Summary
Cullin-RING ligases (CRLs) compose the largest class of E3 ubiquitin ligases. CRLs are modular, multisubunit enzymes, comprising interchangeable substrate receptors dedicated to particular Cullin-RING catalytic cores. Recent structural studies have revealed numerous ways in which CRL E3 ligase activities are controlled, including multimodal E3 ligase activation by covalent attachment of the ubiquitin-like protein NEDD8, inhibition of CRL assembly/activity by CAND1, and several mechanisms of regulated substrate recruitment. These features highlight the potential for CRL activities to be tuned in responses to diverse cellular cues, and for modulating CRL functions through small-molecule agonists or antagonists. As the second installment of a two-review series, this article focuses on recent structural studies advancing our knowledge of how CRL activities are regulated.
Introduction
A predominant form of eukaryotic protein regulation involves alteration of protein function via post-translational covalent attachment of the protein, ubiquitin. In this process, ubiquitin’s C-terminus becomes isopeptide-bonded to a substrate protein’s lysine via the action of E1, E2, and E3 enzymes. E3s generally have at least two domains [1,2]. One domain, usually a RING (Really Interesting New Gene) or HECT (Homologous to E6AP C-Terminus) domain, recruits a labile, thiolester-linked E2~ubiquitin intermediate. The other is a protein-interaction domain that recruits a substrate for ubiquitination. E3-mediated attachment of ubiquitin to substrates is highly regulated in response to cellular cues, and can modulate a target protein’s half-life, localization, interactions with protein or DNA partners, or other functions.
The largest E3 superfamily consists of the multisubunit Cullin-RING ligases (CRLs). CRLs are nucleated by an extended cullin scaffold interacting with a catalytic RING-containing protein, either RBX1 or RBX2 [3]. Structural studies revealed an overall common assembly for the best-studied cullins encoded by the human genome (CUL1, CUL2, CUL3, CUL4A, CUL4B, and CUL5), which form ~300 distinct CRL complexes in different subfamilies (CRL1 containing CUL1, CRL2 containing CUL2, etc.). We recently surveyed overall features of CRL1-CRL5 structures in the first installment of our 2-part review series [4]. Briefly, CRLs adopt an elongated architecture, with the substrate-binding site and E2-binding RING at opposite ends [5]. A cullin’s N-terminal domain (NTD) binds a substrate receptor (SR) either directly or indirectly via an adaptor protein. Each cullin has its own large family of dedicated SRs, which bind a substrate’s “degron” motif. A cullin’s C-terminal domain (CTD) binds the RBX RING protein. The RING domain recruits an E2~ubiquitin intermediate, and promotes ubiquitin transfer from the E2 active site directly to the substrate associated with the SR. In conjunction with CRL1 complexes, the E2 Cdc34 polyubiquitinates a substrate on a millisecond time-scale [6,7].
A fundamental aspect of the ubiquitination process is that it is regulated. In response to signals including daylight, presence of mitogens, low oxygen levels, and pathogenic infections, ubiquitin ligases and/or their substrates can be altered in ways that modulate ubiquitination, and hence the fates of ubiquitinated targets. Here we review structural mechanisms by which CRL activities can be tuned to achieve regulation (Fig. 1).
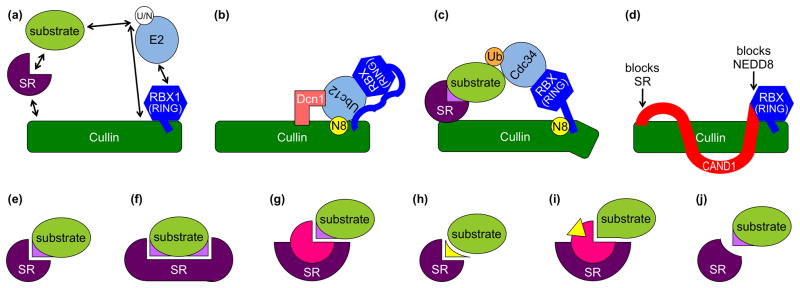
Figure 1. CRL regulation.
The elongated cullin scaffold protein (green) interacts via its N-terminal domain (NTD) with a substrate receptor (SR, purple) that recruits substrate (lime). The cullin C-terminal domain (CTD) tightly associates with an RBX1/2 RING protein (blue) that recruits Ubiquitin (Ub) or NEDD8 (N8)-charged E2 enzymes (sky). (a) Conformations and interactions that are regulated are indicated with arrows. (b) Model for dual E3 mechanism for NEDD8 (yellow) ligation to a cullin, which involves both RBX1 and Dcn1 (salmon) cofunctioning as E3s. (c) Model for NEDD8-activated CRL-mediated ubiquitin (orange) ligation to an SR-associated substrate. (d) CAND1 (red) inhibition of cullin-RBX assembly with SRs, and of NEDD8 ligation. (e–j) Regulation of SR-substrate interactions include (e) SR recognition of a specific substrate post-translational modification (violet), (f) SR recognition of multiple specific post-translational modifications (violet) on a substrate, (g) SR binding a partner protein (pink) to recognize a specific substrate post-translational modification (violet), (h) SR binding a small molecule “glue” (yellow) that mediates interactions between the SR and substrate, (i) SR binding a partner protein (pink) that is allosterically modulated by a small molecule hormone (yellow) to then bind a substrate, and (j) inhibition of SR binding after specific post-translational modification (violet) of a substrate.
