Abstract
Piwi (P-element-induced wimpy testis) first discovered in Drosophila is a member of the Argonaute family of micro-RNA binding proteins with essential roles in germ-cell development. The murine homologue of PiwiL2, also known as Mili is selectively expressed in the testes, and mice bearing targeted mutations of the PiwiL2 gene are male-sterile. PiwiL2 proteins are thought to protect the germ line genome by suppressing retrotransposons, stabilizing heterochromatin structure, and regulating target genes during meiosis and mitosis. Here we report that PiwiL2 and associated piRNAs (piRs) may play similar roles in adult mouse mesenchymal stem cells. We found that PiwiL2 is expressed in the cytoplasm of metaphase mesenchymal stem cells from the bone marrow of adult and aged mice. Knockdown of PiwiL2 with a specific siRNA enhanced cell proliferation, significantly increased the number of cells in G1/S and G2/M cell cycle phases and was associated with increased expression of cell cycle genes CCND1, CDK8, microtubule regulation genes, and decreased expression of tumor suppressors Cables-1, LATS and Cxxc4. The results suggest broader roles for Piwi in genome surveillance beyond the germ-line and a possible role in regulating the cell cycle of mesenchymal stem cells.
Keywords: miRNA, stem cell, stromal cell, cell division, piRNA, cell cycle
INTRODUCTION
MicroRNAs (miRs) are endogenous non-coding ~23 nucleotide RNAs that regulate gene expression in eukaryotic cells at the post-transcriptional level [1]. Over 700 human miRNA sequences have been described and more than 30% of the human genome is thought to be under miRNA control. MiRs are transcribed by RNA polymerases II and III and the primary transcripts are processed to stem-loop-structured miRNA precursors by the RNase III enzyme Drosha (reviewed in [2-5]). Dicer generates 21–25 nt long dsRNAs that separate into 2 strands one of which binds to a member of the Argonaute protein family to form the silencing complex that uses the miR sequence to target specific RNAs for post-translational repression [6, 7]. Depending on the miR and the mRNA target, Ago protein complexes can block transcription or mediate posttranscriptional gene silencing by blocking translation, or by promoting mRNA degradation (reviewed in [8]). MiRs coordinate the expression of multiple genes and play roles in numerous biological contexts including growth, cell division, development and disease.
Piwi (P-element induced wimpy testis) is a subfamily of Argonaute proteins originally described in Drosophila where its expression in germ line cells is essential for spermatogenesis. Like Argonaute miRs, Piwi proteins are targeted to cytoplasmic mRNA after complexing to Piwi-interacting RNAs (piRNAs). These RNAs are slightly longer than the miRs (26-31nts), there are no equivalents of Drosher or Dicer in the Piwi subfamily and with the possible exception of some tumor cells the gene silencer functions are germ line-specific [8, 9]. A role for both fly and mammalian Piwi-piRNAs in germ-line nuclear surveillance has been suggested wherein genomic integrity is preserved by the interception of retrotransposons, regulation of heterochromatin structure, and possibly fine-tuning of selective genes by pretranslational silencing. In the mouse both Miwi and PiwiL2 are essential for spermatogenesis [10, 11]. Male PiwiL2 −/− mice are sterile with smaller testes due to meiosis defects that cause an arrest of spermatogenesis.
Opposite effects have been reported for over-expression of Piwi homologues in different cell types. In Drosophila germ cells and several cancer cells Piwi over-expression increases the rate of cell division [12-16]. Conversely, over-expression of Hiwi in human leukemia KG1 cells was reported to decrease cell proliferation and enhance apoptosis [17]. It has been proposed that Piwi-regulation is restricted to meiotic germ line cells because of a requirement to preserve genomic integrity during the numerous rounds of replication required for spermatogenesis. Few targets for piRNAs have been described and the mechanism for their control of cell cycle is not known.
Mesenchymal stem cells (MSCs) are multipotent primitive cells that may be required for tissue repair and, like germ cells are repopulated throughout life [18-21]. We recently reported profound age-related changes in the expression of differentiation, growth factor, and cell cycle-related genes during aging of mouse MSCs [22]. Here we present evidence that adult and aged MSCs express significant levels of PiwiL2 and its associated piRNA. Expression of PiwiL2 in aged MSCs was restricted to metaphase cells and was amplified by synchronizing cells in metaphase. PiwiL2 knockdown doubled the rate of proliferation and caused an upward shift in the number of cells in G1/S. Microarray and RT-PCR analyses after PiwiL2 knockdown indicated selective regulation of cell cycle and tumor suppressor genes.
