, Sutton C, Patterson LH, Clench MR. Examination of the distribution of the bioreductive drug AQ4N and its active metabolite AQ4 in solid tumours by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:1271–1276. doi: 10.1002/rcm.2952. [DOI] [PubMed] [Google Scholar]
- 24.Hsieh Y, Casale R, Fukuda E, Chen J, Knemeyer I, Wingate J, et al. Matrix-assisted laser desorption/ionization imaging mass spectrometry for direct measurement of clozapine in rat brain tissue. Rapid Communications in Mass Spectrometry. 2006;20:965–972. doi: 10.1002/rcm.2397. [DOI] [PubMed] [Google Scholar]
- 25.Khatib-Shahidi S, Andersson M, Herman J, Gillespie T, Caprioli R. Direct molecular analysis of whole-body animal tissue sections by imaging MALDI mass spectrometry. Analytical Chemistry. 2006;78:6448–6456. doi: 10.1021/ac060788p. [DOI] [PubMed] [Google Scholar]
- 26.Trim P, Henson C, Avery J, McEwen A, Snel M, Claude E, et al. Matrix-Assisted Laser Desorption/Ionization-Ion Mobility Separation-Mass Spectrometry Imaging of Vinblastine in Whole Body Tissue Sections. Analytical Chemistry. 2008;80:8628–8634. doi: 10.1021/ac8015467. [DOI] [PubMed] [Google Scholar]
- 27.Nilsson A, Fehniger TE, Gustavsson L, Andersson M, Kenne K, Marko-Varga G, et al. Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue micro-compartments using imaging mass spectrometry. PLoS ONE. 2010;5:e11411. doi: 10.1371/journal.pone.0011411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vegvari A, Fehniger T, Gustavsson L, Nilsson A, Andren P, Kenne K, et al. Essential tactics of tissue preparation and matrix nano-spotting for successful compound imaging mass spectrometry. Journal of Proteomics. 2010;73:1270–1278. doi: 10.1016/j.jprot.2010.02.021. [DOI] [PubMed] [Google Scholar]
- 29.Dekker L, van Kampen J, Reedijk M, Burgers P, Gruters R, Osterhaus A, et al. A mass spectrometry based imaging method developed for the intracellular detection of HIV protease inhibitors. Rapid Communications in Mass Spectrometry. 2009;23:1183–1188. doi: 10.1002/rcm.3981. [DOI] [PubMed] [Google Scholar]
- 30.Goodwin R, MacIntyre L, Watson D, Scullion S, Pitt A. A solvent-free matrix application method for matrix-assisted laser desorption/ionization imaging of small molecules. Rapid Communications in Mass Spectrometry. 2010;24:1682–1686. doi: 10.1002/rcm.4567. [DOI] [PubMed] [Google Scholar]
- 31.Shroff R, Rulisek L, Doubsky J, Svatos A. Acid-base-driven matrix-assisted mass spectrometry for targeted metabolomics. Proc Natl Acad Sci U S A. 2009;106:10092–10096. doi: 10.1073/pnas.0900914106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altelaar AF, Klinkert I, Jalink K, de Lange RP, Adan RA, Heeren RM, et al. Gold-enhanced biomolecular surface imaging of cells and tissue by SIMS and MALDI mass spectrometry. Anal Chem. 2006;78:734–742. doi: 10.1021/ac0513111. [DOI] [PubMed] [Google Scholar]
- 33.Fitzgerald JJ, Kunnath P, Walker AV. Matrix-enhanced secondary ion mass spectrometry (ME SIMS) using room temperature ionic liquid matrices. Anal Chem. 2010;82:4413–4419. doi: 10.1021/ac100133c. [DOI] [PubMed] [Google Scholar]
- 34.Levisetti R, Hallegot P, Girod C, Chabala JM, Li J, Sodonis A, et al. Critical issues in the application of a gallium probe to high resolution secondary ion imaging. Surf Sci. 1991;246:94–106. [Google Scholar]
- 35.Cheng J, Wucher A, Winograd N. Molecular depth profiling with cluster ion beams. J Phys Chem B. 2006;110:8329–8336. doi: 10.1021/jp0573341. [DOI] [PubMed] [Google Scholar]
- 36.Weibel D, Wong S, Lockyer N, Blenkinsopp P, Hill R, Vickerman JC. A C60 primary ion beam system for time of flight secondary ion mass spectrometry: its development and secondary ion yield characteristics. Anal Chem. 2003;75:1754–1764. doi: 10.1021/ac026338o. [DOI] [PubMed] [Google Scholar]
