Abstract
Protein S-nitrosylation plays a central role in signal transduction by nitric oxide (NO), and aberrant S-nitrosylation of specific proteins is increasingly implicated in disease. Due to their low-abundance and relative instability, S-nitrosylated proteins (SNO-proteins) have been difficult to detect and quantify in biological systems. However, over the last 10 years, the biotin switch technique (BST) and related thiol-based assays have allowed enrichment and identification of specific SNO-proteins from complex biological mixtures as well as the facile identification and quantification of sites of protein S-nitrosylation (SNO-sites), leading to scores of important new discoveries. Structural characterization of SNO-proteins by X-ray crystallography is also increasingly being utilized to understand both the relationships between protein structure and Cys thiol reactivity as well as the consequences of S-nitrosylation on protein structure and function. Here, we review these and other methodologies for the characterization, identification and quantification of SNO-proteins.
1. Introduction
It is increasingly recognized that protein S-nitrosylation, the post-translational modification of Cys thiol by nitric oxide (NO) to generate S-nitrosothiol (SNO), transduces many of the biological effects of NO [1-3]. Aberrant S-nitrosylation in implicated in numerous cardiopulmonary, skeletomuscular and neurodegenerative diseases [4, 5]. Largely driven by methodological limitations, early studies in the field largely focused on: abundant and readily detectable endogenous species, namely SNO-hemoglobin (SNO-Hb) and SNO-albumin (SNO-Alb) [4]; proteins that could be easily obtained in purified form for in vitro analyses (e.g. SNO-GAPDH and SNO-caspase); and the physiological effects of NO that could often be only indirectly ascribed to S-nitrosylation [6]. The development of new techniques for the enrichment and identification of endogenous SNO-proteins, and mapping of sites of S-nitrosylation (SNO-sites) have prompted most of the major discoveries in the field over the last 10 years, many of which are highlighted elsewhere in this Special Issue.
There are now perhaps hundreds of published permutations of assays for SNO-protein characterization, identification and quantification. However, assays generally fall into one of three classes (Table). They involve: 1) direct detection of a NO-modified thiol; 2) chemical reduction or photolytic breakdown of the SNO to a more readily identifiable NO-based species; or 3) tagging of the S-nitrosylated Cys thiol for subsequent enrichment and identification of SNO-proteins as well as facile mapping of SNO-sites. In general, methodologies in class 1 are mostly biophysical techniques—perhaps the most powerful being X-ray crystallography—that are best suited for characterization of single, isolated SNO-proteins. Techniques specific to class 2 detect SNO-derived NO and nitrite and are amenable to absolute quantification of total amounts of protein S-nitrosylation (but not specific proteins) in biological mixtures. The third class of methodologies are particular suited for identification of SNO-proteins and SNO-sites from complex mixtures and relative (but not absolute) quantification of these species across multiple samples. Here we present an overview of both “tried and true” and promising new methodologies for SNO-protein characterization, identification and quantification.
Table 1.
Overview of methodologies for detection of SNO-proteins.
| Class 1: Detection of intact SNOa |
| X-ray crystallography |
| UV-vis spectroscopy |
| NMR spectrocopy |
| Mass spectrometry |
| SNO-specific antibodies |
| Class 2: Detection of SNO-derived nitrite and NOb |
| Saville Assay |
| DAF-2 Assay |
| GC-MS |
| Photolysis chemiluminscence |
| Reductive chemiluminescence |
| NO electrode |
| Class 3: Labeling of SNO-derived Cys thiolc |
| Biotin switch technique (BST) |
| SNO-site identification (SNO-SID) |
| S-nitrosothiol capture (SNOCAP) |
| Resin-assisted capture of SNO-Proteins (SNO-RAC) |
| Spin trapping after UV photolysis |
| Organomercurial-binding |
| Phosphine-based ligation |
These methods, except for SNO-based antibodies, are mostly suitable for the characterization of purified SNO-proteins and are generally low-sensitivity.
These methods are suitable for quantifying total levels of endogenous SNO-proteins but have limited utility for the analysis of specific SNO-proteins from complex mixtures.
These methods are useful for the enrichment, identification and relative quantification of SNO-proteins from complex mixtures and for the facile identification of SNO-sites.
2. Characterization of intact S-nitrosoproteins and protein-derived S-nitrosopeptides
Purified SNO-proteins are amenable to characterization by a number of biophysical techniques, including mass spectrometry (MS) and X-ray crystallography (see below), as well as ultraviolet/visible spectroscopy (UV/Vis) [7, 8] and 15N nuclear magnetic resonance spectroscopy (NMR) [7, 9]. The techniques are mostly applicable to the characterization of isolated SNO-proteins. UV/vis can be used for quantification of low-mass SNOs [10], but SNO-proteins are not generally produced in sufficient quantities to be easily detected, while NMR has little demonstrated utility. SNO-specific antibodies, raised against an S-nitrosocysteine epitope, have also been used for the enrichment and identification of SNO-proteins in situ (see below).
X-ray crystallography
High-resolution crystal structures have been recently solved for a number of SNO-proteins, namely S-nitrosylated hemeproteins [11-13], protein tyrosine phosphatase 1B [14] and thioredoxin (SNO-Trx) [8]. Collectively, these structural analyses have not only enabled SNO-site identification, but also have helped to characterize the effects of S-nitrosylation on protein structure, the “solid-state” conformations of protein-bound SNO (e.g. R-S-N-O dihedral) and potential mechanisms of S-nitrosylation (Fig. 1). Treatment with NO or low-mass S-nitrosothiol has been performed both prior to [8, 13] and after crystallization [11, 12, 14]. SNO-proteins can survive lengthy crystallization protocols, as evidenced by the stability of SNO-myoglobin after one month at room temperature (under light- and metal-free conditions) [13]. However, the S-NO moiety appears to be unstable to synchrotron radiation [13, 15], which is preferred for solving high-resolution structures.
Figure 1. X-ray crystallography of S-nitrosylated proteins.
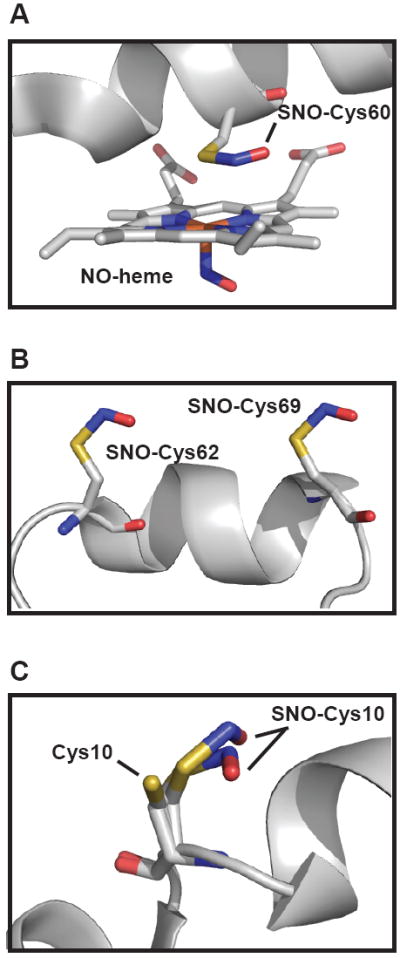
A) Exposure of Cimex lectularius nitrophorin crystal to NO led to distal heme nitrosylation and S-nitrosylation of Cys60 on the proximal side of the heme (PDB ID: 1Y21) [12]. Stopped-flow UV-vis spectroscopy revealed that heme reduction is coupled to S-nitrosylation of the protein by NO. B) Human thioredoxin was S-nitrosylated at two adjacents sites (Cys62 and Cys69) of an α-helix (PDB ID: 2HXK). The S-nitrosylated Cys residues both adopt a cis-planar configuration. Cys62, which is exhibits higher reactivity than Cys69 [8], sits at the helix N-terminus, a newly identified S-nitrosylation motif [55]. Interestingly, SNO-Cys62 is buried and faces the interior of the protein (not shown) [8]. C) The structure of CysNO-treated Blackfin Tuna myoglobin (PDB ID: 2NRM) revealed the presence of both reduced and S-nitrosylated Cys10 [13]. Two conformations of SNO-Cys10, each adopting a cis-planar configuration, were observed. Images were generating using Pymol.