MATERIALS AND METHODS
Cell isolation and culture
MSCs were isolated from C57BL/6 WT mice aged 2, 8 and 26 months (4 mice per group) as described in Ref 34. Cells were cultured in mouse MSC basal media with supplements (Stem Cell Technologies, Va) for 10 passages before harvest. Details of marker expression and phenotypic characterization are reported in [22].
Flow cytometry
MSCs were fixed in 4% formaldehyde, permeabilized with Triton X-100 (0.1% in PBS) and incubated with anti-mouse Piwil2 antibody (Abcam) in 2% calf serum and 0.05% sodium azide for 60 minutes at 4°C. After rinsing the cells were incubated with 0.5μl FITC conjugated anti-rabbit antibody for 30 minutes at 4°C. Cells were analyzed by flow cytometry using a FACSCalibur with CellQuest Pro 4.0.2 software (Becton Dickinson, San Jose, CA). For negative control the primary antibody was substituted with an irrelevant IgG.
Microarray
These analyses are described in detail in [22].
Quantitative RT-PCR
RNA was reverse-transcribed using TaqMan Reverse Transcription Reagents (Applied Biosystems, Foster City, CA). Piwil2 expression levels were measured by real-time PCR with an ABI/PRISM 7700 sequence detection system (Applied Biosystems). The expression level of Piwil2 mRNA relative to hypoxanthine–guanine phosphoribosyl-transferase (HPRT) was calculated using the comparative threshold cycle (CT) method (2-ΔΔCT).
Immunostaining of Piwil2
MSCs were seeded in chamber slides, fixed in 4% formaldehyde, permeabilized with 0.1% Triton X-100, and incubated with anti-mouse Piwil2 antibody in at 4°C overnight followed by PE-anti-goat secondary antibody. Imaging was achieved using a Zeiss LSM-510 Confocal Laser Scanning Microscope. Substitution of the primary antibody with irrelevant IgG isotopes served as negative controls.
Immunoprecipitation
Cells were lysed in buffer containing 1% Triton X-100, 25mM β-glycerophosphate, and protease inhibitors. Equal amounts of protein were incubated with anti-Piwil2 antibody and A/G agarose beads (Santa Cruz). Proteins were analyzed by 10% SDS-PAGE as described [23].
Metaphase Synchronization
Exponentially-growing MSCs were incubated in complete Mesencult medium with 1200 ng/ml Nocodazole at 37°C for 12 hr. Loosely attached, rounded mitotic cells were harvested and resuspended in complete Mesencult medium.
SiRNA knockdown and microarray analysis
Piwil2 siRNA (Ambion) or random sequence siRNA were delivered by Lipfectamine™ 2000 following the manufacturer’s instructions (Invitrogen, CA). Cultures were incubated at 37° for 48 hours and transfected cells harvested for proliferation, RNA or protein assays. Proliferation: Transfected cells were trypsinized and counted in a Vi-CELL™ XR cell viability analyzer (Beckman coulter Inc.). Cell cycle: Cells were fixed in 70% ethanol at -20°C overnight, washed and resuspended in PI/RNase Staining Buffer (BD Pharmingen). Cells were analyzed by flow cytometry using a FACS Calibur with CellQuest Pro 4.0.2 software, (Becton Dickinson, San Jose, CA).
Northern blot
Total RNA was isolated from cells or tissue using Trizol and small RNAs separated by 15% denaturing PAGE. Gels were stained with SybrGold (Molecular Probes) and electroblotted onto Hybond-N+membranes (Amersham). Membranes were UV crosslinked and hybridized with biotin end-labeled oligonucleotide probes, sequence: AAAGCTATCTGAGCACCTGTGTTCAT TCA (Girard et al). A North2South Chemiluminescent Hybridization and Detection Kit (Thermo Fisher Scientific Inc) was used to detect the piRNA. Blots were exposed to streptadivin-HRP and developed with an ECL reagent.
