Abstract
The application of hydrostatic pressure generally leads to protein unfolding, implying, in accordance to LeChatelier’s principle, that the unfolded state has smaller molar volume than the folded state. However, the origin of the volume change upon unfolding, ΔVu, has yet to be determined. We have examined systematically the effects of protein size and sequence on the value of ΔVu using as a model system a series of deletion variants of the ankyrin repeat domain of the Notch receptor. The results provide strong evidence in support of the notion that the major contributing factor to pressure effects on proteins is their imperfect internal packing in the folded state. These packing defects appear to be specifically localized in the 3D structure, in contrast to the uniformly distributed effects of temperature and denaturants which depend upon hydration of exposed surface area upon unfolding. Given its local nature, the extent to which pressure globally affects protein structure can inform on the degree of cooperativity and long range coupling intrinsic to the folded state. We also show that the energetics of the protein’s conformations can significantly modulate their volumetric properties, providing further insight into protein stability.
Keywords: protein folding, pressure, repeat protein, volume change
Introduction
It has been known since the beginning of the last century that the application of hydrostatic pressure to proteins in solution generally leads to their unfolding1. However, despite a fair amount of work in the field since the first observation of the phenomenon by Bridgeman, our understanding of the basis for the volume change upon unfolding, ΔVu, has progressed very little. Elucidating the basis for pressure effects on proteins merits considerable effort, as pressure is an important intensive thermodynamic variable. Moreover, understanding the mechanisms underlying pressure perturbation of proteins will lead to novel insights concerning their folding, stability and dynamics.
The pressure-temperature phase diagram for two-state protein folding is approximated by an ellipse2–5, and in addition to the standard enthalpy, entropy and heat capacity changes upon unfolding, the shape and position of the ellipse in the P–T plane is determined by the standard volume change upon unfolding, the change in thermal expansivity between the folded and unfolded states, and to a much lesser degree, the difference in their isothermal compressibilities. The volume change of unfolding, ΔVu, is negative over most of the accessible temperature range for most proteins, i.e., the molar volume of the unfolded state is smaller than that of the folded state6. Hence the application of pressure to proteins leads to their unfolding.
The molar volume of folded proteins in aqueous solutions includes the volume of their constitutive atoms, the internal solvent-excluded void volume due to imperfect packing and the volume of the interacting water molecules. It is clear that changes in this molar volume that accompany changes in protein conformation arise from changes in the last two terms, since the volume of the atoms is constant. However, the relative contributions of each, and indeed even the sign of these contributions, have remained controversial. It has recently been proposed that hydration of the peptide backbone is responsible for the bulk of the volume change upon unfolding7, while studies on cavity mutants8–11 have suggested that it is the internal void volume that is dominant. However, these studies, either based on calculations from model compound data or rather limited experimental evidence, have not advanced the debate considerably.
Examination of the ensemble of values for ΔVu obtained from pressure induced unfolding of monomeric proteins at or near 20 degrees Celsius and neutral pH, and reported in the literature10,12–18 reveals a rather weak correlation (r2 =0.6) between the measured ΔVu values and the size of the protein, evaluated as molecular volume, accessible surface area or number of residues (Figure S1). This is in contrast to the strong correlation (r2 > 0.95) between the heat capacity change upon unfolding or m-value (susceptibility to denaturant), with the number of residues and the change in solvent exposed surface area upon unfolding19. For single domain proteins only, a stronger correlation (r2 = 0.85) is found between ΔVu and ratio of the total internal void volume of each protein relative to its total van der Waals volume (Figure S1). This suggests that for a given size, less efficient packing results in a larger negative ΔVu. However, these estimations of internal solvent excluded volume do not take into account any internal water molecules beyond those that have been determined by x-ray crystallography20. Moreover, the coordinates obtained from crystallographic analysis represent only one conformation of the folded state ensemble.