Activation by the ubiquitin-like protein NEDD8: springing a cullin-RING into action
Original CRL structures revealed the molecular basis for assembly of cullin-RBX1-SR complexes, but represented catalytically-inactive versions of the E3s [5,8-11]. In original CRL-substrate-E2 models, it was unclear how an E2 active site would become juxtaposed with the substrate for ubiquitin transfer, or how an E2 could add a ubiquitin onto the growing end of a polyubiquitin chain (Fig. 2a). A view of active CRLs came from examining effects of covalent ligation of a ubiquitin-like protein, NEDD8, to a conserved lysine near a cullin’s C-terminus (Fig. 2b, c). NEDD8 attachment stimulates multiple CRL ubiquitin E3 activities, including binding to E2~ubiquitin, enhancing ubiquitin transfer from the E2 active site, and positioning the E2 active site adjacent to the substrate [12,13]. Structural studies revealed how NEDD8 ligation could transform a CRL’s structure for these diverse effects. Duda et al. compared crystal structures and small-angle X-ray scattering (SAXS) data for CUL5CTD-RBX1 and CUL1CTD-RBX1 in both unmodified and NEDD8-modified forms [12]. The isolated unmodified cullin CTDs resemble the corresponding regions of full-length CRLs, and adopt a compact “closed” architecture, where the RBX1 RING domain contacts a portion of the cullin protein. In the NEDD8-modified structures, the NEDD8-ligated cullin subdomain is dramatically reoriented and the RBX1 RING domain is essentially sprung from its interaction with the cullin (Fig. 1a, c, 2c). In this open form, the RBX1 RING is free to rotate and adopt multiple orientations, two of which were trapped crystallographically (Fig. 2c) [12]. Biochemical data support the idea that freeing the RBX1 RING from interactions with a cullin activates CRL ubiquitin ligase activity, and that hindering rotation of the RING domain hinders CRL-mediated polyubiquitination [12,14].
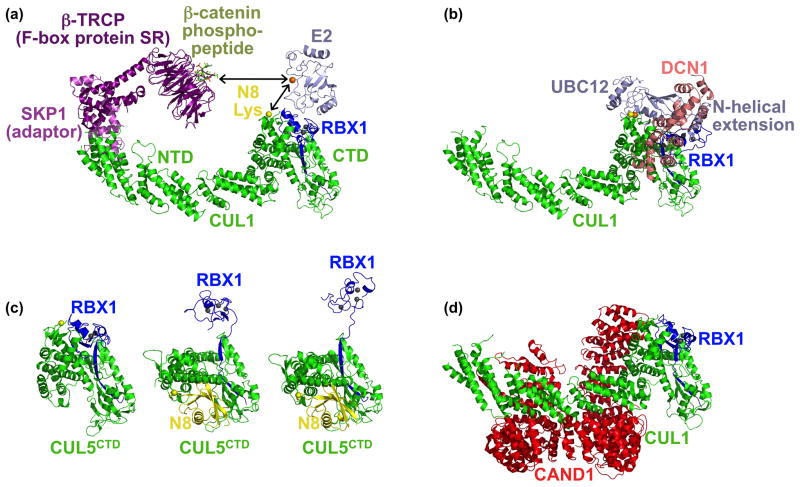
Figure 2. Conformational control of CRL E3 ligase activities.
(a) Structural model of a representative CRL1 complex, obtained by superimposing the Skp1 (violet)-β-TRCP (purple)-β-catenin phosphodegron peptide (lime) and SKP1-F-box domain-CUL1 (green)-RBX1 (blue) structures [5,10]. An E2 (sky) is modeled by superimposing RBX1 RING domain with the UbcH7-bound c-Cbl RING domain [8]. Zinc atoms are shown as grey spheres. Arrows indicate that conformational changes allow the E2 active site cysteine (orange sphere) to approach the substrate peptide or a cullin’s NEDD8 acceptor lysine (yellow sphere). (b) Structural model for the dual E3 mechanism for NEDD8 ligation to cullins [20]. After docking the structure of full-length yeast Ubc12 [20] on the E2-RBX1 model in (a), the RING domain was rotated so that the Ubc12 active site and NEDD8 ligation site are juxtaposed. The second E3 for NEDD8, Dcn1 (salmon), was docked on one side with CUL1 based on a complex structure, and on the other with Ubc12’s N-terminal helical extension based on mutational data [20]. (c) Structures of CUL5CTD-RBX1 and NEDD8~ CUL5CTD-RBX1 showing subdomain rearrangement, including “freeing” of the RBX1 RING domain, in the NEDD8 ligated CRL structures [12]. (d) Structural basis for CAND1-mediated inhibition of CUL1-RBX1. CAND1 (red) locks CRLs in a rigid state, and prevents assembly with SRs and NEDD8 ligation [30].