- 37.Cheng J, Kozole J, Hengstebeck R, Winograd N. Direct comparison of Au(3)(+) and C(60)(+) cluster projectiles in SIMS molecular depth profiling. J Am Soc Mass Spectrom. 2007;18:406–412. doi: 10.1016/j.jasms.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kertesz V, Van Berkel G, Vavrek M, Koeplinger K, Schneider B, Covey T. Comparison of drug distribution images from whole-body thin tissue sections obtained using desorption electrospray ionization tandem mass spectrometry and autoradiography. Analytical Chemistry. 2008;80:5168–5177. doi: 10.1021/ac800546a. [DOI] [PubMed] [Google Scholar]
- 39.Nemes P, Barton A, Vertes A. Three-Dimensional Imaging of Metabolites in Tissues under Ambient Conditions by Laser Ablation Electrospray Ionization Mass Spectrometry. Analytical Chemistry. 2009;81:6668–6675. doi: 10.1021/ac900745e. [DOI] [PubMed] [Google Scholar]
- 40.Troendle F, Reddick C, Yost R. Detection of pharmaceutical compounds in tissue by matrix-assisted laser desorption/ionization and laser desorption/chemical ionization tandem mass spectrometry with a quadrupole ion trap. Journal of the American Society for Mass Spectrometry. 1999;10:1315–1321. [Google Scholar]
- 41.Krutchinsky AN, Chait BT. On the nature of the chemical noise in MALDI mass spectra. Journal of the American Society for Mass Spectrometry. 2002;13:129–134. doi: 10.1016/s1044-0305(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 42.Benabdellah F, Touboul D, Brunelle A, Laprevote O. In Situ Primary Metabolites Localization on a Rat Brain Section by Chemical Mass Spectrometry Imaging. Analytical Chemistry. 2009;81:5557–5560. doi: 10.1021/ac9005364. [DOI] [PubMed] [Google Scholar]
- 43.Bouslimani A, Bec N, Glueckmann M, Hirtz C, Larroque C. Matrix-assisted laser desorption/ionization imaging mass spectrometry of oxaliplatin derivatives in heated intraoperative chemotherapy (HIPEC)-like treated rat kidney. Rapid Communications in Mass Spectrometry. 2010;24:415–421. doi: 10.1002/rcm.4408. [DOI] [PubMed] [Google Scholar]
- 44.Bunch J, Clench M, Richards D. Determination of pharmaceutical compounds in skin by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Communications in Mass Spectrometry. 2004;18:3051–3060. doi: 10.1002/rcm.1725. [DOI] [PubMed] [Google Scholar]
- 45.Drexler DM, Garrett TJ, Cantone JL, Diters RW, Mitroka JG, Prieto Conaway MC, et al. Utility of imaging mass spectrometry (IMS) by matrix-assisted laser desorption ionization (MALDI) on an ion trap mass spectrometer in the analysis of drugs and metabolites in biological tissues. J Pharmacol Toxicol Methods. 2007;55:279–288. doi: 10.1016/j.vascn.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Goodwin R, Scullion P, MacIntyre L, Watson D, Pitt A. Use of a Solvent-Free Dry Matrix Coating for Quantitative Matrix-Assisted Laser Desorption Ionization Imaging of 4-Bromophenyl-1,4- diazabicyclo(3.2.2)nonane-4-carboxylate in Rat Brain and Quantitative Analysis of the Drug from Laser Microdissected Tissue Regions. Analytical Chemistry. 2010;82:3868–3873. doi: 10.1021/ac100398y. [DOI] [PubMed] [Google Scholar]
- 47.Signor L, Varesio E, Staack RF, Starke V, Richter WF, Hopfgartner G. Analysis of erlotinib and its metabolites in rat tissue sections by MALDI quadrupole time-of-flight mass spectrometry. J Mass Spectrom. 2007;42:900–909. doi: 10.1002/jms.1225. [DOI] [PubMed] [Google Scholar]
- 48.Stoeckli M, Staab D, Schweitzer A. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. International Journal of Mass Spectrometry. 2007;260:195–202. [Google Scholar]
- 49.Wang H, Jackson S, McEuen J, Woods A. Localization and analyses of small drug molecules in rat brain tissue sections. Analytical Chemistry. 2005;77:6682–6686. doi: 10.1021/ac050868d. [DOI] [PubMed] [Google Scholar]
- 50.Northen TR, Yanes O, Northen MT, Marrinucci D, Uritboonthai W, Apon J, et al. Clathrate nanostructures for mass spectrometry. Nature. 2007;449:1033–1036. doi: 10.1038/nature06195. [DOI] [PubMed] [Google Scholar]