In the present study we have sought to examine in a systematic fashion the factors affecting the value of ΔVu for proteins. We have used deletion variants of the ankyrin repeat domain of the Notch receptor, Nank (residues 1901–2148), as a model system (Figure 1A)21. The folding equilibrium and kinetics of full-length Nank (Nank1–7) and several of its deletion constructs are well-documented for denaturant melts at atmospheric pressure22. These studies showed that its modular architecture results in cooperative unfolding consistent with an Ising model that includes energetic terms for the stability of each repeat and interfacial free energy terms resulting from interactions between sets of adjacent repeats22. Moreover, our recent study of the pressure-induced unfolding of full-length Nank1–723 revealed that the volume change of unfolding for this protein at 20 C is rather small, given its size. With 248 amino acids, the weak correlation of volume change with size predicts a ΔVu of ~−125ml/mol, whereas the measured value is only −44 ml/mol. We postulated that the small volume change might be related to the organization and packing of the protein, which is quite different from the globular proteins. To test the correlation between the size and sequence of the protein and ΔVu, while maintaining the same fold, we have investigated the effects of pressure on constructions of Nank with N-terminal and C-terminal deletions of one or more ankyrin repeats. We combined experimental approaches with molecular dynamics simulations to characterize the packing of side chains in an ensemble of configurations in the folded state of the protein. Our approach presents the advantage that the overall fold of the protein remains the same, while the size changes significantly, and the effects of sequence can likewise be examined.
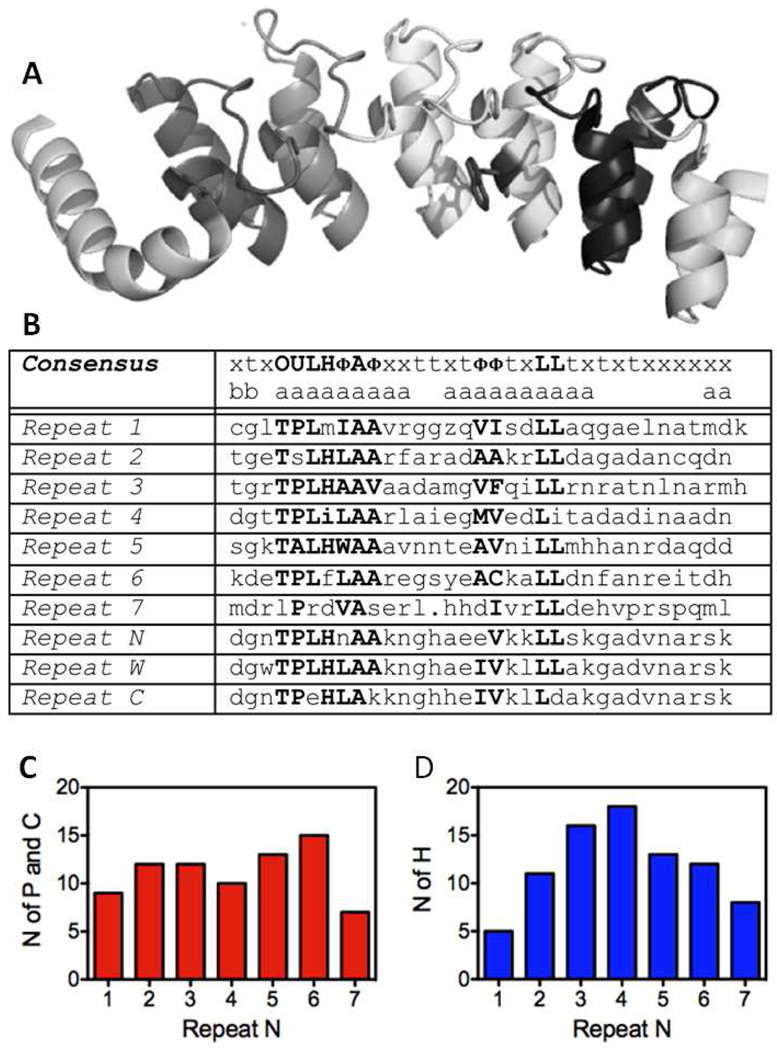
Figure 1.