Notably, the activating function of NEDD8 itself can be altered. Certain pathogenic bacteria produce effector proteins called Cif or CHBP, which deamidate glutamine 40 of NEDD8 and inhibit NEDD8-mediated CRL activation and host cell division [15–17]. In the NEDD8~ CUL5CTD-RBX1 structure, NEDD8’s glutamine 40 is positioned near the isopeptide linkage to cullin. It is possible that deamidation could alter the conformation of NEDD8-ligated CRLs. It will be interesting to gain further structural insights into how Cif abrogates CRL activation by NEDD8.
CRL neddylation: two steps forward and one step back
NEDD8 becomes ligated to a conserved cullin lysine by the sequential action of its own set of E1, E2, and E3 enzymes. In higher eukaryotes, there are distinct NEDD8 conjugation pathways for different cullins: two NEDD8-specific E2s, Ubc12 and UBE2F, function with the two different RBX proteins to modify specific cullins. Ubc12 and RBX1 neddylate CUL1, 2, 3, and 4, whereas UBE2F and RBX2 neddylate CUL5 [18]. It is likely that RBX1/2 bind Ubc12/Ube2F with an interface analogous to that observed in other RING-E2 structures, via a RING hydrophobic surface binding a surface from the E2 catalytic domain comprising the N-terminal helix and two loops [8]. However, a structural model of a cullin-RBX1-Ubc12 complex based on the “closed” CRL architecture is incompatible with juxtaposition of the Ubc12 active site and the cullin acceptor lysine for the neddylation reaction (Fig. 2a). This problem was partly reconciled by SAXS data on CUL1CTD-RBX1, which indicated some conformational flexibility within the cullin-RING complex that likely allows neddylation to occur [12].
Recent structural, biophysical, and enzymological studies suggest that a second E3 for NEDD8, called Dcn1 [19], harnesses the flexible cullin-RING structure to enhance neddylation [20]. Dcn1 adopts an ovoid structure composed of a UBA domain and two EF-hands and shares no resemblance to RING or HECT E3s [21,22]. An acidic patch near the Dcn1 C-terminus binds a basic surface on the cullin WHB on the opposite side from the NEDD8 acceptor lysine. The groove between the two Dcn1 EF-hands binds a unique Ubc12 helix, which is N-terminal of Ubc12’s catalytic core domain [20]. Thus, Dcn1 uses novel mechanisms to bind both a cullin and Ubc12. Through these interactions, Dcn1 positions the RBX1-bound Ubc12 adjacent to the cullin NEDD8 acceptor lysine and enhances neddylation (Fig. 1b, 2b) [20].
NEDD8 is removed from cullins by a ~320 kDa 8-subunit enzyme, the COP9 Signalosome (CSN) [23]. The catalytic component, CSN5, is a member of the JAMM family of zinc metalloproteinases [24]. Although no high-resolution structure is available for a CSN5-NEDD8~cullin complex, insights into CSN’s catalytic mechanism may be extrapolated from the crystal structure of a JAMM-family deubiquitinase, AMSH-LP, complexed with a diubiquitin chain [25]. CSN5 likely uses a parallel thermolysin-like mechanism, in which a zinc-coordinated water mediates cleavage of the isopeptide bond between NEDD8 and the cullin. AMSH-LP binds the distal ubiquitin via its Leu8/Ile44/Val70 hydrophobic patch conserved in NEDD8. This NEDD8 hydrophobic patch contacts CUL5 in the NEDD8~CUL5CTD-RBX1 crystal structure [12]. Thus, it seems likely that additional structural flexibility in NEDD8-modified cullin complexes, perhaps induced by CSN-binding, allows NEDD8 recognition for removal from cullins.
Interestingly, CSN5 is inactive in isolation, and becomes catalytic in the CSN complex. CSN5 interactions with other subunits may cause an activating conformational change. Mass spectrometric [26] and cryo-Electron Microscopy [27] data on active recombinant CSN complexes indicate that CSN5 contacts other subunits within the CSN. CSN forms a clamshell-like structure with a large groove in the middle, with CSN5 at the edge of the shell [27]. Perhaps a cullin binds within the groove, making NEDD8 accessible to activated CSN5. Future studies will be required to elucidate details of CSN-mediated removal of NEDD8 from cullins.
The CAND1 inhibitor: a two-pronged attack on CRL activity
A cellular inhibitor, CAND1 (Cullin-Associated and Neddylation-Dissociated 1), binds specifically to free cullin-RBX complexes lacking NEDD8 modification, and inhibits CRL assembly and NEDD8 activation [28,29]. A CAND1-CUL1-RBX1 crystal structure revealed that CAND1 adopts a solenoid structure that wraps around CUL1-RBX1 to form a 2-pronged clamp (Fig. 1d, 2d) [30]. One prong comes from CAND1’s C-terminus, which binds the same site of CUL1 that recruits an adaptor/SR complex for substrate recognition. Thus, CAND1 and SR association with cullins are mutually exclusive. The second prong comes from CAND1’s N-terminus, which binds at the junction of the RBX1 RING and cullin that forms only in the closed conformation adopted by unmodified CRLs. At this interface, CAND1 binds directly to CUL1’s NEDD8 acceptor lysine. Thus, CAND1 inhibits neddylation by (1) locking the cullin-RING complex into a closed conformation, and (2) blocking access to the cullin’s NEDD8 acceptor lysine. The converse is also true: NEDD8-modification hinders CAND1 binding to CRLs. Comparing structures of CAND1-CUL1-RBX1 and NEDD8~CUL5CTD-RBX1 suggests that NEDD8-induced reorientation of cullin-RING subdomains eliminates the CAND1 binding surface at the cullin-RBX RING interface [12,30].