RESULTS
Quantification of PiwiL2 expression by microarray, RT-PCR and FACs
Microarray analyses revealed that MSCs from mice at age 8 and 26 months contained significantly more Piwil2 mRNA transcripts than those from 2-month mice. These results were confirmed by RT-PCR (Figure 1a). To confirm these results at the protein level MSCs from each age group were labeled with anti-Piwi2L antibody and permeabilized cells quantified by flow cytometry. FACS analysis confirmed a significant increase of Piwil2 expression in MSCs derived from 8-month compared with 2-month old mice (Fig. 1b). To relate the level of expression of PiwiL2 in 8-month MSCs to that in germ line cells PiwiL2 protein was immunoprecipitated from mouse testis and MSCs. As shown in Figure 1c PiwiL2 was expressed in the 8-month MSCs at a level about 10% of that in testes. To detect piRNA expression, MSCs from 2-month mice were transfected with Piwi2L cDNA as described in Methods and RNAs from 2- and 8-month mice were analyzed by Northern blot. As shown in Figure 1d, piRNA was faintly detected in 2-month MSCs, this expression was strongly augmented by PiwiL2 gene transfection, and 8-month MSCs expressed piRNA at a level about one third of that in the transfected 2-month MSCs. To compare piRNA from MSCs with that from testes RNA was purified from 8-month MSCs and mouse testes and analyzed by Northern blot. As shown in Figure 1e MSCs displayed distinct RNA bands at 32 and 26 nucleotides, sizes predicted for miRNA and piRNA respectively. The level was about 10% of that in testes. These results are consistent with significant expression of PiwiL2 and probably its piRNA in adult mouse MSCs.
Figure 1. Selective expression of PiwiL2 in adult and aged bone marrow MSCs.
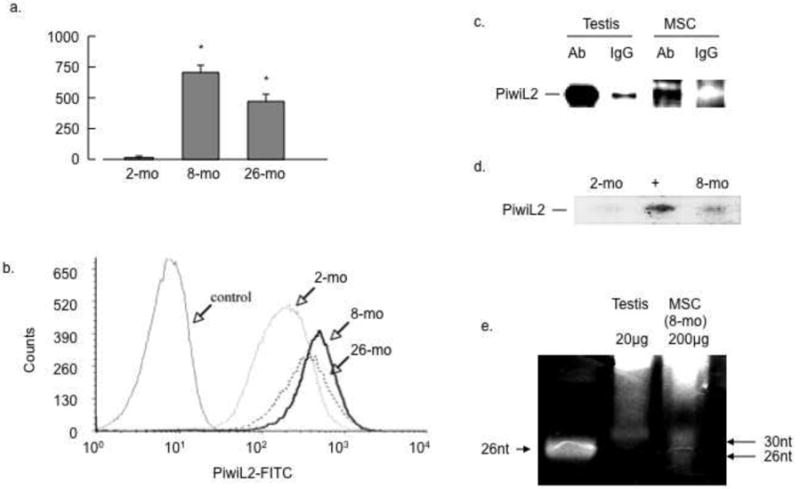
(a) RT-PCR quantification of PiwiL2 transcripts: RT-PCR was implemented on RNA from passage 12 bone marrow MSCs from mice at the indicated age groups as described in Methods (n=3; p<0.05). (b) FACS quantification of PiwiL2 expression: MSCs from each age group at passage 14 were analyzed by FACS as described in Methods, results are representative of 3 separate experiments. (c) PiwiL2 expression in testis and MSCs: protein lysates were prepared from mouse testis or MSCs from 8-month mice, immunoprecipitated and analyzed by Western blots as described in Methods. (d) Identification of micro-RNAs: MSCs from 2-month mice were transfected with PiwiL2 cDNA; RNA extracted from 2 months ± transfection or 8 months was analyzed by Northern blot and probed with anti-piRNA oligonucleotide as described in Methods. (e) Northern Blot comparison of testis and MSC piRNA: RNA was purified from mouse testes or MSCs as indicated. Testis (20μg) and MSC (200μg) RNAs were electrophoresed on 15% denaturing PAGE gels and stained with SybrGold (Molecular Probes). Results are representative of at least 3 separate experiments.
Piwil2 expression is confined to the cytoplasm of mitotic MSCs
Previous studies have shown that PiwiL2 expression in germ line cells is cytoplasmic and mostly confined to mitotic cells [14]. To determine the distribution of PiwL2 in MSCs, semi-confluent cells from 8-month mice were immunostained with anti-PiwiL2 antibody and analyzed by fluorescence microscopy. As shown in Figure 2, fluorescence was perinuclear and selective for actively dividing cells with only weak diffuse expression in non-dividing cells. The expression and intracellular location were similar to that reported for mouse germ line cells [14]. These results suggest that PiwiL2 expression and intracellular localization in MSCs are similar to those in mitotic germ line cells.
Figure 2. Intracellular localization of PiwiL2 by immunostaining.