A) Ribbon diagram of Nank1–721 (pdb code 1OT8). Each ankyrin repeat is a different grey scale color, B) Consensus sequence and specific sequences for the Nank repeats 1–7 and the sequences of the N, W and C repeats in the two designed consensus repeat constructs, NW and NWC, C) and D) Bar graphs of the number of polar (red) and hydrophobic (blue) residues, respectiveley in the Nank repeats 1–7 that bury more than 50% of their surface area (calculated as as described in the Mathods section) in the crystal structure of the full-length Nank1–7. The residues, R, K, H, D, E, G, N, Y, Q, S, T, W were considered to be polar, the rest apolar
Materials and Methods
Protein constructions
Plasmids pET15b (Novagen, Madison, WI) were used to express and purify the various deletion constructs, as previously described22. Purification of some constructs from inclusion bodies involved an overnight dialysis was performed against Tris 50 mM, NaCl 150 mM pH 7.5 buffer after purification. Proteins were stored in aliquots at − 80°C.
Fluorescence data acquisition
High-pressure fluorescence measurements were carried out as previously described23. Briefly, protein solutions at 40–50 µM in Tris 50mM NaCl 150 mM pH 7.5 were loaded into a 500 uL cell fitted with a DuraSeal cap held in place by an o-ring. The cell is placed in a stainless steel high-pressure vessel, equipped with 4 sapphire windows, and connected to a high pressure pump and temperature bath. Pressure inside the vessel is monitored via a gauge connected in line, and temperature maintained stable at 20°C by the thermostat. The pressure transducing liquid is 18 MOhm MilliQ water. Excitation light is provided from a Xenon lamp and monochromator to the vessel via an optical fiber, and emission is collected on a PMT in photon counting mode at 90° through a monochromator (ISS, Champaign, IL). For each experiment, a tryptophan emission spectrum was collected at equilibrium, from 320 to 400 nm, using an excitation wavelength of 295 nm. Pressure jumps, either positive or negative, were approximately 120 bars each to remain close to equilibrium. The fluorescence intensity at 340 nm was collected as a function of time after the jump, until complete equilibration.
Fluorescence data Analysis
As previously described, we calculated the average emission wavelength <λ> of each tryptophan emission spectrum at equilibrium (320–400 nm), for each pressure.
| (1) |
The values reported previously for the full-length construct23 were recalculated for the same spectral scan range (320– 400 nm, rather than 320–450 nm as previously done) in order to make a direct comparison. Data were fitted to a two state unfolding equilibrium as a function of pressure for values of ΔGou and ΔVou, using the BioEQS program, as previously described23. For pressure-induced equilibrium unfolding analysis, the free energy of unfolding is assumed to evolve linearly with the pressure, p:
| (2) |
where:
| (3) |
and:
| (4) |
Unfolding profiles were weighted for the quantum yield ratio of the folded and unfolded states.
Initial analysis of the unfolding profiles at each temperature for each construct yielded values for the ΔVou which were equivalent within uncertainty. Hence for each construct at a given temperature the ensemble of the curves at different urea concentrations were fit globally, linking the values of <λ>f of the folded state and the ΔVou across the different urea concentrations. The value of <λ>u of the unfolded state was not only a floating parameter, but remained unlinked as separate parameters across urea concentrations in the fits. The uncertainty in the recovered ΔVou values was evaluated by rigorous confidence limit testing in which the data sets were reanalyzed at multiple values of the tested parameter (ΔVou), with all other parameters allowed to float. This allows taking into account any correlation between fitting parameters. Pressure and denaturant unfolding of the NW and NWC constructs was analyzed globally as described in supplementary methods.
The p-jump kinetics data were analyzed as previously described for the full length construct23. Briefly, the pressure jump kinetic relaxation fluorescence intensity vs time profiles obtained at 340 nm at multiple urea concentrations at 20°C and at multiple pressures were fit according to a single exponential intensity decay function to obtain the relaxation time τ, at each pressure
| (5) |
.