- 51.Wiseman J, Ifa D, Venter A, Cooks R. Ambient molecular imaging by desorption electrospray ionization mass spectrometry. Nature Protocols. 2008;3:517–524. doi: 10.1038/nprot.2008.11. [DOI] [PubMed] [Google Scholar]
- 52.Wiseman J, Ifa D, Zhu Y, Kissinger C, Manicke N, Kissinger P, et al. Desorption electrospray ionization mass spectrometry: Imaging drugs and metabolites in tissues. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18120–18125. doi: 10.1073/pnas.0801066105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Belu A, Davies M, Newton J, Patel N. TOF-SIMS characterization and imaging of controlled-release drug delivery systems. Analytical Chemistry. 2000;72:5625–5638. doi: 10.1021/ac000450+. [DOI] [PubMed] [Google Scholar]
- 54.Mahoney C, Fahey A. Three-dimensional compositional analysis of drug eluting stent coatings using cluster secondary ion mass spectrometry. Analytical Chemistry. 2008;80:624–632. doi: 10.1021/ac701644j. [DOI] [PubMed] [Google Scholar]
- 55.Belu A, Mahoney C, Wormuth K. Chemical imaging of drug eluting coatings: Combining surface analysis and confocal Raman microscopy. Journal of Controlled Release. 2008;126:111–121. doi: 10.1016/j.jconrel.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 56.Fisher G, Belu A, Mahoney C, Wormuth K, Sanada N. Three-Dimensional Time-of-Flight Secondary Ion Mass Spectrometry Imaging of a Pharmaceutical in a Coronary Stent Coating as a Function of Elution Time. Analytical Chemistry. 2009;81:9930–9940. doi: 10.1021/ac901587k. [DOI] [PubMed] [Google Scholar]
- 57.Leuthold L, Mandscheff J, Fathi M, Giroud C, Augsburger M, Varesio E, et al. Desorption electrospray ionization mass spectrometry: direct toxicological screening and analysis of illicit Ecstasy tablets. Rapid Communications in Mass Spectrometry. 2006;20:103–110. doi: 10.1002/rcm.2280. [DOI] [PubMed] [Google Scholar]
- 58.Nyadong L, Harris G, Balayssac S, Galhena A, Malet-Martino M, Martino R, et al. Combining Two- Dimensional Diffusion-Ordered Nuclear Magnetic Resonance Spectroscopy, Imaging Desorption Electrospray Ionization Mass Spectrometry, and Direct Analysis in Real-Time Mass Spectrometry for the Integral Investigation of Counterfeit Pharmaceuticals. Analytical Chemistry. 2009;81:4803–4812. doi: 10.1021/ac900384j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Earnshaw C, Carolan V, Richards D, Clench M. Direct analysis of pharmaceutical tablet formulations using matrix-assisted laser desorption/ionisation mass spectrometry imaging. Rapid Communications in Mass Spectrometry. 2010;24:1665–1672. doi: 10.1002/rcm.4525. [DOI] [PubMed] [Google Scholar]
- 60.Cha S, Song Z, Nikolau B, Yeung E. Direct Profiling and Imaging of Epicuticular Waxes on Arabidopsis thaliana by Laser Desorption/Ionization Mass Spectrometry Using Silver Colloid as a Matrix. Analytical Chemistry. 2009;81:2991–3000. doi: 10.1021/ac802615r. [DOI] [PubMed] [Google Scholar]
- 61.Holscher D, Shroff R, Knop K, Gottschaldt M, Crecelius A, Schneider B, et al. Matrix-free UV-laser desorption/ionization (LDI) mass spectrometric imaging at the single-cell level: distribution of secondary metabolites of Arabidopsis thaliana and Hypericum species. Plant J. 2009;60:907–918. doi: 10.1111/j.1365-313X.2009.04012.x. [DOI] [PubMed] [Google Scholar]
- 62.Anderson D, Carolan V, Crosland S, Sharples K, Clench M. Examination of the distribution of nicosulfuron in sunflower plants by matrix-assisted laser desorption/ionisation mass spectrometry imaging. Rapid Communications in Mass Spectrometry. 2009;23:1321–1327. doi: 10.1002/rcm.3973. [DOI] [PubMed] [Google Scholar]