Mass spectrometry (MS)
MS has been applied to the analysis of both intact and protease-digested SNO-proteins. SNOs are unstable to analysis by matrix-assisted laser desorption ionization MS (MALDI-MS) but can be detected by electrospray ionization MS (ESI-MS) [16]. Deconvoluted spectra of intact SNO-proteins exhibit a mass shift of +29 amu per bound NO and can suggest SNO:protein stoichiometry [16-19]. Alternatively, SNO-containing peptides have been identified from proteolyzed proteins using a variety of digestion conditions, instrumentation and acquisition methods [20-22]. SNO-Hb was characterized by pepsin digestion (50:1 protein:protease, 5% formic acid, 37 °C, 1.5 h) followed by reverse phase-LC-MS, varying the cone voltage between 20-80V across multiple injections [20]. A voltage-dependent shift of -29 amu was observed for a peptide corresponding to residues 89-105 of β-Hb (Cys93 is the known SNO-site). A SNO-site in argininosuccinate synthetase (AS) was localized by neutral loss scanning using a triple quadrupole mass spectrometer [21]. SNO-AS was digested with trypsin (50:1, pH 8.0, 37 °C, 4 h) and infused into the ESI source. Parent ions that lost 30 or 15 m/z (singly or doubly charged ions, respectively) in the first quadropole were allowed to pass into the second quadrupole for mass analysis. More recently, a different technique was used to map a SNO-site on Arabidopsis thaliana NPR1 after digestion with trypsin (20:1, pH 5.5, 37 °C, 1 h) and analysis by nano-RP-LC-MS3 on a LTQ-Orbitrap [22]. Peptides were first subjected to a normalized collision energy of 35V, and ions which had a loss of 9.7 or 14.5 m/z (from 3+ and 2+ ions respectively) were fragmented for sequence identification. Low pH and short digestion conditions might favor preservation of SNO, but this has not been adequately examined, nor has the ability of these methods to identify SNO-sites in multi-protein mixtures.
SNO-specific antibodies
Circulating immunoglobulins (IgM) recognizing a SNO epitope (SNO-Cys cross-linked to BSA; SNO-Cys-BSA) were first identified in sera of patients with MS [23], raising the idea that SNO-specific antibodies could be developed as a research tool. Gow et al. demonstrated proof-of-concept by raising a rabbit polyclonal Ab to SNO-Cys-BSA and used it for ELISA-, western blotting- and immunohistochemistry-based analysis of SNO proteins, including the demonstration of cytokine- and GPCR agonist-stimulated S-nitrosylation [24]. Similar antibodies have been used for co-immunoprecipitation and immunofluorescence of endogenous SNO-proteins [25, 26]. However, there are several limitations: the SNO must be stable throughout the course of the assay, which can involve tissue fixation or immunoblotting; SNO antibodies are raised against S-nitrosocysteine, suggesting that only solvent-accessible sites may be good epitopes; and the specificities of commercially-available antibodies (for SNO-Cys versus Cys) are not well-validated.
3. Quantification of protein-bound SNO using nitrite- and NO-based assays
The absolute quantification of protein-bound SNO in isolated proteins or in complex mixtures is most easily achieved through the cleavage of SNO to a more easily assayable species, namely nitrous acid/nitrite or nitric oxide. These methodologies are generally medium to high throughput and can have adequate sensitivity for quantifying endogenous S-nitrosylation. However, immunoprecipitation (IP) or affinity purification is required for detection of specific SNO-proteins from complex biological mixtures [27, 28], the main exceptions being S-nitrosohemogobin (SNO-Hb) and S-nitrosoalbumin (SNO-Alb) which are the major reactive thiol- and SNO-containing species in their respective milieus (erythrocytes and plasma, respectively) [29, 30].
Nitrite-based assays
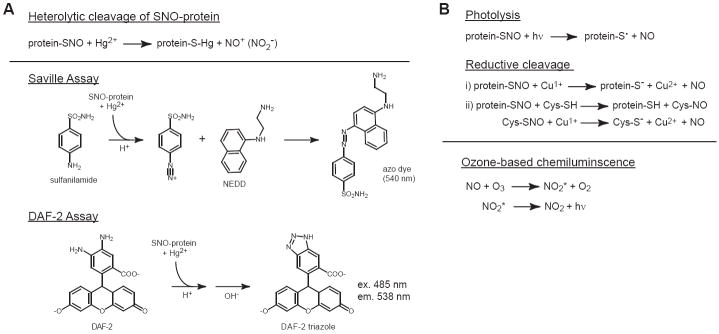
Saville, DAF and GC-MS
The heterolytic cleavage of RSNO by mercury(II) to mercury-thiolate and nitrous acid/nitrite is the basis for a number of SNO-specific assays (Fig. 2). Mercury(II) nitrate has been shown to react with low-mass mass SNOs ~103 more rapidly than mercury(II) chloride due to preferential ionization of the nitrate salt in water [31]. It is assumed that Hg2+ reacts quantitatively with protein-bound SNO, although it is not known whether or how solvent accessibility of the SNO-Cys effects this reaction. However, Hg2+ does not appear to react with the related Cys-coordinated dinitrosyliron complexes [32]. In general, standard curves are generated using low-mass SNOs (e.g. S-nitrosoglutathione) which can be synthesized and quantified in a highly reproducible fashion [10]. Metal chelators, which are often utilized to prevent SNO decomposition, should be present at sub-stoichiometric amounts relative to Hg2+.
Figure 2. Nitrite- and NO-based quantification of SNO-proteins.
A) Nitrite-based assays. Heterolytic cleavage of SNO-protein by Hg2+ generates nitrite/nitrous acid. In the Saville assay, SNO-proteins are treated with HgCl2 in acid to yield nitrous acid, which reacts with sulfanilamide to form diazonium ion. Addition of N-(1-napthyl)ethyelenediamine generates and azo dye with a λmax of 540 nm; In the DAF assay, SNO-proteins are treated with HgCl2 at neutral pH to generate nitrite. The addition of diaminofluorescein and acid yields a triazole which exhibits strong fluorescence upon neutralization of the reaction mixture. B) Nitric oxide-based quantification of SNO-proteins. Nitric oxide is generated from SNO-protein by UV photoloysis or Cu-dependent reductive cleavage (Cu/Cys). NO is quantified by ozone-based cheluminiescence. See text for description of instrumentation used for photolysis-chemiluminescence versus Cu/Cys-based reduction-chemiluminscence.
The colorimetric Saville assay (Fig. 2) [33] is the simplest Hg2+-based assay. SNO-proteins are first mixed with sulfanilamide and HgCl2 in acid, and after incubation are treated with N-(1-naphthyl)ethylenediamine to give an azo dye [34]. The Saville assay is suitable for quantifying ~1-100 μM SNO and thus lacks sensitivity for detection of endogenous SNO-proteins. More sensitive methods utilize diaminonapthalene, diaminofluorescein (DAF) or analogous compounds which form fluorescent triazole species upon reaction with nitrous acid [35]. In a typical DAF assay (Fig. 2), cleavage of SNO-protein by Hg2+ is performed at neutral pH, followed by addition of DAF and acid (to affect nitrosation) and subsequent neutralization (as the DAF triazole fluorescence is highly pH sensitive). The DAF assay exhibits low nM sensitivity and it has been applied to the quantification of SNO-Hb from human erythrocytes [30] and in the activity-based purification of thrioredoxin as a SNO-caspase-3-denitrosylase [36].