Then, assuming that this relaxation time corresponds to the inverse sum of the folding and unfolding rate constants at each pressure,
| (6) |
| (7) |
| (8) |
the plots of ln τ vs pressure at each urea concentration, and for each deletion mutant were fit for the activation volume and rate constant for folding at atmospheric pressure, constrained by the equilibrium volume change of unfolding and the equilibrium constant at atmospheric pressure and the corresponding urea concentration.
| (9) |
| (10) |
The reported activation volumes are averages of all of the urea concentrations tested. Relaxation times were at least 100-fold longer than the dead time of the p-jumps (1–2 s) and no burst phase was ever observed.
Molecular dynamics simulations and cavity and hydration estimations
The folded state ensemble was characterized by performing molecular dynamics simulations of the folded state of the proteins. Molecular dynamics simulations were run with Gromacs 4.424 using OPLS-AA force field25 which was shown to accurately describe helical content in proteins and disordered peptides26,27. A 214-residue (from N25 to H239) configuration of Nank 1–7 was generated using the crystallographic structure 1OT8.pdb and MODELLER software28. The protein was inserted in a cubic box such that there was 1 nm from the box boundary to any protein atom. The box was then hydrated with 32916 SPC/E water molecules29 and 15 Na+ ions were added to neutralize the system. H-bonds for protein and water were constrained respectively with LINCS and SETTLE30,31. After steepest descent energy minimization and 10 ps equilibration, this configuration was used as starting configuration for 5 nslong MD simulation using 2fs time step. Configurations were saved every 5 ps. Long-range electrostatic interactions were calculated using particle-mesh Ewald32 with a grid spacing of 0.12 nm and cubic interpolation. Van der Waals interactions were cut off at 1nm. The non-bonded list was updated every 10 integration steps. Temperature was controlled using a Nosé-Hoover thermostat33,34 and pressure was controlled using a Parinello-Rahman barostat.35,36 Both were used with a 5 ps coupling time. Simulations were performed at 293K and 1bar with system compressibility set to 4.6 × 10−5 bar−1. The 1000 generated configurations were analyzed using the McVol algorithm37. First the protein surface is defined using a 0.11 nm radius rolling probe and discretized with a maximum of 2500 surface points per atoms. This particular radius probe was shown to accurately reproduce the experimentally determined cavities in the hen-egg lysozyme example37
To characterize the unoccupied volume (cavities) inside and around the protein we did a MC integration in which points are selected at random and probe for occupancy by protein or solvent atoms. If unoccupied, this volume is assigned to cavities. A first Monte Carlo (MC) integration step is realized with 50k MC points per nm3 box volume. Each MC point is attributed to either solvent, protein atoms or cavities, allowing calculation of van der Waals volume, molecular volume and solvent-excluded volume of the protein. A second MC integration step is specifically used to refine the cavity volume. The void density is defined as the mean number of MC cavity points found within a 0.4 nm radius sphere around each amino-acid Cα atom. Correspondingly, the water density is defined as the mean number of water oxygen atoms found within a 0.4 nm radius sphere around each amino-acid Cα. A 0.4 nm probe radius was found after testing several values to appropriately minimize overlap and maximize coverage. Similar parameters, solvation, and neutralization methods were used for simulations of Nank NW and NWC based on homology models from the crystal structure of Nank1–7 (1OT8.pdb).
The number of polar and apolar residues in each repeat that bury more than 50% of their surface area were calculated with the algorithm developed by Sarai and co-workers38 available online at http://gibk26.bse.kyutech.ac.jp/jouhou/shandar/netasa/asaview/. The residues, R, K, H, D, E, G, N, Y, Q, S, T, W were considered to be polar, the rest apolar.