Substrates: to bind or not to bind
Even when a CRL is assembled into an active complex through liberation from CAND1, binding to an SR, and ligation to NEDD8, additional regulation may occur at the level of substrate recognition. An associated substrate’s binding motif is often referred to as a “degron”, due to E3 interactions often directing substrates for ubiquitin-mediated degradation. In order to achieve precise control of substrate ubiquitination, degron binding to CRL SRs is regulated. Structural studies have revealed numerous mechanisms by which such regulation is achieved.
Many CRLs bind substrates only after the degron motif has been post-translationally modified (Fig. 1e–g, 3). Early structural studies of CRLs showed how modifications such as phosphorylation (Fig. 3a), glycosylation (Fig. 3b), and prolyl hydroxylation (Fig. 3c) served as “knobs” that fit into complementary “holes” in cognate SRs [10,11,31–35]. There are also many variations on SR recognition of post-translationally modified substrates. For example, recognition can be amplified by SRs preferentially binding doubly phosphorylated degrons [10,36], or by SR dimerization [36–38]. Also, an SR can lack its own phosphopeptide binding site, but instead can import phosphate recognition via partnering with a protein harboring an anion-binding site (Fig. 3d) [39]. Recently, crystallographic studies revealed how substrate post-translational modification can also block interaction with a CRL SR: a groove on the CRL3 SR SPOP intimately binds a Ser/Thr-rich degron conserved among substrates [38]. Phosphorylation of the degron would clash with the SPOP groove and repel an aspartate central to the binding site (Fig. 1j).
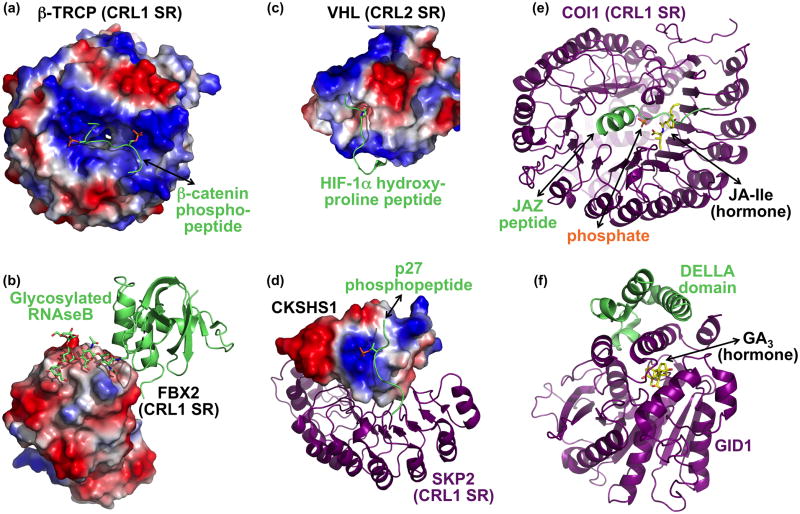
Figure 3. Selected examples of regulated substrate binding to CRLs.
(a) Two precisely positioned basic patches and a peptide binding groove in the CRL1 SR β-TRCP (surface showing electrostatic potential) recruit a doubly-phosphorylated β-catenin (lime, with phosphates as orange/red sticks) [10]. (b) Sugar recognition on a glycosylated substrate (lime) by the CRL1 SR FBX2 (electrostatic surface) [35]. The sugar moiety is depicted in sticks. (c) Recognition of HIF-1α hydroxyproline (peptide in lime, with hydroxyproline in sticks) by the CRL2 SR VHL (electrostatic surface) [32,33]. Only a portion of the VHL structure is shown to highlight substrate recognition. (d) The CRL1 SR Skp2 (purple) imports phosphopeptide recognition by binding to CKSHS1 (electrostatic surface), whose anion binding pocket recognizes the phosphate moiety on the substrate phospho-p27 (lime, with phosphate in orange/red sticks) [39]. (e) The plant hormone jasmonic acid-isoleucine (JA-Ile) functions in a multicomponent molecular glue mechanism to recruit substrates to the CRL1 SR COI-1 (purple). JA-Ile (yellow) mediates binding to the substrate JAZ degron peptide (lime), but the JAZ degron helix also clamps JA-Ile into the COI-1 binding site. A nearby phosphate most likely resembles part of inositol pentakisphosphate, which potentiates interaction between COI-1, JA-Ile, and a JAZ substrate. (f) Gibberellic acid (GA, yellow) uses an indirect allosteric mechanism to promote interactions between DELLA transcriptional repressors (lime) and a multiprotein CRL1 SR that includes GID1 (purple) [42]. GA serves as part of a hydrophobic core required for folding of the GID1 N-terminus. This portion of GID1, in turn, helps recruit DELLA substrates for ubiquitination.