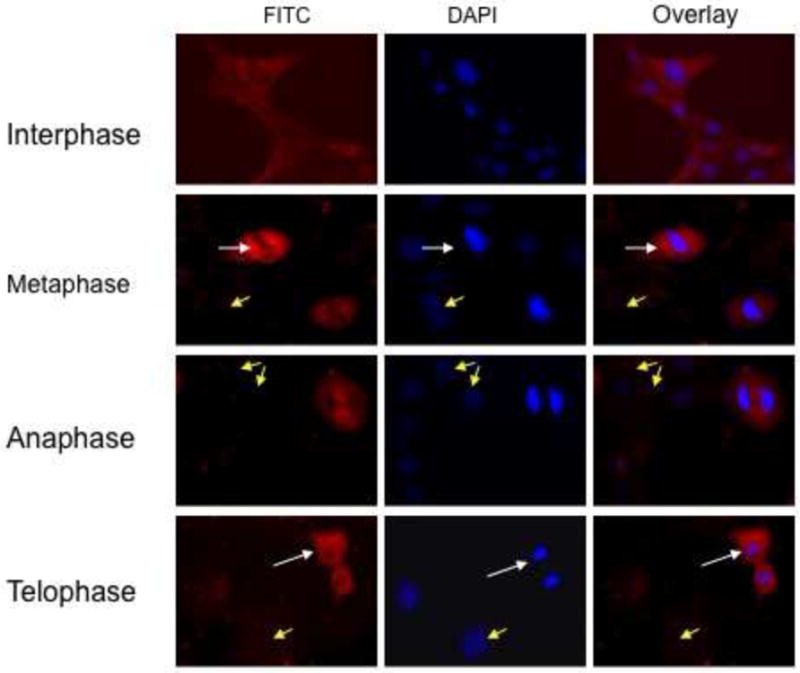
Bone marrow MSCs from 8-month old mice were cultured on chamber slides, fixed and stained as described in Methods. No staining was evident when the primary antibody was replaced with irrelevant IgG antiserum (not shown).
Enhanced Piwi2L expression by synchronizing cells in metaphase
To further explore the relationship between Piwi2L and cell division we synchronized cells in metaphase (Fig. 3). As shown in Figure 3a there was a significant enrichment of metaphase cells and FACS analysis revealed a ~ 3-fold increase of PiwiL2 expression (Fig. 3b). This suggest that PiwiL2 expression is associated with cell division.
Figure 3. PiwiL2 expression in Metaphase Synchronized Cultures and effects of knockdown on proliferation.
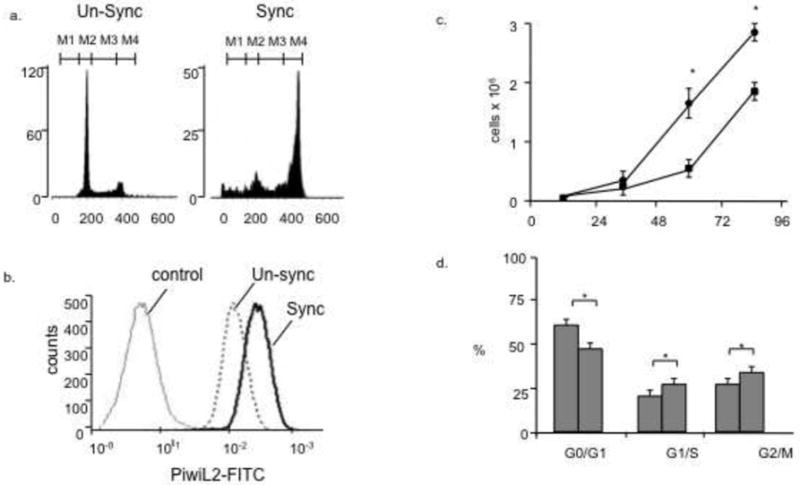
MSCs from 8-month mice were synchronized, labeled with PiwiL2-FITC antibody and analyzed by FACS as described in Methods. (a) Metaphase cells before and after synchronization (b) PiwiL2-FITC stain displays a rightward shift to higher PiwiL2 expression in synchronized cells. (c) MSCs from 8-month old mice were transfected with an optimal concentration of Piwil2 siRNA. PiwiL2 mRNA was undetectable in cells after siRNA knockdown (not shown). Cells were quantified at each time point as described in Methods. Each point is the results of 3 separate experiments in triplicate (p<0.05). (d) Cells were treated as described in 3(c) and cell cycle determined as described in Methods. Results from 3 separate experiments (n=3; p<0.05).
Cell proliferation affected by siRNA knockdown
To investigate a possible role for PiwiL2 in the regulation of cell division we knocked down (KD) the mRNA using a PiwiL2-sepective siRNA and measured proliferation in parallel cultures after treatments with PiwiL2 or random sequence siRNA as described in Methods. As shown in Figure 3c, cultures with PiwiL2 KD showed a significant 2-fold increase in cell number compared with random sequence siRNA. In support of these results we found that PiwiL2 KD also caused a significant increase in the number of cells in G1/S (Fig. 3d).