Results
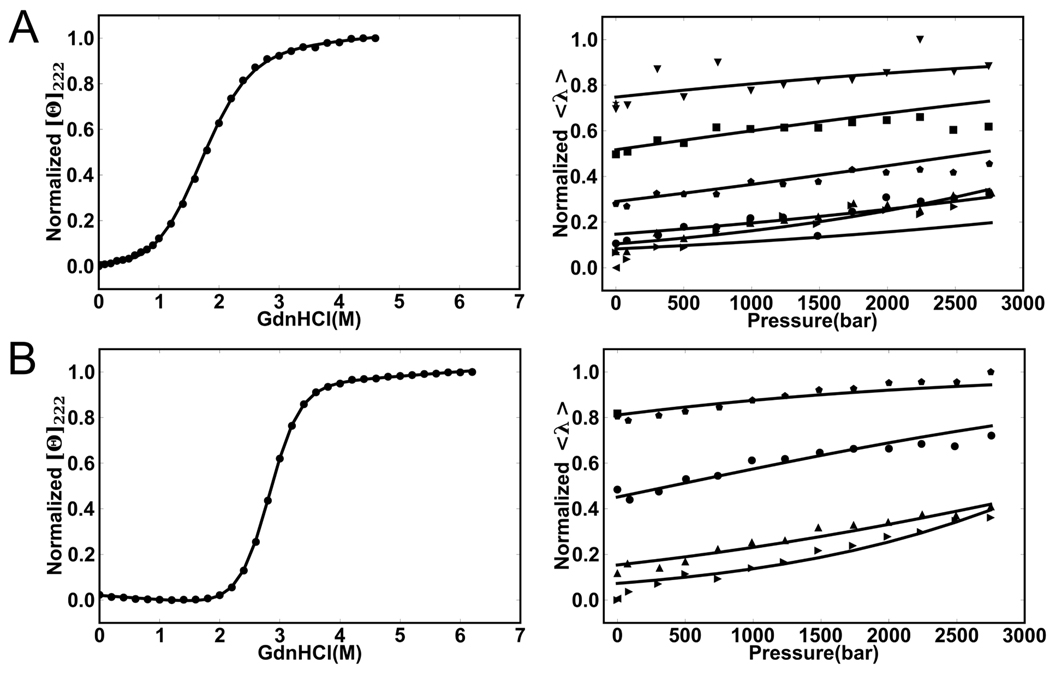
As in the prior pressure study on the full-length ankyrin repeat domain, Nank1–723, pressure dependent unfolding profiles of the deletion constructs were measured by monitoring the (quantum yield weighted) red shift in the fluorescence of the unique tryptophan residue located at the interface between repeats four and five (Figure 1A). We examined pressure effects on two C-terminal (Nank1–6 and Nank1–5) and three N-terminal (Nank2–7, Nank3–7 and Nank4–7) deletion constructs (for repeat sequences see Figure 1B). Urea was used in all cases to bring the pressure unfolding transition into our observable range (< 3 kbar), and for the Nank4–7 construct, TMAO and urea were used in conjunction to populate the folded state at atmospheric pressure to approximately the same extent as in previous unfolding studies of this construct39. The fluorescence-based pressure unfolding profiles (Figure 2) for all of the constructs are very similar to that previously observed for Nank1–7 (shown also in Figure 2 for comparison). Increasing urea concentration lowered the pressure unfolding midpoint, as expected. Indeed the ΔGuo,atm and m-values obtained from linear extrapolation of the values of ΔGui,atm at each urea concentration, i, were found to be in good agreement with the previously published results from urea melts (Table S1)22, particularly given the small number of points and long extrapolation of the pressure experiments. This indicates that the pressure induced transition monitored using the central tryptophan emission corresponds energetically to the global unfolding equilibrium. However, as with the full-length protein, the value of the average emission wavelength at the high pressure plateau also shifted to longer wavelengths with increasing urea, indicating that urea works to increase tryptophan exposure to solvent in the pressure unfolded state. For Nank1–7, high pressure SAXS and FTIR revealed that urea also increases the radius of gyration and decreases the residual helical content in the pressure unfolded state23. However, as observed for other systems, urea had no effect on the volume change of unfolding12,23,40, indicating that the region around the tryptophan does not contribute to the value of ΔVu. Accordingly, the fits in Figure 2 were carried out by linking the ΔVu across all urea concentrations, as was previously done for Nank1–7.