Tsikas and coworkers have developed an elegant nitrite-based gas chromatography-MS (GC-MS) method for absolute quantification of S-nitrosoalbumin [37, 38]. First, an internal standard (15N-labelled SNO-albumin) is synthesized by treating albumin with 15N-tert-butyl nitrite or similar isotope-labeled S-nitrosylating agent. After spiking freshly isolated plasma with S15NO-albumin, total albumin is affinity-purified from plasma with HiTrap Blue Sepharose and treated with Hg2+. Following derivitization with pentafluorbenzyl bromide, nitrite (14N and 15N) is extracted, separated by capillary GC and quantified by selective ion monitoring of NO2- (m/z 46) or 15NO2- (m/z 47). This procedure has enabled quantification of SNO-albumin in human plasma [38] and has been used to assess the fate of SNO-albumin in vivo [39].
Nitric oxide detection using ozone-based chemiluminescence and NO-specific electrodes
Nitric oxide-based assays for SNO quantification rely on the chemical reduction or UV-catalyzed homolytic cleavage of SNOs and subsequent quantification of NO using ozone-based chemiluminescence detection or NO-specific electrodes. Compared to nitrite-based assays, NO-based assays provide equal or better sensitivity for detection of endogenous SNOs and in some cases are able to differentiate total protein-bound NO versus SNO.
Hg2+-coupled photolysis-chemiluminscence (P-C) was the first SNO-specific assay that had high sensitivity for detection of endogenous SNOs (Fig. 2) [29]. Samples are injected via a low-pressure pump and delivered via inert gas-purged water into a glass capillary coil. As it travels around the coil, the sample is photolyzed by a Hg vapor lamp, resulting in quantitative release of protein-bound NO which is carried via inert gas under negative pressure to a chemiluminescence-based NO analyzer (Thermal Energy Analyzer; TEA). SNO is defined as the difference in NO signal (area under the curve) in untreated versus Hg2+-treated samples, and the NO that is not displaced by Hg2+ is classified as X-NO (where X=metal, N, O and/or C) [40, 41]; GSNO standards are used for absolute quantification, and the relationship between low-mass and protein-bound SNO is determined by desalting or ultrafiltration [40, 41]. Nitrite and nitrate are generally non-interfering at physiological concentrations [42, 43]. The photolysis apparatus (Nitrolite™, Thermo Orion) is no longer commercially available, making this technique among the least accessible.
NO-based SNO quantification can also be accomplished using a more common chemiluminescence-based NO analyzer (e.g. Sievers NOA) via metal-catalyzed reduction. In the “Cu/Cys” method (Fig. 3) [44], samples are injected into an inert gas purge vessel containing a mixture of CuCl and cysteine in PBS, and the resulting NO is detected. This assay is insensitive to contaminating nitrite, but unlike photolysis-chemiluminescence, only detects SNO. SNO-protein is thought to be first denitrosylated by Cys to CysNO, which is then reduced to NO by Cu1+ (although Cu1+ may also reduce SNO-protein directly). However, it is unclear whether either Cu/Cys can access all protein-bound SNO (only ~1/3 of albumin SNO was recovered as NO [44]). To prevent autocapture of NO by hemeproteins, carbon monoxide can be introduced into the inert gas stream [45]. Triiodide has also been used as a reductant, particularly for the measurement of SNO-Hb [46-48]. However, this requires numerous manipulations: I3- is not specific for NO2- versus total protein-bound NO, so samples must be treated with sulfanilamide to remove background nitrite; “subtractive” analysis of SNO versus X-NO is further accomplished by treatment with Hg2+ and sulfanilamide; and ferricyanide is used to prevent NO autocapture. In part because of these requirements, the triiodide method may underestimate SNO-protein levels [42] and the interpretation of data obtained using this assay should be viewed with caution.
Figure 3. Ascorbate-based enrichment and identification of SNO-proteins and -peptides.
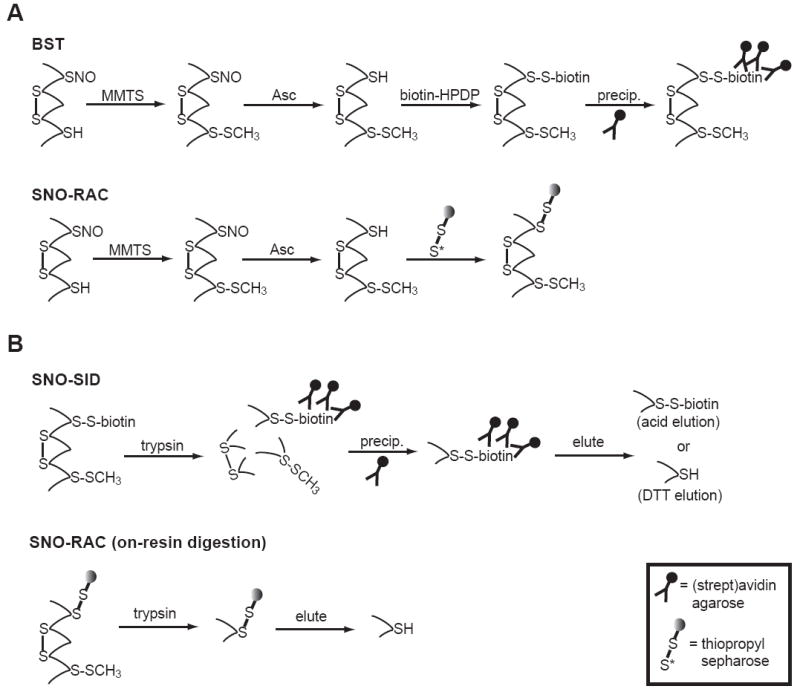
A) In the biotin switch technique (BST) and resin-assisted capture of SNO-proteins (SNO-RAC), free thiols are first S-methylthiolated with S-methyl methanethiosulfonate (MMTS) under denaturing conditions (SDS, heat). In a second step, SNO-Cys residues are denitrosylated with Asc and nascent thiols are biotinylated (BST) or captured using a thiol-reactive resin (SNO-RAC). Note that the BST requires an additional precipitation step. Bound proteins are commonly eluted with reductant and analyzed by SDS-PAGE with staining or western blotting. B) SNO-site identification (SNO-SID) is a modification of the BST, where biotinylated protein is trypsinized, and biotinylated peptides are precipitated and analyzed by LC-MS/MS. Acid elution from (strept)avidin agarose preserves bound biotin. SNO-RAC offers a simplified approach. Trypsin disgestion is performed on-resin to isolate SNO-site containing peptides. Elution must be performed with reductant. BST/SNO-SID and SNO-RAC are amenable to relative quantification by LC-MS/MS (see text).
NO-selective electrodes have also been employed for quantifying S-nitrosothiols in plasma and whole blood. However, in contrast to NOA-based methods, NO electrodes measure steady-state dissolved NO in an open system and can be sensitive to factors that influence NO solubility and diffusion (e.g. temperature, rate of stirring, surface area of the air-liquid interface). These factors may make absolute quantification a challenge. Nonetheless, addition of CuSO4 and ascorbic acid (i.e. Cu1+) to SNO-Alb and to plasma has been shown to generate a quantifiable NO signal [49]; as a negative control, samples were pre-photolyzed by UV irradiation (>330 nm). An immobilized polymeric organoselenium catalyst has also been incorporated into the distal tip of an NO-selective electrode for the specific detection of S-nitrosothiols from whole blood [50]. However, because the catalyst is immobilized to a dialysis membrane, the measurement of SNO-protein requires transnitrosylation to a low mass thiol such as cysteine.