Downstream signals affected by Piwil2
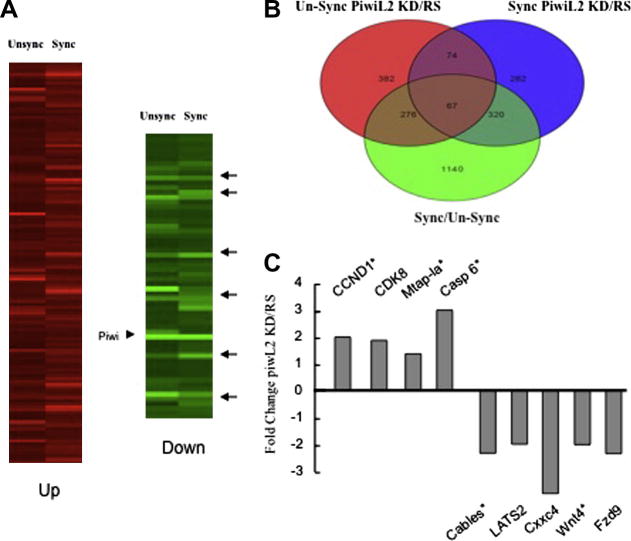
To examine possible target genes of PiwiL2, synchronized and unsynchronized MSCs from 8-month old mice were transfected with PiwiL2 siRNA or random sequence siRNA and subjected to microarray analyses as described in Methods. Heat maps are shown in Figure 4a, the Piwi arrowhead indicates strong KD of Piwil2 by the siRNA treatment in both synchronized and unsynchronized groups, and arrows indicate commonly affected target genes. There was significant overlap of putative PiwiL2-regulated genes between synchronized groups and unsynchronized groups with over 25% of down-regulated genes common to both groups. These results suggest multiple gene-specific effects of Piwl2 KD. When we compared genes that were affected by both synchronization and PiwiL2 knockdown, 1803 transcripts were increased or decreased by >2-fold in synchronize cells, and of these 663 (37%) overlapped with genes that were also up- or down-regulated 2-fold or more by PiwiL2 knockdown (Fig. 4b). GeneGo process network software analysis wherein genes are grouped with respect to their biological functions revealed multiple cell cycle-related genes as potential targets for PiwiL2, including genes linked to mitotic chromosome condensation, DNA packaging, chromatin assembly, mitotic cell cycle process, and M phase, (data not shown). Microarray and RT-PCR analyses identified a series of putative PiwL2 regulated cell cycle/proliferation-associated genes. CCND1 (cyclin D1), CDK8, KIF15, and microtubule associated protein 1a (Mtap-1a) expression increased when PiwiL2 was knocked down, while transcripts of Cables 1, LATS2, and Wnt-pathway genes Cxxc4 and Fzd9 decreased. CCND1 and CDK8 regulate cell cycle transition while KIF15 and Mtap-1a regulate microtubule assembly [24, 25]. Cxxc4 is a negative regulator of the Wnt pathway and a tumor suppressor that causes decreased cell proliferation [26, 27]. Cables-1 and LATS2 are also tumor suppressors [28-30]. These changes are consistent with a mechanism whereby PiwiL2 suppresses proliferation through the selective targeting and reciprocal regulation of cell cycle genes and tumor suppressors. PiwiL2 knockdown was also associated with increased expression of caspase 6.
Figure 4. Microarray analyses and RT-PCR of putative PiwiL2-regulated genes.
(a) MSCs from 8-month mice were synchronized, RNA extracted and subjected to microarray as described in Methods. Significantly expressed genes were clustered using GeneSpring software (Silicon Genetics, Redwood City, CA). Arrows in (a) indicate down-regulated genes common to unsynchronized and synchronized cells. (b) Overlapping down and up-regulated genes common to synchronized and unsynchronized cells (c) Gene expression quantified by microarray or RT-PCR of RNA from 8-month MSCs transfected with PiwiL-2 siRNA or random sequence (RS) siRNA. Transcript changes for each gene shown were significant (n=3; p<0.05). * Indicates quantification by RT-PCR.