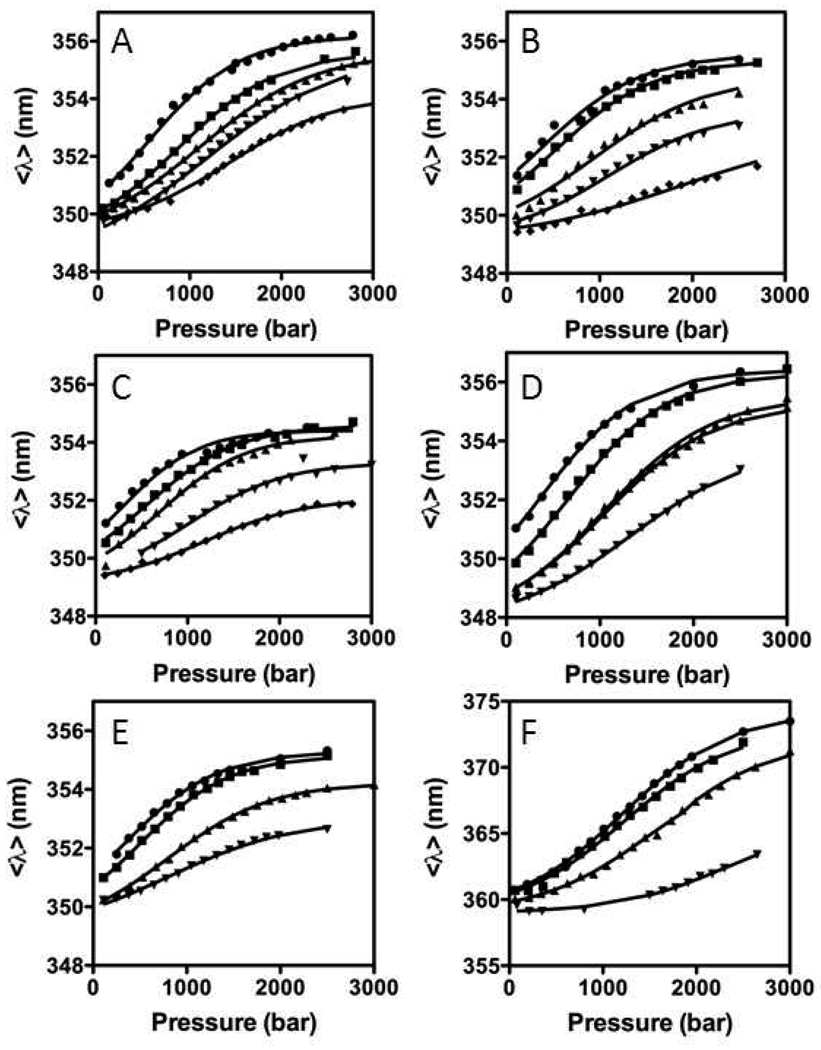
Figure 2.
Average emission wavelength of the tryptophan fluorescence as a function of pressure for the indicated urea concentrations at 20°C for A) Nank1–7 (2, 2.1, 2.2, 2.3, 2.35 M urea), B) Nank 1–6 (1, 1.4, 1.6, 1.8, 2 M urea), C) Nank1–5 (1.1, 1.3, 1.5, 1.7, 1.9 M urea), D) Nank 2–7 (1.9, 2.1, 2.3, 2.4 M urea), E), Nank3–7 (1.1, 1.3, 1.5, 1.7 M urea), F) Nank4–7 (1, 1.5, 1.75, 1.85 M urea). Urea concentrations are indicated from the highest (●), and then in decreasing order (■,▲ ,▼), until the lowest concentration (♦).