- 63.Shroff R, Vergara F, Muck A, Svatos A, Gershenzon J. Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc Natl Acad Sci U S A. 2008;105:6196–6201. doi: 10.1073/pnas.0711730105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang YL, Xu Y, Straight P, Dorrestein PC. Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol. 2009;5:885–887. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schafer R. ultrafleXtreme: Redefining MALDI Mass Spectrometry Performance. Lc Gc N Am. 2009:14–15. [Google Scholar]
- 66.Moskovets E, Preisler J, Chen HS, Rejtar T, Andreev V, Karger BL. High-throughput axial MALDI-TOF MS using a 2-kHz repetition rate laser. Anal Chem. 2006;78:912–919. doi: 10.1021/ac051393t. [DOI] [PubMed] [Google Scholar]
- 67.Simmons D. Improved MALDI-MS imaging performance using continuous laser rastering. MDS Analytical Technologies. 2008 [Google Scholar]
- 68.Vestal C, Parker K, Hayden K, et al. Tissue imaging by 5 kHz high-performance MALDI-TOF. Presented at: American Society for Mass Spectrometry Conference on Mass Spectrometry and Applied Topics; 31 May – 4 June, 2009; Philadelphia, Pennsylvania, USA. [Google Scholar]
- 69.Spengler B, Hubert M. Scanning microprobe matrix-assisted laser desorption ionization (SMALDI) mass spectrometry: instrumentation for sub-micrometer resolved LDI and MALDI surface analysis. J Am Soc Mass Spectrom. 2002;13:735–748. doi: 10.1016/S1044-0305(02)00376-8. [DOI] [PubMed] [Google Scholar]
- 70.Bouschen W, Schulz O, Eikel D, Spengler B. Matrix vapor deposition/recrystallization and dedicated spray preparation for high-resolution scanning microprobe matrix-assisted laser desorption/ionization imaging mass spectrometry (SMALDI-MS) of tissue and single cells. Rapid Commun Mass Spectrom. 2010;24:355–364. doi: 10.1002/rcm.4401. [DOI] [PubMed] [Google Scholar]
- 71.Cornett DS, Frappier SL, Caprioli RM. MALDI-FTICR imaging mass spectrometry of drugs and metabolites in tissue. Anal Chem. 2008;80:5648–5643. doi: 10.1021/ac800617s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Manicke NE, Dill AL, Ifa DR, Cooks RG. High-resolution tissue imaging on an orbitrap mass spectrometer by desorption electrospray ionization mass spectrometry. J Mass Spectrom. 2010;45:223–226. doi: 10.1002/jms.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen R, Jiang X, Conaway MC, Mohtashemi I, Hui L, Viner R, et al. Mass spectral analysis of neuropeptide expression and distribution in the nervous system of the lobster Homarus americanus. J Proteome Res. 2010;9:818–832. doi: 10.1021/pr900736t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fenn LS, McLean JA. Biomolecular structural separations by ion mobility-mass spectrometry. Anal Bioanal Chem. 2008;391:905–909. doi: 10.1007/s00216-008-1951-x. [DOI] [PubMed] [Google Scholar]
- 75.McLean JA, Ridenour WB, Caprioli RM. Profiling and imaging of tissues by imaging ion mobility-mass spectrometry. J Mass Spectrom. 2007;42:1099–1105. doi: 10.1002/jms.1254. [DOI] [PubMed] [Google Scholar]
- 76.Carado A, Passarelli MK, Kozole J, Wingate JE, Winograd N, Loboda AV. C60 secondary ion mass spectrometry with a hybrid-quadrupole orthogonal time-of-flight mass spectrometer. Anal Chem. 2008;80:7921–7929. doi: 10.1021/ac801712s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fletcher JS, Rabbani S, Henderson A, Blenkinsopp P, Thompson SP, Lockyer NP, et al. A new dynamic in mass spectral imaging of single biological cells. Anal Chem. 2008;80:9058–9064. doi: 10.1021/ac8015278. [DOI] [PubMed] [Google Scholar]
- 78.Lomenick B, Hao R, Jonai N, Chin R, Aghajan M, Warburton S, et al. Target identification using drug affinity responsive target stability (DARTS) Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21984–21989. doi: 10.1073/pnas.0910040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lomenick B, Olsen RW, Huang J. Identification of Direct Protein Targets of Small Molecules. ACS Chemical Biology. 2011;6:34–46. doi: 10.1021/cb100294v. [DOI] [PMC free article] [PubMed] [Google Scholar]