DISCUSSION
We demonstrate that adult murine mesenchymal stem cells express PiwiL2 protein and associated piRNAs selectively in the cytoplasm of mitotic cells. MSCs from both groups of older mice constituting a total of 8 mice expressed higher levels of PiwL2 than those isolated from the young group. The characteristics of these cells were described previously [23]. Cells from each age group were isolated and cultured in parallel using identical procedures. All cells were analyzed at the same passage number, they expressed similar patterns of cell surface markers and displayed a similar potential for osteogenic differentiation. The main difference between groups was the age of the donors and whereas we cannot entirely eliminate other variables it seems possible that PiwiL2 expression in bone marrow MSCs is age-related. Immunostaining (Fig. 2) demonstrated intense cytoplasmic expression of PiwL2 protein only in dividing cells identical to the pattern seen in mouse germ line cells (14). Selective expression during mitosis is consistent with the 3-fold increase observed when cells were synchronized in metaphase (Fig 3). Knockdown of PiwiL2 in MSCs from 8-month mice resulted in a 2-fold increase in cell proliferation compared with cells transfected with random sequence RNA and PiwiL2 knockdown increased the fraction of cells in G1/S. Microarray analyses comparing the effects of PiwiL2 knockdown in synchronized and non-synchronized cultures revealed multiple genes that were commonly regulated. Out of more than 35,000 probes on the microarray, about 1000 showed >1.5 fold down-regulation after PiwiL2 knockdown, and of these >25% were common to synchronized and unsynchronized cells. Similarly almost one half of the genes that were altered by synchronization, were also affected by PiwiL2 knock down. These results suggest non-random targeting of metaphase-related genes by PiwiL2. As discussed below putative PiwiL2-targets include cell cycle checkpoint and tumor suppressor genes.
Roles for Piwi family proteins in regulating germ line stem cell self-renewal and spermatogenesis is well established. In Drosophila loss of Piwi function results in depletion of germ line stem cells in both males and females whereas Piwi over-expression increases mitosis [10, 11]. It is not clear whether Piwi proteins play direct roles in cell cycle regulation, or whether their deficiency triggers secondary events that result in abortion of the program. Mice with mutations of the Miwi and PiwiL2 genes accumulate DNA damage suggesting that Piwi proteins are required for DNA repair [12, 31, 32]. Miwi has been shown to complex with the activator of the cAMP-responsive element modulator, a key transcription activator of genes required for spermatogenesis [11]. Miwi and PiwiL2 also form complexes with the mouse Vasa homologue, essential for spermatogenesis [33]. During spermatogenesis Miwi and PiwL2 form complexes with eIF4E and positively regulate translation of multiple genes [34]. In mammals only about 20% of piRNAs correspond to transposon encoding regions, compared with >70% for Drosophila, suggesting that forms of gene regulation distinct from transposon protection may be a priority in mammals [33].
The germ line restriction of Piwi-family proteins has recently been questioned and an emerging field of investigation implicates Piwi homologues in the regulation of cell cycle and proliferation of germ line and non-germ line tumors, and pre-cancer stem cells [13-18]. Over-expression of the human homologue Hiwi was shown to activate Stat3 and BclxL and increase proliferation of multiple cancers [13-16]. In most cancer cells expression of Piwi family homologues is associated with increased oncogenicity [16, 17]. In an apparent exception to this rule, Sharma et al [18] reported that Hiwi was expressed in primitive human CD34+ hematopoietic progenitor cells, but not in immortalized leukemia cell lines. They found that Hiwi over expression in human myeloid KG1 cancer cells resulted in markedly reduced cell proliferation and increased apoptosis.
We present the first evidence that adult murine mesenchymal cells express PiwiL2 and this is associated with decreased proliferation. Expression of PiwiL2 was age-dependent; there was negligible expression of PiwiL2 in cells from 2-month mice but strong expression at ages 8 and 26 months. Eight month old mice are equivalent to mid-aged humans whereas a 26-month old mouse is equivalent to >70 human years [35]. Our observation that PiwiL2 expression correlates with decreased proliferation is consistent with that of Sharma et al [18] and suggest cell specific effects of PiwiL2 on cell division. In our studies increased proliferation by PiwiL2 knockdown correlated with increased expression of cyclin-D and CDK8 and decreased expression of tumor suppressors. The three most strongly down regulated transcripts were Cables 1, LATS2 and Cxxc4. Cables 1 interacts with CDKs 2, 3 and 5, p53 and p73, and its expression slows cell proliferation and stimulates apoptosis [28, 29, 36-38]. LATS2 and Cxxc4 are linked with p53 and Wnt signaling pathways respectively and negatively regulate proliferation and apoptosis [26-30]. Transcript levels of each of these putative tumor suppressors decreased about 3-fold after PiwiL2 knockdown. These effects that could contribute to the increased proliferation seen after PiwiL2 knockdown.
CONCLUSIONS
PiwiL2 expression in mouse MSCs is associated with decreased expression of cyclin D1, CDK8 and microtubule associated proteins, increased expression of tumor suppressors and decreased proliferation. It is unknown whether this regulation involves direct or indirect effects of PiwL2. Our results support an extended role for PiwiL2 in the regulation of cell division and perhaps genome integrity outside of germ line cells that maybe associated with aging.