None of the deletion constructs tested showed a significantly smaller absolute value for ΔVu than Nank1–7 (Figures 5A and 6, Table S2). In fact, the N-terminal Nank2–7 deletion construct exhibited a slightly larger absolute value for ΔVu value than that obtained for the full-length Nank1–7. We note that the volume change for the Nank4–7 variant is slightly lower than that of the Nank2–7 variant, indicating loss of a small amount of volume with the deletion of repeats 2 and 3. To extend the range of sizes tested, we also examined two consensus ankyrin repeat constructs bearing two and three ankyrin repeats (NW and NWC; Figure 1B). These constructs were very stable, and their volume changes were much smaller than those observed for the larger constructs. Hence a more efficient denaturant, guanidinium hydrochloride, was used to observe their unfolding under pressure (Figure 3). The values of ΔVu obtained from analysis of the pressure dependence of the average emission wavelength at all GuHCl concentrations yielded values for the ΔVu of −8 ± 2.4 and −11.8±1.8 ml/mol for the two and three domain constructs, NW and NWC, respectively.
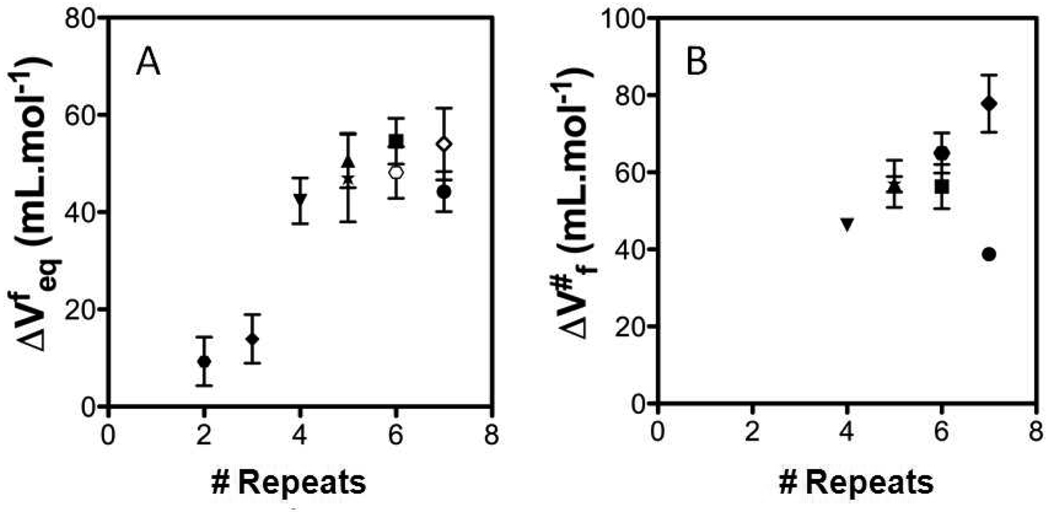
Figure 5.
Volume changes obtained at 20°C for the Nank variants and the NW and NWC consensus constructs as a function of the number of repeats. A) Absolute value of ΔVu (=ΔVf), the volume change of unfolding and B) the activation volume for folding ΔVf* for Nank1–7 (●), Nank2–7 (■), Nank3–7 (▲), Nank4–7 (▼), Nank1–6 (○), Nank1–5 (*), the alanine to glycine mutant in domain 2, NAG2 (◊), the double consensus repeat, MW (●) and the triple consensus repeat, MWC (♦). The data for the full-length Nank1–7 construct is were previously reported23.