4. Thiol-based assays utilizing ascorbate as a denitro(sy)lating agent
A pivotal moment in analysis of SNO-proteins came 10 years ago with the publication of the biotin switch technique (BST) by Synder and coworkers [51]. The BST was the first assay designed to detect the S-nitrosylated Cys thiol rather than SNO-derived nitrite or NO, and consequently, it made routine the identification of endogenously S-nitrosylation proteins from cells and tissues as well as the proteomic identification of tens to hundreds of SNO-proteins (mostly following treatment with low-mass SNOs) in a single analysis. Subsequent modifications to the assay have enabled identification [52] and relative quantification of SNO-sites by mass spectrometry [53, 54], as well as global S-nitrosylation analysis on protein microarrays [55].
Biotin Switch Technique (BST)
As originally described [51], the BST consists of two sequential steps (Fig. 3A). First, reduced protein thiols are reversibly blocked under denaturing conditions with the S-thiomethylating agent, S-methyl methanethiosulfonate (MMTS). Next, proteins are treated with ascorbate (Asc) to remove the thiol-bound NO, and the newly liberated thiol is simultaneously labeled with a reversible biotinylating agent (biotin-HPDP). These steps are performed in the dark and in the presence of metal chelators. Biotinylated proteins are visualized directly using avidin-HRP after SDS-PAGE and western blotting; alternatively, biotinylated proteins are precipitated with immobilized monomeric avidin or streptavidin and identified by western blotting for protein(s)-of-interest or mass spectrometry. Critical to the success of the assay is the apparent stability of SNO-protein during the blocking step (2.5% SDS, 50 °C) and the specificity of the ascorbate reaction for S-nitrosylation versus S-oxidized or S-thiolated Cys residues (see below).
Thiol blocking step
In the BST and related methods, blocking is typically performed with MMTS, which generates a mixed disulfide. Alternatively, for SNO-site identification (see below), irreversible blocking with reagents such as N-ethylmaleimide [56] can help to identify the site of S-nitrosylation in peptides that contain more than one Cys residue (the tryptic peptide containing the active site of GAPDH, IVSNAS*CTTNCLAPLAK, where the asterisk denotes the SNO-site, is a notable example [56]). It is also important to remember that false positives can arise from incomplete blocking of free thiols. Thus, a true positive cannot be assigned based solely on a signal in the BST, and it is critical to include a negative control such as pre-photolysis or NOS inhibition (also see below).
Specificity and sensitivity of Asc-mediated denitrosation
In the presence of metal chelators (i.e. under the BST reaction conditions), Asc does not act as a reductant but rather undergoes transnitrosation with SNO to yield thiol and O-nitrosoascorbate [57]. However, it has been suggested that Asc can reduce small molecule and protein disulfides under these same conditions [58, 59], observations that were not consistent with the previously demonstrated specificity of the BST [51, 60] and would essentially render the assay useless. In addressing this controversy, Forrester et al. found that exposure to indirect sunlight from a laboratory window (and not fluorescent light) could reduce the activated disulfide in biotin-HPDP and facilitate biotinylation of MMTS-blocked protein thiol [61], providing a mechanism for generation of Asc-dependent artifacts. In this scenario, it is presumed that UV light causes generation of the semi-dehydroascorbate radical, reduction of the reactive disulfide biotin-HPDP and thiol-disulfide exchange with S-methylated protein thiol to form biotinylated protein thiol [61, 62]. Prior to these studies, Asc-dependence was often utilized to demonstrate assay specificity, but it has become clear that additional controls are needed. NOS inhibition, deletion or knockdown can be utilized in some instances to address specificity, but these tools are not amenable to all studies. Alternatively, UV-photolysis of samples, to destroy the SNO prior to analysis, has been used in conjunction with the BST [61, 63, 64] and related assays [65, 66] to identify true positives.
The efficiency of Asc-mediated denitrosation has also been a subject of investigation. It has been reported that at 1 mM Asc (the concentration originally used by Jaffrey et al. [51]) only a fraction of SNO may be converted to free thiol within the timescale of the assay [67]. Concentrations of Asc up to 100 mM increase protein biotinylation and do not appear to sacrifice assay specificity [60, 61, 67]. These findings are consistent with the kinetics of Asc denitrosation, which shows first-order dependence for SNO and Asc in the presence of metal chelators [57]. The rate of Asc-mediated denitrosation can also vary over three orders of magnitude between pH 7-12, although to our knowledge [57], the relationship between pH and sensitivity of the BST has not been tested. Whereas Asc acts as a nucleophile in the absence of added metal ion, Asc can also catalyze the Cu-dependent reduction of S-nitrosothiols [57]. As an alternative to the standard BST, the combination of Cu and 1 mM Asc has been shown to greatly potentiate the specific biotinylation of SNO-protein compared to 1 mM Asc alone [68].
Identification of SNO-sites
It has been a tractable approach to combine site-directed mutagenesis with the BST for identification of SNO-sites. Mass spectrometry-based methods, however, are more direct and amenable to large-scale analyses. Peptide-level Cys biotinylation and capture had been utilized previously for expression proteomics [69], but Gross and coworkers first described the use of a similar methodology for SNO-site identification (SNOSID; Fig. 3B) [52]. Simply, following the BST, biotinylated proteins are proteolyzed with trypsin, and the resulting peptides are affinity-purified with immobilized monomeric avidin or streptavidin, eluted and analyzed by LC-MS/MS. Elution can be performed with reductant [52] or acid [63, 70]; the latter results in a peptide with disulfide-linked biotin.
The SNO-RAC methodology substitutes a thiol-reactive resin (e.g. thiopropyl sepharose) for biotin-HPDP(Fig. 3A) [54]. The elimination of biotin removal and avidin-based enrichment steps appears to result in improved sensitivity for detection of high-mass SNO-proteins. SNO-RAC also simplifies SNO-site identification: “captured” proteins are proteolyzed on resin, and SNO-site-containing peptides are eluted with reductant and identified by LC-MS/MS (Fig. 3B) [54].
Relative protein and SNO-site quantification
Relative levels of SNO-proteins can be assessed across multiple conditions using western blotting and densitometry [55, 71] or by fluorescence-based quantification using either thiol-reactive fluorophores [72-75] or fluorescent secondary antibodies [55]. Measurements can be normalized to total amounts of protein-of-interest in each sample [71]. However, note that the linearity of Asc-based SNO-protein detection across a range of concentrations has not been critically evaluated.
Mass spectrometry has also been employed for relative quantification of SNO-proteins and SNO-site following enrichment by the BST, SNO-RAC and related methodologies. The SNO-CAP method utilizes light and heavy isotope-labeled thiol biotinylating agents to perform relative quantification of SNO-sites between two conditions and was used to identify glutathione-reversible/unstable versus non-reducible/stable S-nitrosylated proteins in vitro and in cultured cells [53]. Alternatively, stable isotope labeling of amino acids in cell culture (SILAC) [76], which employs light and heavy isotope-labeled Arg and/or Lys for relative quantification of tryptic peptides, has been combined with the BST for relative quantification of SNO-proteins in unstimulated versus cytokine-stimulated mouse macrophages [77] and to identify targets of the denitrosylase, thioredoxin [78]. iTRAQ-based isobaric quantification [79] has been used in conjunction with the SNO-RAC method [54]. The amine-reactive iTRAQ tags each produce unique product ions (e.g. 114-117 amu for 4-plex iTRAQ) after peptide fragmentation that are used for relative quantification [79]. To examine kinetics of intracellular S-nitrosylation and denitrosylation, Forrester et al. performed SNO-RAC at 10, 30 and 50 min following treatment of HEK cells with CysNO; a fourth sample was a negative control (i.e. untreated) [54]. After on-resin digestion, samples were labeled with one of 4 amine-reactive isobaric tags, and after elution, samples were mixed and analyzed by LC-MS/MS, resulting in ~350 quantifiable SNO-sites. S-nitrosylation at most sites peaked at 10 min and decayed rapidly, but about one quarter of quantified SNO-sites were relatively stable over the time course of the assay.