Acknowledgments
Supported by grants # HL072924 and #HL44578 from the NIH, grant # 07KT-02 from the James and Esther King Biomedical Research Program, State of Florida, and by a Walter G. Ross Distinguished Chair in Vascular Biology (K.A.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis Elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- 2.Lee R, Feinbaum R, Ambros V. A short history of a short RNA. Cell. 2004;116:S89–92. doi: 10.1016/s0092-8674(04)00035-2. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 4.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–9. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 5.Zhang H, Kolb FA, Jaskiewicz L, et al. Single processing center models for human Dicer and bacterial RNase III. Cell. 2004;118:57–68. doi: 10.1016/j.cell.2004.06.017. [DOI] [PubMed] [Google Scholar]
- 6.Hammond SMS, Boettcher AA, Caudy R, et al. Argonaute2, a link between genetic and biochemical analyses of RNAi. Science. 2001;293:1146–1150. doi: 10.1126/science.1064023. [DOI] [PubMed] [Google Scholar]
- 7.Martinez J, Patkaniowska A, Urlaub H, et al. Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell. 2002;110:563–74. doi: 10.1016/s0092-8674(02)00908-x. [DOI] [PubMed] [Google Scholar]
- 8.Seto AG, Kingston RE, Lau NC. The coming of age for Piwi proteins. Mol Cell. 2007;26:603–9. doi: 10.1016/j.molcel.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 9.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol Cell. 2007;26:611–23. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Thomson T, Lin H. The Biogenesis and Function PIWI Proteins and piRNAs: Progress and Prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng W, Lin H. Miwi, a murine homolog of Piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell. 2002;2:819–830. doi: 10.1016/s1534-5807(02)00165-x. [DOI] [PubMed] [Google Scholar]
- 12.Kuramochi-Miyagawa S, Kimura T, Ijiri TW, et al. Mili, a mammalian member of Piwi family gene, is essential for spermatogenesis. Development. 2004;131:839–849. doi: 10.1242/dev.00973. [DOI] [PubMed] [Google Scholar]
- 13.Qiao D, Zeeman AM, Deng W, Looijenga LH, Lin H. Molecular characterization of Hiwi, a human member of the Piwi gene family whose overexpression is correlated to seminomas. Oncogene. 2002;21:3988–3999. doi: 10.1038/sj.onc.1205505. [DOI] [PubMed] [Google Scholar]
- 14.Lee JH, Schutte D, Wulf G, et al. Stem-cell protein Piwil2 is widely expressed in tumors and inhibits apoptosis through activation of Stat3/Bcl-XL pathway. Hum Mol Genet. 2006;15:201–211. doi: 10.1093/hmg/ddi430. [DOI] [PubMed] [Google Scholar]
- 15.Liu X, Sun Y, Guo J, et al. Expression of Hiwi gene in human gastric cancer was associated with proliferation of cancer cells. Int J Cancer. 2006;118:1922–1929. doi: 10.1002/ijc.21575. [DOI] [PubMed] [Google Scholar]
- 16.Chen Y, Pane A, Schupbach T. Cutoff and aubergine mutations result in retrotransposon upregulation and checkpoint activation in Drosophila. Curr Biol. 2007;17:637–642. doi: 10.1016/j.cub.2007.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leedham SJ, Wright NA. Expansion of a mutated clone: from stem cell to tumour. J Clin Pathol. 2008;61:164–171. doi: 10.1136/jcp.2006.044610. [DOI] [PubMed] [Google Scholar]
- 18.Sharma AK, Mary C, Nelson JE, et al. Human CD34+ stem cells express the hiwi gene, a human homologue of the Drosophila gene Piwi. Blood. 2001;97:426–434. doi: 10.1182/blood.v97.2.426. [DOI] [PubMed] [Google Scholar]
- 19.da Silvaz Meirelles L, Chagastelles PC, Nardi N. Mesenchymal stem cells reside in virtually all post natal organs and tissues. J Cell Sci. 2006;119:2201–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- 20.Zou Z, Zhang Y, Hao L, et al. More insight into mesenchymal stem cells and their effects inside the body. Expert Opin Biol Ther. 2010;10:215–30. doi: 10.1517/14712590903456011. [DOI] [PubMed] [Google Scholar]
- 21.Ohishi M, Schipani E. Bone marrow mesenchymal stem cells. J Cell Biochem. 2010;109:277–82. doi: 10.1002/jcb.22399. [DOI] [PubMed] [Google Scholar]