Distinct mechanisms regulating substrate recruitment to CRLs were recently elucidated by studies of the plant hormones auxin, jasmonate, and gibberellin, which promote interactions of particular transcriptional repressors with the CRL1 SRs TIR1, COI1, and SLY1/GID2, respectively (Fig. 1h, i). Structures revealed that auxin and jasmonate function via related 2-sided “molecular glue” mechanisms [40,41]. On one side, the hormones fit in a fixed pocked in the TIR1 or COI1 substrate-binding domains, which are comprised of 18 leucine-rich repeats (LRRs) that form alternating β-strands and α-helices packing in a horseshoe-like shape. The other side of auxin and jasmonate bind degron peptides, thereby recruiting substrates to their SRs. The recent structure of a COI1-jasmonic acid/Ile (hormone)-JAZ (degron) complex showed that the converse is also true: the substrate can also mediate binding to the hormone, with a conserved JAZ degron helix interacting with a concave surface inside the COI1 LRR horseshoe and clamping hormone into place (Fig. 3e) [41]. Additional regulation may be achieved by TIR1 and COI1 binding tightly to inositol-phosphates near their hormone/degron binding sites [40,41]. Indeed, crystallographic and mutational data revealed how one of the phosphates interacts directly with jasmonate-binding COI1 and JAZ residues [41].
Gibberellic acid (GA) uses a completely different allosteric mechanism to promote interactions between DELLA transcriptional repressors and a multiprotein CRL1 SR. Here, the F-box protein SLY1 (Arabidopsis; GID2 in rice) on its own does not bind either GA or DELLA substrates. Rather SLY1 interacts with the GA receptor GID1 to import GA-sensitivity and substrate binding. A GA-GID1-DELLA structure showed that GA promotes a conformational change in GID1: the GID1 tri-helix N-terminal region folds only in the presence of GA [42]. GA “glues” the GID1 N- and C-terminal regions together, to form a platform for DELLA substrate recruitment (Fig. 3f) [42]. Future studies will be required to reveal how SLY1/GID2 recruits GA-GID1-DELLA for ubiquitination.
Synthetic small molecule modulation of CRL-substrate interactions: from direct agonists to indirect allosteric inhibitors
The numerous mechanisms regulating CRL activities provide abundant opportunities for manipulation by synthetic small molecules. A founding example is the interaction between TIR1 and auxin. Building on structural data, Hayashi et al. synthesized compounds related to auxin, but differing in the α-alkyl moiety, to design both agonists that mimicked the auxin “molecular glue” effect that mediates substrate binding, and antagonists blocking substrate interaction [43]. Structures and modeling of TIR1 with these compounds revealed that they bind the native auxin-binding pocket [43]. The α-alkyl groups of crystallized antagonists extend away from TIR1, apparently to clash with substrate. Accordingly, the potency of the antagonist correlates with the length of the interfering alkyl chain.
In contrast to the direct antagonist for TIR1, a recent screen identified a small molecule, SCF-I2, which inhibits the SR Cdc4 via an indirect allosteric mechanism (Fig. 4) [44]. Remarkably, SCF-I2 blocks a phosphodegron-containing substrate from binding the Cdc4 WD40 β-propeller without binding to the same site as a substrate phosphodegron. Rather, SCF-I2 intercalates between two blades of the propeller. This leads to local distortion, which propagates via a continuum of side-chain rearrangements over 25 Å and ultimately perturbs the substrate-binding pocket (Fig. 4). This mode of inhibition could not have been designed or predicted based on previous structures, as SCF-I2 binding relies on flexibility and dynamics within the SR; the SCF-I2 binding site is wholly lacking in the absence of the small molecule. As β-propellers are prominent among substrate interaction domains in CRL SRs, this may represent a broad means for inhibiting many CRLs.
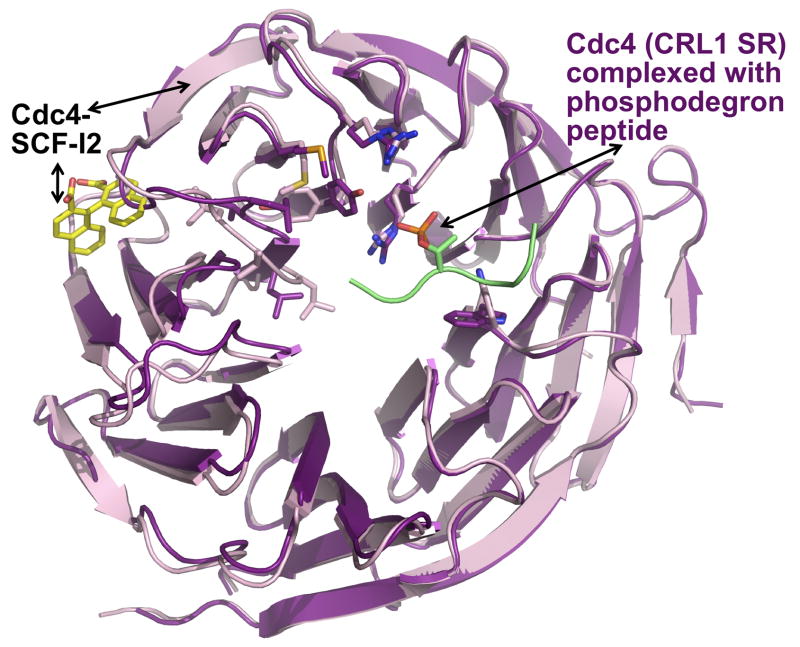
Figure 4. SCF-I2 allosterically inhibits the Cdc4 WD40 β-propeller from binding to a phosphodegron.