5. Alternative chemistries for thiol-based SNO assays
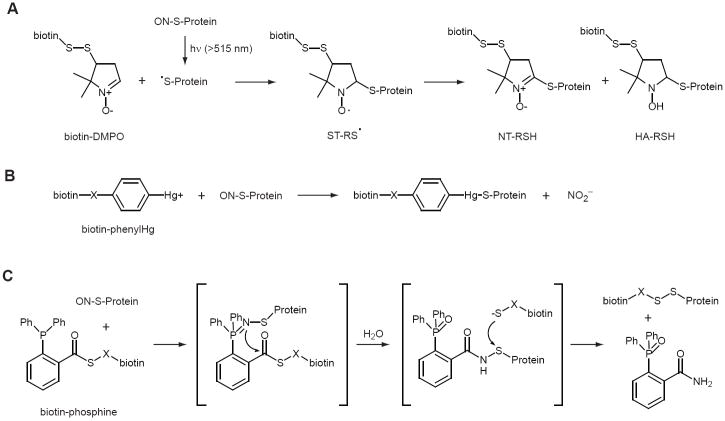
Despite the almost universal acceptance of the BST, several other approaches have emerged as potentially feasible alternatives to Asc-based methodologies (Fig. 4). The radical spin trap 5,5-dimethyl-1-pyrroline N-oxide (DMPO) reacts with S-nitrosothiols after photolysis to thiyl radical [80] (Fig. 4A), and this reaction has recently been used to label protein-bound SNO without the requirement of a thiol-blocking step [81]. Specificity for S-NO versus photolyzable C-NO and N-NO was achieved by irradiation at >515 nm. DMPO adducts were initially visualized by western blotting using a anti-DMPO antibody, but the potential for enrichment and identification of protein-DMPO conjugates has been made easier by the recent development of a disulfide-linked, biotinylated DMPO [82]. In order positively to identify SNO-sites using this assay, it will be critical that there be minimal intramolecular electron transfer between other redox-sensitive sidechains (e.g. Cys, Tyr, Trp); the use of excess radical trap appears to prevent such crosstalk [83]. The initial spin trap/thiyl adduct (ST-RS·), has been shown to disproportionate to nitrone-thiol (N-RSH) and hydroxylamine-thiol (HA-RSH) [84]; the N-RSH adduct is unstable and its decomposition may compromise yield of tagged Cys thiol.
Figure 4. New methods for thiol-based detection of SNO-protein.
Three new methods have recently been reported for tagging of SNO-Cys and biotin-avidin-based detection of SNO-proteins. A) Spin-trapping [81]. SNO-proteins are photolyzed to NO and thiyl radical in the presence of the radical spin trap, DMPO. The initial spin trap-thiyl conjugate undergoes disproportionation to nitrone-thiol and hydroxylamine-thiol (see text for discussion) [84]. A disulfide-containing biotin-DMPO conjugate may facilitate enrichment and identification of SNO-proteins [82]. B) Organomercurial trapping [65]. Free thiols are blocked, and SNO-proteins are reacted with biotin- or resin-conjugated phenylHg. Performic acid can be used to cleave Hg-S, generating cysteine sulfonic acid (Cys-SO3H). C) Reductive ligation [88]. Free thiols are blocked, and SNO-proteins are reacted with a biotin-conjugate phosphine thioester to yield sulfenamide and thiolate intermediates which further react to form disulfide. Reaction scheme is as published in ref. 92.
A SNO-specific assay has also been based on the reaction of SNO-proteins and phenylmercury compounds, which result in covalent Hg-S conjugates (Fig. 4B) [65]. An initial blocking is step is required to prevent reaction of reduced Cys thiol with the organomercurial, which is followed by reaction with either biotin-conjugated or resin-bound phenylmercury compounds for SNO-specific protein enrichment. Performic acid was used to elute and oxidize bound Cys thiol to the oxidized sulfonic acid form, thus providing a unique SNO-site signature for MS analysis. This method identified numerous endogenously S-nitrosylated proteins in both mouse liver and thymus [65, 66], suggesting that it is highly sensitive.
In a series of elegant studies, Xian and coworkers have taken advantage of a neglected reaction between SNO and triphenylphosphine (TPP) [85] to develop novel reagents that may have immense utility for general SNO detection and for the affinity capture of SNO proteins [86]. They recognized the similarity between the SNO/TPP chemistry and that of the Staudinger ligation, which has been exploited for the specific labeling of azide-conjugated molecules (include cell-surface glycans) [87]. Through a series of iterations [86, 88-92] they arrived at biotinylated phosphine capable of trapping S-nitrosylated proteins under aqueous conditions (Fig. 4C). Although they showed that the biotin-phosphine did not react with thiol, Cys thiol blocking was nonetheless employed as a first step. They have also developed a SNO-specific sensor, a coumarin-phosphine, which fluoresces upon oxidation by low-mass SNO [93]. The specificity of this sensor to SNO versus other cellular oxidants, as well as it applicability to the detection of protein-bound SNO, have to yet to be shown.
6. Remaining Challenges
Despite the recent progress in methods for characterization and quantification of SNO-proteins, several challenges remain. Some techniques, namely X-ray crystallography, are underutilized for the characterization of SNO-dependent effects on protein structure and function, and consequently there is still little understanding how S-nitrosylation regulates protein function (although the same might be said for phosphorylation). Furthermore, the BST and related assays have enabled the high-throughput identification of hundreds to thousands of novel targets of exogenous S-nitrosylating compounds but have has had limited success [52, 65, 75] in the de novo identification of endogenous SNO-proteins. More is not always better. The physiological relevance of SNO-proteomics has often been questioned, and the argument that S-nitrosylation is comparable to O-phosphorylation in its ubiquitousness is not well substantiated. Improvements in the sensitivity and efficiency of thiol-based assays are needed to make improvements in these arenas. Finally, as it is increasingly recognized that altered levels of specific SNO-proteins may be symptomological of disease, clinical assays that can robustly quantify SNO-proteins from biological fluids or tissues may represent the next logical step forward for SNO-based assays; it remains to be seen if exisiting methodologies can meet the necessary criteria for clinical utility or whether substantial additional method development will be required.