- 22.Wilson AM, Shehedah L, Yu H, Webster KA. Age-related molecular genetic changes of murine bone marrow mesenchymal stem cells. BMC Genomics. 2010;11:229. doi: 10.1186/1471-2164-11-229. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubasiak L, Bishopric N, Webster KA. Hypoxia and Acidosis Activate Cardiac Myocyte Death through the Bcl-2 Family Protein Bnip3. Proc Natl Acad Sci. 2002;99:12825–30. doi: 10.1073/pnas.202474099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vanneste D, Takagi M, Imamoto N, Vernos I. The role of Hklp2 in the sdtabilization and maintenance of spindle bipolarity. Curr Biol. 2009;19:1712–1717. doi: 10.1016/j.cub.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 25.Witzel II, Koh LF, Perkins ND. Regulation of cyclin D1 expression. Biochem Soc Trans. 2010;38:217–22. doi: 10.1042/BST0380217. [DOI] [PubMed] [Google Scholar]
- 26.Kojima T, Shimazui T, Hinotsu S, et al. Decreased expression of CXXC4 promotes a malignant phenotype in renal cell carcinoma by activating Wnt signaling. Oncogene. 2009;28:297–305. doi: 10.1038/onc.2008.391. [DOI] [PubMed] [Google Scholar]
- 27.Filleur S, Hirsch J, Wille A, et al. INTS6/DICE1 inhibits growth of human androgen-independent prostate cancer cells by altering the cell cycle profile and Wnt signaling. Cancer Cell Int. 2009;9:28. doi: 10.1186/1475-2867-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sakamoto H, Friel AM, Wood AW, et al. Mechanisms of Cables 1 gene inactivation in human ovarian cancer development. Cancer Biology & Therapy. 2008;7:180–188. doi: 10.4161/cbt.7.2.5253. [DOI] [PubMed] [Google Scholar]
- 29.Aylon Y, Yabuta N, Besserglick H, et al. Silencing of the Lats2 tumor suppressor overrides a p53-dependent oncogenic stress checkpoint and enables mutant H-Ras-driven cell transformation. Oncogene. 2009;28:4469–79. doi: 10.1038/onc.2009.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cho WJ, Shin JM, Kim JS, et al. miR-372 regulates cell cycle and apoptosis of AGS human gastric cancer cell line through direct regulation of LATS2. Mol Cells. 2009;28:521–7. doi: 10.1007/s10059-009-0158-0. [DOI] [PubMed] [Google Scholar]
- 31.Carmell MA, Girard A, van de Kant HJ, et al. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell. 2007;12:503–14. doi: 10.1016/j.devcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Klattenhoff C, Bratu DP, McGinnis-Schultz N, et al. Drosophila rasiRNA pathway mutations disrupt embryonic axis specification through activation of an ATR/Chk2 DNA damage response. Dev Cell. 2007;12:45–55. doi: 10.1016/j.devcel.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Thomson T, Lin H. The Biogenesis and Function PIWI Proteins and piRNAs: Progress and Prospect. Annu Rev Cell Dev Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Unhavaithaya Y, Hao Y, Beyret E, et al. MILI, a piRNA binding protein, is required for germline stem cell self-renewal and appears to positively regulate translation. J Biol Chem. 2009;284:6507–19. doi: 10.1074/jbc.M809104200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flurkey K, Currer JM, Harrison DE. The Mouse in Aging Research. In: Fox JG, et al., editors. The Mouse in Biomedical Research. 2. American College Laboratory Animal Medicine (Elsevier); Burlington, MA: 2007. pp. 637–672. [Google Scholar]
- 36.Wu CL, Kirley SD, Xiao H, et al. Cables enhances cdk2 tyrosine 15 phosphorylation by Wee1, inhibits cell growth, and is lost in many human colon and squamous cancers. Cancer Res. 2001;61:7325–32. [PubMed] [Google Scholar]
- 37.Zukerberg LR, DeBernardo RL, Kirley SD, et al. Loss of Cables, a cyclin-dependent kinase regulatory protein, is associated with the development of endometrial hyperplasia and endometrial cancer. Cancer Res. 2004;64:202–8. doi: 10.1158/0008-5472.can-03-2833. [DOI] [PubMed] [Google Scholar]
- 38.Debernardo RL, Littell RD, Luo H, et al. Defining the extent of Cables loss in endometrial cancer subtypes and its effectiveness as an inhibitor of cell proliferation in malignant endometrial cells in vitro and in vivo. Cancer Biol Ther. 2005;4:103–7. doi: 10.4161/cbt.4.1.1433. [DOI] [PubMed] [Google Scholar]