Superposition of the CRL1 SR Cdc4 WD40 β-propeller from structures of complexes with a phosphodegron peptide (Cdc4 – purple; phosphodegron lime) or with SCF-I2 (Cdc4 – pink; SCF-I2 – yellow). SCF-I2 (yellow) intercalates between two blades of the propeller (pink) that leads to a continuum of structural distortions over 25 Å, ultimately preventing interaction with a phosphodegron.
Conclusions
Numerous structural mechanisms regulating CRLs have been elucidated in recent years. It is now known that CRL structures are dynamic, and their conformations can be regulated to control CRL assembly and E3 ligase activities. At one end of the conformational spectrum, CAND1 maintains CRLs in a rigid off-state, and prevents assembly with SRs and dynamic activation by cullin modification by NEDD8 [30]. At the opposite extreme, NEDD8 ligation favors open conformations, where the RBX RING domain samples multiple orientations during substrate polyubiquitination [12]. In the middle, another E3, Dcn1, harnesses the flexible CRL structure for NEDD8 ligation to cullins [20]. SR-substrate interactions are also highly regulated, by numerous mechanisms that include positive or negative modulation by post-translational modifications or natural or synthetic small molecules.
Despite these advances, it seems the existing structures have touched only the tip of the iceberg in terms of understanding CRL regulation. Outstanding questions include how CRLs interact with molecular machines. In addition to regulation by CSN, CRLs are also recruited to p97, apparently for processing of ubiquitinated substrates as a prelude to their degradation by the 26S Proteasome [45]. Furthermore, many other forms of SR-substrate interactions remain to be structurally elucidated. For example, SRs, themselves, can be post-translationally modified to regulate binding to substrates. A prime example is the CRL3 SR Keap1, whose oxidation prevents binding and ubiquitin-mediated destruction of the anti-oxidant transcription factor Nrf2 [46]. We also now know that small molecule modulation of SR-substrate interactions is not limited to plants: dioxin promotes formation of one CRL4-based E3 [47], and thalidomide inhibits activity of another [48]. We anticipate many exciting and novel structural mechanisms regulating CRL activities to be discovered for years to come.
Acknowledgments
D.M.D., D.C.S., M.C., and B.A.S. are supported by ALSAC, the St. Jude Cancer Center Core grant (P30CA021765), the National Institutes of Health (R01 GM069530), and the Damon-Runyon Cancer Research Foundation (postdoctoral fellowship to M.C.). E.S.Z. and N.Z. are supported by National Institutes of Health (R01 CA107134 and F32 GM093497), the Burroughs Wellcome Fund, and National Science Foundation. B.A.S. and N.Z. are investigators of the Howard Hughes Medical Institute.