Acknowledgments
This work was supported in part by a grant from the National Institutes of Health (HL106121). The author thanks J. Will Thompson and Zacchary Kelleher for helpful comments.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 2.Sen N, Snyder SH. Protein modifications involved in neurotransmitter and gasotransmitter signaling. Trends Neurosci. 2010;33:493–502. doi: 10.1016/j.tins.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura T, Lipton SA. S-Nitrosylation of Critical Protein Thiols Mediates Protein Misfolding and Mitochondrial Dysfunction in Neurodegenerative Diseases. Antioxid Redox Signal. 2010 doi: 10.1089/ars.2010.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster MW, McMahon TJ, Stamler JS. S-nitrosylation in health and disease. Trends Mol Med. 2003;9:160–168. doi: 10.1016/s1471-4914(03)00028-5. [DOI] [PubMed] [Google Scholar]
- 5.Foster MW, Hess DT, Stamler JS. Protein S-nitrosylation in health and disease: a current perspective. Trends Mol Med. 2009;15:391–404. doi: 10.1016/j.molmed.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stamler JS. Redox signaling: nitrosylation and related target interactions of nitric oxide. Cell. 1994;78:931–936. doi: 10.1016/0092-8674(94)90269-0. [DOI] [PubMed] [Google Scholar]
- 7.Stamler JS, Simon DI, Osborne JA, Mullins ME, Jaraki O, Michel T, Singel DJ, Loscalzo J. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Natl Acad Sci U S A. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weichsel A, Brailey JL, Montfort WR. Buried S-nitrosocysteine revealed in crystal structures of human thioredoxin. Biochemistry. 2007;46:1219–1227. doi: 10.1021/bi061878r. [DOI] [PubMed] [Google Scholar]
- 9.Simon DI, Mullins ME, Jia L, Gaston B, Singel DJ, Stamler JS. Polynitrosylated proteins: characterization, bioactivity, and functional consequences. Proc Natl Acad Sci U S A. 1996;93:4736–4741. doi: 10.1073/pnas.93.10.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hart TW. Some observations concerning the S-nitroso and S-phenylsulfonyl derivatives of L-cysteine and glutathione. Tetrahedron Lett. 1985;26:2013–2016. [Google Scholar]
- 11.Chan NL, Rogers PH, Arnone A. Crystal structure of the S-nitroso form of liganded human hemoglobin. Biochemistry. 1998;37:16459–16464. doi: 10.1021/bi9816711. [DOI] [PubMed] [Google Scholar]
- 12.Weichsel A, Maes EM, Andersen JF, Valenzuela JG, Shokhireva T, Walker FA, Montfort WR. Heme-assisted S-nitrosation of a proximal thiolate in a nitric oxide transport protein. Proc Natl Acad Sci U S A. 2005;102:594–599. doi: 10.1073/pnas.0406549102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schreiter ER, Rodriguez MM, Weichsel A, Montfort WR, Bonaventura J. S-nitrosylation-induced conformational change in blackfin tuna myoglobin. J Biol Chem. 2007;282:19773–19780. doi: 10.1074/jbc.M701363200. [DOI] [PubMed] [Google Scholar]
- 14.Chen YY, Chu HM, Pan KT, Teng CH, Wang DL, Wang AH, Khoo KH, Meng TC. Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J Biol Chem. 2008;283:35265–35272. doi: 10.1074/jbc.M805287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosenfeld RJ, Bonaventura J, Szymczyna BR, MacCoss MJ, Arvai AS, Yates JR, 3rd, Tainer JA, Getzoff ED. Nitric-oxide synthase forms N-NO-pterin and S-NO-cys: implications for activity, allostery, and regulation. J Biol Chem. 2010;285:31581–31589. doi: 10.1074/jbc.M109.072496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaneko R, Wada Y. Decomposition of protein nitrosothiolsin matrix-assisted laser desorption/ionization and electrospray ionization mass spectrometry. J Mass Spectrom. 2003;38:526–530. doi: 10.1002/jms.466. [DOI] [PubMed] [Google Scholar]
- 17.Ferranti P, Malorni A, Mamone G, Sannolo N, Marino G. Characterisation of S-nitrosohaemoglobin by mass spectrometry. FEBS Lett. 1997;400:19–24. doi: 10.1016/s0014-5793(96)01258-6. [DOI] [PubMed] [Google Scholar]
- 18.Upmacis RK, Hajjar DP, Chait BT, Mirza UA. Direct Observation of Nitrosylated Heme in Myoglobin and Hemoglobin by Electrospray Ionization Mass Spectrometry. J Am Chem Soc. 1997;119:10424–10429. [Google Scholar]
- 19.Wu C, Liu T, Chen W, Oka S, Fu C, Jain MR, Parrott AM, Baykal AT, Sadoshima J, Li H. Redox regulatory mechanism of transnitrosylation by thioredoxin. Mol Cell Proteomics. 2010;9:2262–2275. doi: 10.1074/mcp.M110.000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ferranti P, Malorni A, Mamone G, Sannolo N, Marino G. Characterization of S-nitrosohemoglobin by mass spectrometry. FEBS Lett. 1997;400:19–24. doi: 10.1016/s0014-5793(96)01258-6. [DOI] [PubMed] [Google Scholar]
- 21.Hao G, Xie L, Gross SS. Argininosuccinate synthetase is reversibly inactivated by S-nitrosylation in vitro and in vivo. J Biol Chem. 2004;279:36192–36200. doi: 10.1074/jbc.M404866200. [DOI] [PubMed] [Google Scholar]
- 22.Lindermayr C, Sell S, Muller B, Leister D, Durner J. Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell. 2010;22:2894–2907. doi: 10.1105/tpc.109.066464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boullerne AI, Petry KG, Meynard M, Geffard M. Indirect evidence for nitric oxide involvement in multiple sclerosis by characterization of circulating antibodies directed against conjugated S-nitrosocysteine. J Neuroimmunol. 1995;60:117–124. doi: 10.1016/0165-5728(95)00061-6. [DOI] [PubMed] [Google Scholar]
- 24.Gow AJ, Chen Q, Hess DT, Day BJ, Ischiropoulos H, Stamler JS. Basal and stimulated protein S-nitrosylation in multiple cell types and tissues. J Biol Chem. 2002;277:9637–9640. doi: 10.1074/jbc.C100746200. [DOI] [PubMed] [Google Scholar]
- 25.Wang G, Moniri NH, Ozawa K, Stamler JS, Daaka Y. Nitric oxide regulates endocytosis by S-nitrosylation of dynamin. Proc Natl Acad Sci U S A. 2006;103:1295–1300. doi: 10.1073/pnas.0508354103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwakiri Y, Satoh A, Chatterjee S, Toomre DK, Chalouni CM, Fulton D, Groszmann RJ, Shah VH, Sessa WC. Nitric oxide synthase generates nitric oxide locally to regulate compartmentalized protein S-nitrosylation and protein trafficking. Proc Natl Acad Sci U S A. 2006;103:19777–19782. doi: 10.1073/pnas.0605907103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mannick JB, Hausladen A, Liu L, Hess DT, Zeng M, Miao QX, Kane LS, Gow AJ, Stamler JS. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 28.Haendeler J, Hoffmann J, Tischler V, Berk BC, Zeiher AM, Dimmeler S. Redox regulatory and anti-apoptotic functions of thioredoxin depend on S-nitrosylation at cysteine 69. Nat Cell Biol. 2002;4:743–749. doi: 10.1038/ncb851. [DOI] [PubMed] [Google Scholar]
- 29.Stamler JS, Jaraki O, Osborne J, Simon DI, Keaney J, Vita J, Singel D, Valeri CR, Loscalzo J. Nitric oxide circulates in mammalian plasma primarily as an S-nitroso adduct of serum albumin. Proc Natl Acad Sci U S A. 1992;89:7674–7677. doi: 10.1073/pnas.89.16.7674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMahon TJ, Moon RE, Luschinger BP, Carraway MS, Stone AE, Stolp BW, Gow AJ, Pawloski JR, Watke P, Singel DJ, Piantadosi CA, Stamler JS. Nitric oxide in the human respiratory cycle. Nat Med. 2002;8:711–717. doi: 10.1038/nm718. [DOI] [PubMed] [Google Scholar]