Footnotes
Conflict of interest
The authors declare no competing interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
•• of special interest
•• of outstanding interest
- 1.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 2.Kee Y, Huibregtse JM. Regulation of catalytic activities of HECT ubiquitin ligases. Biochem Biophys Res Commun. 2007;354:329–333. doi: 10.1016/j.bbrc.2007.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petroski MD, Deshaies RJ. Function and regulation of cullin-RING ubiquitin ligases. Nat Rev Mol Cell Biol. 2005;6:9–20. doi: 10.1038/nrm1547. [DOI] [PubMed] [Google Scholar]
- 4.Zimmerman ES, Schulman BA, Zheng N. Structural assembly of cullin-RING ubiquitin ligase complexes. Curr Opin Struct Biol. 2010;20:714–721. doi: 10.1016/j.sbi.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
- 6.Kleiger G, Saha A, Lewis S, Kuhlman B, Deshaies RJ. Rapid E2-E3 assembly and disassembly enable processive ubiquitylation of cullin-RING ubiquitin ligase substrates. Cell. 2009;139:957–968. doi: 10.1016/j.cell.2009.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pierce NW, Kleiger G, Shan SO, Deshaies RJ. Detection of sequential polyubiquitylation on a millisecond timescale. Nature. 2009;462:615–619. doi: 10.1038/nature08595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 9.Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- 10.Wu G, Xu G, Schulman BA, Jeffrey PD, Harper JW, Pavletich NP. Structure of a beta-TrCP1-Skp1-beta-catenin complex: destruction motif binding and lysine specificity of the SCF(beta-TrCP1) ubiquitin ligase. Mol Cell. 2003;11:1445–1456. doi: 10.1016/s1097-2765(03)00234-x. [DOI] [PubMed] [Google Scholar]
- 11.Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 12••.Duda DM, Borg LA, Scott DC, Hunt HW, Hammel M, Schulman BA. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell. 2008;134:995–1006. doi: 10.1016/j.cell.2008.07.022. This study revealed the structures of NEDD8-ligated cullins as open and flexible. The data indicate that the RBX1 RING domain samples multiple orientations during cullin neddylation and subsequent substrate ubiquitination. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saha A, Deshaies RJ. Multimodal activation of the ubiquitin ligase SCF by Nedd8 conjugation. Mol Cell. 2008;32:21–31. doi: 10.1016/j.molcel.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamoah K, Oashi T, Sarikas A, Gazdoiu S, Osman R, Pan ZQ. Autoinhibitory regulation of SCF-mediated ubiquitination by human cullin 1’s C-terminal tail. Proc Natl Acad Sci U S A. 2008;105:12230–12235. doi: 10.1073/pnas.0806155105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui J, Yao Q, Li S, Ding X, Lu Q, Mao H, Liu L, Zheng N, Chen S, Shao F. Glutamine deamidation and dysfunction of ubiquitin/NEDD8 induced by a bacterial effector family. Science. 2010;329:1215–1218. doi: 10.1126/science.1193844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morikawa H, Kim M, Mimuro H, Punginelli C, Koyama T, Nagai S, Miyawaki A, Iwai K, Sasakawa C. The bacterial effector Cif interferes with SCF ubiquitin ligase function by inhibiting deneddylation of Cullin1. Biochem Biophys Res Commun. 401:268–274. doi: 10.1016/j.bbrc.2010.09.048. [DOI] [PubMed] [Google Scholar]
- 17.Jubelin G, Taieb F, Duda DM, Hsu Y, Samba-Louaka A, Nobe R, Penary M, Watrin C, Nougayrede JP, Schulman BA, et al. Pathogenic bacteria target NEDD8-conjugated cullins to hijack host-cell signaling pathways. PLoS Pathog. 2010;6:e1001128. doi: 10.1371/journal.ppat.1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, et al. E2-RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurz T, Ozlu N, Rudolf F, O’Rourke SM, Luke B, Hofmann K, Hyman AA, Bowerman B, Peter M. The conserved protein DCN-1/Dcn1p is required for cullin neddylation in C. elegans and S. cerevisiae. Nature. 2005;435:1257–1261. doi: 10.1038/nature03662. [DOI] [PubMed] [Google Scholar]
- 20•.Scott DC, Monda JK, Grace CR, Duda DM, Kriwacki RW, Kurz T, Schulman BA. A dual E3 mechanism for Rub1 ligation to Cdc53. Mol Cell. 2010;39:784–796. doi: 10.1016/j.molcel.2010.08.030. A novel “Dual E3 mechanism” was dissected for ligation of the yeast ortholog of NEDD8 (Rub1) ligation to its cullin target. Structural and biophysical studies indicated how two E3s (RBX1 and Dcn1) function with a single E2 (Ubc12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurz T, Chou YC, Willems AR, Meyer-Schaller N, Hecht ML, Tyers M, Peter M, Sicheri F. Dcn1 functions as a scaffold-type E3 ligase for cullin neddylation. Mol Cell. 2008;29:23–35. doi: 10.1016/j.molcel.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 22.Yang X, Zhou J, Sun L, Wei Z, Gao J, Gong W, Xu RM, Rao Z, Liu Y. Structural basis for the function of DCN-1 in protein Neddylation. J Biol Chem. 2007;282:24490–24494. doi: 10.1074/jbc.C700038200. [DOI] [PubMed] [Google Scholar]
- 23.Lyapina S, Cope G, Shevchenko A, Serino G, Tsuge T, Zhou C, Wolf DA, Wei N, Deshaies RJ. Promotion of NEDD-CUL1 conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 24.Cope GA, Suh GS, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 25.Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature. 2008;455:358–362. doi: 10.1038/nature07254. [DOI] [PubMed] [Google Scholar]