- 31.Swift HR, Williams DLH. Decomposition of S-nitrosothiols by mercury(II) and silver salts. J Chem Soc Perkin Trans. 1997;2:1933–1935. [Google Scholar]
- 32.Bosworth CA, Toledo JC, Jr, Zmijewski JW, Li Q, Lancaster JR., Jr Dinitrosyliron complexes and the mechanism(s) of cellular protein nitrosothiol formation from nitric oxide. Proc Natl Acad Sci U S A. 2009;106:4671–4676. doi: 10.1073/pnas.0710416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liddell HF, Saville B. Colorimetric determination of cysteine. Analyst. 1959;84:188–190. [Google Scholar]
- 34.Feelisch M, Stamler JS, editors. Methods in Nitric Oxide Research. 1996. [Google Scholar]
- 35.Park JK, Kostka P. Fluorometric detection of biological S-nitrosothiols. Anal Biochem. 1997;249:61–66. doi: 10.1006/abio.1997.2159. [DOI] [PubMed] [Google Scholar]
- 36.Benhar M, Forrester MT, Hess DT, Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsikas D, Sandmann J, Rossa S, Gutzki F-M, Frolich JC. Measurement of S-nitrosoalbumin by gas chromatography-mass spectrometry. I. Preparation, purification, isolation, characterization and metabolism of S-[15N]nitrosoalbumin in human blood in vitro. J Chromatogr B: Biomed Sci Appl. 1999;726:1–12. [PubMed] [Google Scholar]
- 38.Tsikas D, Sandmann J, Gutzki F-M, Stichtenoth DO, Frolich JC. Measurement of S-nitrosoalbumin by gas chromatography-mass spectrometry. II. quantitative determination of S-nitrosoalbumin in human plasma using S-[15N]nitrosoalbumin as internal standard. J Chromatogr B: Biomed Sci Appl. 1999;726:13–24. [PubMed] [Google Scholar]
- 39.Warnecke A, Luessen P, Sandmann J, Ikic M, Rossa S, Gutzki F-M, Stichtenoth DO, Tsikas D. Application of a stable-isotope dilution technique to study the pharmacokinetics of human 15N-labelled S-nitrosoalbumin in the rat: Possible mechanistic and biological implications. J Chromatogr B: Anal Technol Biomed Life Sci. 2009;877:1375–1387. doi: 10.1016/j.jchromb.2008.11.035. [DOI] [PubMed] [Google Scholar]
- 40.Eu JP, Liu L, Zeng M, Stamler JS. An apoptotic model for nitrosative stress. Biochemistry. 2000;39:1040–1047. doi: 10.1021/bi992046e. [DOI] [PubMed] [Google Scholar]
- 41.Foster MW, Liu L, Zeng M, Hess DT, Stamler JS. A genetic analysis of nitrosative stress. Biochemistry. 2009;48:792–799. doi: 10.1021/bi801813n. [DOI] [PubMed] [Google Scholar]
- 42.Hausladen A, Rafikov R, Angelo M, Singel DJ, Nudler E, Stamler JS. Assessment of nitric oxide signals by triiodide chemiluminescence. Proc Natl Acad Sci U S A. 2007;104:2157–2162. doi: 10.1073/pnas.0611191104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bennett-Guerrero E, Veldman TH, Doctor A, Telen MJ, Ortel TL, Reid TS, Mulherin MA, Zhu H, Buck RD, Califf RM, McMahon TJ. Evolution of adverse changes in stored RBCs. Proc Natl Acad Sci U S A. 2007;104:17063–17068. doi: 10.1073/pnas.0708160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang K, Ragsdale NV, Carey RM, MacDonald T, Gaston B. Reductive assays for S-nitrosothiols: implications for measurements in biological systems. Biochem Biophys Res Commun. 1998;252:535–540. doi: 10.1006/bbrc.1998.9688. [DOI] [PubMed] [Google Scholar]
- 45.Doctor A, Platt R, Sheram ML, Eischeid A, McMahon T, Maxey T, Doherty J, Axelrod M, Kline J, Gurka M, Gow A, Gaston B. Hemoglobin conformation couples erythrocyte S-nitrosothiol content to O2 gradients. Proc Natl Acad Sci U S A. 2005;102:5709–5714. doi: 10.1073/pnas.0407490102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Samouilov A, Zweier JL. Development of chemiluminescence-based methods for specific quantitation of nitrosylated thiols. Anal Biochem. 1998;258:322–330. doi: 10.1006/abio.1998.2609. [DOI] [PubMed] [Google Scholar]
- 47.Rogers SC, Khalatbari A, Gapper PW, Frenneaux MP, James PE. Detection of human red blood cell-bound nitric oxide. J Biol Chem. 2005;280:26720–26728. doi: 10.1074/jbc.M501179200. [DOI] [PubMed] [Google Scholar]
- 48.Wang X, Bryan NS, MacArthur PH, Rodriguez J, Gladwin MT, Feelisch M. Measurement of nitric oxide levels in the red cell: validation of tri-iodide-based chemiluminescence with acid-sulfanilamide pretreatment. J Biol Chem. 2006;281:26994–27002. doi: 10.1074/jbc.M603953200. [DOI] [PubMed] [Google Scholar]
- 49.Gandley RE, Tyurin VA, Huang W, Arroyo A, Daftary A, Harger G, Jiang J, Pitt B, Taylor RN, Hubel CA, Kagan VE. S-nitrosoalbumin-mediated relaxation is enhanced by ascorbate and copper: effects in pregnancy and preeclampsia plasma. Hypertension. 2005;45:21–27. doi: 10.1161/01.HYP.0000150158.42620.3e. [DOI] [PubMed] [Google Scholar]
- 50.Cha W, Anderson MR, Zhang F, Meyerhoff ME. Amperometric S-nitrosothiol sensor with enhanced sensitivity based on organoselenium catalysts. Biosens Bioelectron. 2009;24:2441–2446. doi: 10.1016/j.bios.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaffrey SR, Erdjument-Bromage H, Ferris CD, Tempst P, Snyder SH. Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat Cell Biol. 2001;3:193–197. doi: 10.1038/35055104. [DOI] [PubMed] [Google Scholar]
- 52.Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci U S A. 2006;103:1012–1017. doi: 10.1073/pnas.0508412103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Paige JS, Xu G, Stancevic B, Jaffrey SR. Nitrosothiol reactivity profiling identifies S-nitrosylated proteins with unexpected stability. Chem Biol. 2008;15:1307–1316. doi: 10.1016/j.chembiol.2008.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol. 2009;27:557–559. doi: 10.1038/nbt.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Foster MW, Forrester MT, Stamler JS. A protein microarray-based analysis of S-nitrosylation. Proc Natl Acad Sci U S A. 2009;106:18948–18953. doi: 10.1073/pnas.0900729106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kohr MJ, Aponte AM, Sun J, Wang G, Murphy E, Gucek M, Steenbergen C. Characterization of Potential S-nitrosylation Sites in the Myocardium. Am J Physiol Heart Circ Physiol. 2011 doi: 10.1152/ajpheart.00997.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holmes AJ, Williams DLH. Reaction of ascorbic acid with S-nitrosothiols: clear evidence for two distinct reaction pathways. Perkin. 2000;2:1639–1644. [Google Scholar]
- 58.Huang B, Chen C. An ascorbate-dependent artifact that interferes with the interpretation of the biotin switch assay. Free Radic Biol Med. 2006;41:562–567. doi: 10.1016/j.freeradbiomed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 59.Landino LM, Koumas MT, Mason CE, Alston JA. Ascorbic acid reduction of microtubule protein disulfides and its relevance to protein S-nitrosylation assays. Biochem Biophys Res Commun. 2006;340:347–352. doi: 10.1016/j.bbrc.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 60.Foster MW, Stamler JS. New insights into protein S-nitrosylation. Mitochondria as a model system. J Biol Chem. 2004;279:25891–25897. doi: 10.1074/jbc.M313853200. [DOI] [PubMed] [Google Scholar]
- 61.Forrester MT, Foster MW, Stamler JS. Assessment and application of the biotin switch technique for examining protein S-nitrosylation under conditions of pharmacologically induced oxidative stress. J Biol Chem. 2007;282:13977–13983. doi: 10.1074/jbc.M609684200. [DOI] [PubMed] [Google Scholar]
- 62.Walmsley TA, Abernethy MH, Fitzgerald HP. Clin Chem. Vol. 33. Winston-Salem, N C: 1987. Effect of daylight on the reaction of thiols with Ellman’s reagent, 5,5’-dithiobis(2-nitrobenzoic acid) pp. 1928–1931. [PubMed] [Google Scholar]
- 63.Whalen EJ, Foster MW, Matsumoto A, Ozawa K, Violin JD, Que LG, Nelson CD, Benhar M, Keys JR, Rockman HA, Koch WJ, Daaka Y, Lefkowitz RJ, Stamler JS. Regulation of beta-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell. 2007;129:511–522. doi: 10.1016/j.cell.2007.02.046. [DOI] [PubMed] [Google Scholar]
- 64.Into T, Inomata M, Nakashima M, Shibata K, Hacker H, Matsushita K. Regulation of MyD88-dependent signaling events by S nitrosylation retards toll-like receptor signal transduction and initiation of acute-phase immune responses. Mol Cell Biol. 2008;28:1338–1347. doi: 10.1128/MCB.01412-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Doulias PT, Greene JL, Greco TM, Tenopoulou M, Seeholzer SH, Dunbrack RL, Ischiropoulos H. Structural profiling of endogenous S-nitrosocysteine residues reveals unique features that accommodate diverse mechanisms for protein S-nitrosylation. Proc Natl Acad Sci U S A. 2010;107:16958–16963. doi: 10.1073/pnas.1008036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang Z, Wang ZE, Doulias PT, Wei W, Ischiropoulos H, Locksley RM, Liu L. Lymphocyte development requires S-nitrosoglutathione reductase. J Immunol. 2010;185:6664–6669. doi: 10.4049/jimmunol.1000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Keszler A, Broniowska KA, Hogg N. Characterization and application of the biotin-switch assay for the identification of S-nitrosated proteins. Free Radic Biol Med. 2005;38:874–881. doi: 10.1016/j.freeradbiomed.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 68.Wang X, Kettenhofen NJ, Shiva S, Hogg N, Gladwin MT. Copper dependence of the biotin switch assay: modified assay for measuring cellular and blood nitrosated proteins. Free Radic Biol Med. 2008;44:1362–1372. doi: 10.1016/j.freeradbiomed.2007.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 70.Greco TM, Hodara R, Parastatidis I, Heijnen HF, Dennehy MK, Liebler DC, Ischiropoulos H. Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc Natl Acad Sci U S A. 2006;103:7420–7425. doi: 10.1073/pnas.0600729103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kelleher ZT, Matsumoto A, Stamler JS, Marshall HE. NOS2 regulation of NF-kappaB by S-nitrosylation of p65. J Biol Chem. 2007;282:30667–30672. doi: 10.1074/jbc.M705929200. [DOI] [PubMed] [Google Scholar]
- 72.Kettenhofen NJ, Wang X, Gladwin MT, Hogg N. In-gel detection of S-nitrosated proteins using fluorescence methods. Methods Enzymol. 2008;441:53–71. doi: 10.1016/S0076-6879(08)01204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Santhanam L, Gucek M, Brown TR, Mansharamani M, Ryoo S, Lemmon CA, Romer L, Shoukas AA, Berkowitz DE, Cole RN. Selective fluorescent labeling of S-nitrosothiols (S-FLOS): a novel method for studying S-nitrosation. Nitric Oxide. 2008;19:295–302. doi: 10.1016/j.niox.2008.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tello D, Tarin C, Ahicart P, Breton-Romero R, Lamas S, Martinez-Ruiz A. A “fluorescence switch” technique increases the sensitivity of proteomic detection and identification of S-nitrosylated proteins. Proteomics. 2009;9:5359–5370. doi: 10.1002/pmic.200900070. [DOI] [PubMed] [Google Scholar]
- 75.Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res. 2007;101:1155–1163. doi: 10.1161/CIRCRESAHA.107.155879. [DOI] [PubMed] [Google Scholar]
- 76.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 77.Zhou X, Han P, Li J, Zhang X, Huang B, Ruan HQ, Chen C. ESNOQ, proteomic quantification of endogenous S-nitrosation. PLoS One. 2010;5:e10015. doi: 10.1371/journal.pone.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Benhar M, Thompson JW, Moseley MA, Stamler JS. Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry. 2010;49:6963–6969. doi: 10.1021/bi100619k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 80.Singh RJ, Hogg N, Joseph J, Kalyanaraman B. Mechanism of nitric oxide release from S-nitrosothiols. J Biol Chem. 1996;271:18596–18603. doi: 10.1074/jbc.271.31.18596. [DOI] [PubMed] [Google Scholar]
- 81.Sengupta R, Billiar TR, Stoyanovsky DA. Studies toward the analysis of S-nitrosoproteins. Org Biomol Chem. 2009;7:232–234. doi: 10.1039/b817981f. [DOI] [PubMed] [Google Scholar]
- 82.Lardinois OM, Chatterjee S, Mason RP, Tomer KB, Deterding LJ. Biotinylated analogue of the spin-trap 5,5-dimethyl-1-pyrroline-N-oxide for the detection of low-abundance protein radicals by mass spectrometry. Anal Chem. 2010;82:9155–9158. doi: 10.1021/ac1023183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bhattacharjee S, Deterding LJ, Jiang J, Bonini MG, Tomer KB, Ramirez DC, Mason RP. Electron transfer between a tyrosyl radical and a cysteine residue in hemoproteins: spin trapping analysis. J Am Chem Soc. 2007;129:13493–13501. doi: 10.1021/ja073349w. [DOI] [PubMed] [Google Scholar]
- 84.Potapenko DI, Bagryanskaya EG, Tsentalovich YP, Reznikov VA, Clanton TL, Khramtsov VV. Reversible Reactions of Thiols and Thiyl Radicals with Nitrone Spin Traps. J Phys Chem B. 2004;108:9315–9324. [Google Scholar]
- 85.Haake M. Deoxygenation of trityl thionitrite. Tetrahedron Lett. 1972:3405–3408. [Google Scholar]
- 86.Wang H, Xian M. Fast reductive ligation of S-nitrosothiols. Angew Chem Int Ed. 2008;47:6598–6601. doi: 10.1002/anie.200801654. [DOI] [PubMed] [Google Scholar]
- 87.Saxon E, Armstrong JI, Bertozzi CR. A “traceless” Staudinger ligation for the chemoselective synthesis of amide bonds. Org Lett. 2000;2:2141–2143. doi: 10.1021/ol006054v. [DOI] [PubMed] [Google Scholar]
- 88.Zhang J, Li S, Zhang D, Wang H, Whorton AR, Xian M. Reductive ligation mediated one-step disulfide formation of S-nitrosothiols. Org Lett. 2010;12:4208–4211. doi: 10.1021/ol101863s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang J, Wang H, Xian M. Exploration of the “traceless” reductive ligation of S-nitrosothiols. Org Lett. 2009;11:477–480. doi: 10.1021/ol802663q. [DOI] [PubMed] [Google Scholar]
- 90.Zhang J, Wang H, Xian M. An unexpected Bis-ligation of S-nitrosothiols. J Am Chem Soc. 2009;131:3854–3855. doi: 10.1021/ja900370y. [DOI] [PubMed] [Google Scholar]
- 91.Wang H, Zhang J, Xian M. Facile formation of dehydroalanine from S-nitrosocysteines. J Am Chem Soc. 2009;131:13238–13239. doi: 10.1021/ja905558w. [DOI] [PubMed] [Google Scholar]
- 92.Wang H, Xian M. Chemical methods to detect S-nitrosation. Curr Opin Chem Biol. 2010 doi: 10.1016/j.cbpa.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pan J, Downing JA, McHale JL, Xian M. A fluorogenic dye activated by S-nitrosothiols. Mol Biosyst. 2009;5:918–920. doi: 10.1039/b822283e. [DOI] [PubMed] [Google Scholar]