- 26.Sharon M, Mao H, Boeri Erba E, Stephens E, Zheng N, Robinson CV. Symmetrical modularity of the COP9 signalosome complex suggests its multifunctionality. Structure. 2009;17:31–40. doi: 10.1016/j.str.2008.10.012. [DOI] [PubMed] [Google Scholar]
- 27.Enchev RI, Schreiber A, Beuron F, Morris EP. Structural insights into the COP9 signalosome and its common architecture with the 26S proteasome lid and eIF3. Structure. 2010;18:518–527. doi: 10.1016/j.str.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, Furukawa M, Matsumoto T, Xiong Y. NEDD8 modification of CUL1 dissociates p120(CAND1), an inhibitor of CUL1-SKP1 binding and SCF ligases. Mol Cell. 2002;10:1511–1518. doi: 10.1016/s1097-2765(02)00783-9. [DOI] [PubMed] [Google Scholar]
- 29.Zheng J, Yang X, Harrell JM, Ryzhikov S, Shim EH, Lykke-Andersen K, Wei N, Sun H, Kobayashi R, Zhang H. CAND1 binds to unneddylated CUL1 and regulates the formation of SCF ubiquitin E3 ligase complex. Mol Cell. 2002;10:1519–1526. doi: 10.1016/s1097-2765(02)00784-0. [DOI] [PubMed] [Google Scholar]
- 30•.Goldenberg SJ, Cascio TC, Shumway SD, Garbutt KC, Liu J, Xiong Y, Zheng N. Structure of the Cand1-Cul1-Roc1 complex reveals regulatory mechanisms for the assembly of the multisubunit cullin-dependent ubiquitin ligases. Cell. 2004;119:517–528. doi: 10.1016/j.cell.2004.10.019. This work showed how CAND1 binds CUL1-RBX1 to inhibit assembly of an active CRL. [DOI] [PubMed] [Google Scholar]
- 31.Babon JJ, McManus EJ, Yao S, DeSouza DP, Mielke LA, Sprigg NS, Willson TA, Hilton DJ, Nicola NA, Baca M, et al. The structure of SOCS3 reveals the basis of the extended SH2 domain function and identifies an unstructured insertion that regulates stability. Mol Cell. 2006;22:205–216. doi: 10.1016/j.molcel.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 32.Min JH, Yang H, Ivan M, Gertler F, Kaelin WG, Jr, Pavletich NP. Structure of an HIF-1alpha -pVHL complex: hydroxyproline recognition in signaling. Science. 2002;296:1886–1889. doi: 10.1126/science.1073440. [DOI] [PubMed] [Google Scholar]
- 33.Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- 34.Mizushima T, Hirao T, Yoshida Y, Lee SJ, Chiba T, Iwai K, Yamaguchi Y, Kato K, Tsukihara T, Tanaka K. Structural basis of sugar-recognizing ubiquitin ligase. Nat Struct Mol Biol. 2004;11:365–370. doi: 10.1038/nsmb732. [DOI] [PubMed] [Google Scholar]
- 35.Mizushima T, Yoshida Y, Kumanomidou T, Hasegawa Y, Suzuki A, Yamane T, Tanaka K. Structural basis for the selection of glycosylated substrates by SCF(Fbs1) ubiquitin ligase. Proc Natl Acad Sci U S A. 2007;104:5777–5781. doi: 10.1073/pnas.0610312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- 37.Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 38.Zhuang M, Calabrese MF, Liu J, Waddell MB, Nourse A, Hammel M, Miller DJ, Walden H, Duda DM, Seyedin SN, et al. Structures of SPOP-substrate complexes: insights into molecular architectures of BTB-Cul3 ubiquitin ligases. Mol Cell. 2009;36:39–50. doi: 10.1016/j.molcel.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 41••.Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu FF, Sharon M, Browse J, et al. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010;468:400–405. doi: 10.1038/nature09430. This comprehensive analysis revealed a bidirectional “molecular glue” mechanism for hormone-mediated substrate recruitment to the CRL1 SR COI1, and vice-versa. An inositol phosphate moiety was discovered to further potentiate SR-substrate interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Murase K, Hirano Y, Sun TP, Hakoshima T. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature. 2008;456:459–463. doi: 10.1038/nature07519. This article describes how the plant hormone gibberellic acid functions as a true hormone, allosterically modulating interactions between the CRL1 component GID1 and a DELLA substrate to be ubiquitinated. [DOI] [PubMed] [Google Scholar]
- 43.Hayashi K, Tan X, Zheng N, Hatate T, Kimura Y, Kepinski S, Nozaki H. Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling. Proc Natl Acad Sci U S A. 2008;105:5632–5637. doi: 10.1073/pnas.0711146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol. 2010;28:733–737. doi: 10.1038/nbt.1646. The authors identify and determine the structural basis for activity of the first synthetic small molecule allosteric inhibitor of substrate recognition by a CRL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alexandru G, Graumann J, Smith GT, Kolawa NJ, Fang R, Deshaies RJ. UBXD7 binds multiple ubiquitin ligases and implicates p97 in HIF1alpha turnover. Cell. 2008;134:804–816. doi: 10.1016/j.cell.2008.06.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Itoh K, Ishii T, Wakabayashi N, Yamamoto M. Regulatory mechanisms of cellular response to oxidative stress. Free Radic Res. 1999;31:319–324. doi: 10.1080/10715769900300881. [DOI] [PubMed] [Google Scholar]
- 47.Ohtake F, Baba A, Takada I, Okada M, Iwasaki K, Miki H, Takahashi S, Kouzmenko A, Nohara K, Chiba T, et al. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature. 2007;446:562–566. doi: 10.1038/nature05683. [DOI] [PubMed] [Google Scholar]
- 48.Ito T, Ando H, Suzuki T, Ogura T, Hotta K, Imamura Y, Yamaguchi Y, Handa H. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]