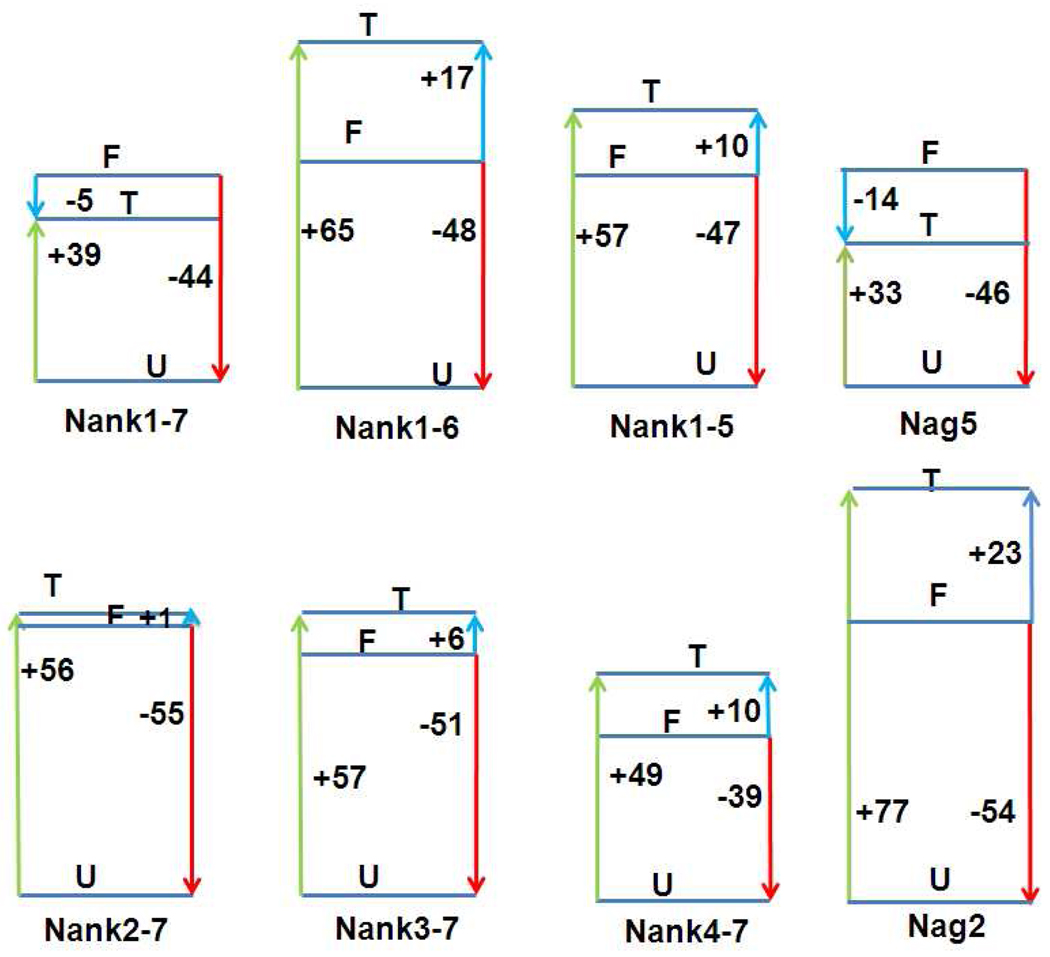
Figure 6.
Volume diagrams for the Nank deletion constructs, as indicated. F, U and T correspond to the folded, unfolded and transition states. Volume values are given in units of ml/mol. Red arrows correspond to the equilibrium unfolding volume, green arrows to the activation volume for the folding transition and blue arrows to the activation volume for the unfolding transition.
Figure 3.
Results of the equilibrium pressure unfolding of A) NW and B) NWC at multiple GuHCl concentrations. Figures in the left column present the guanidine melts at atmospheric pressure, while those on the right correspond to the pressure profiles. Curves from bottom to top correspond to increasing concentrations of guanidine for A) NW (◀:0.0, ▶:0.5, ▲:1.0, ●:1.5, ⬟:2.0, ■:2.5, ▼:3.0, ⭑:6.0 M GuHCl) and B) NWC (◀:0.0, ▶:1.7, ▲:2.5, ●:3.0, ⬟:3.5, ■:6.0 M GuHCl). Lines through the points correspond to the fits of the data as described in the Supplementary Methods section.
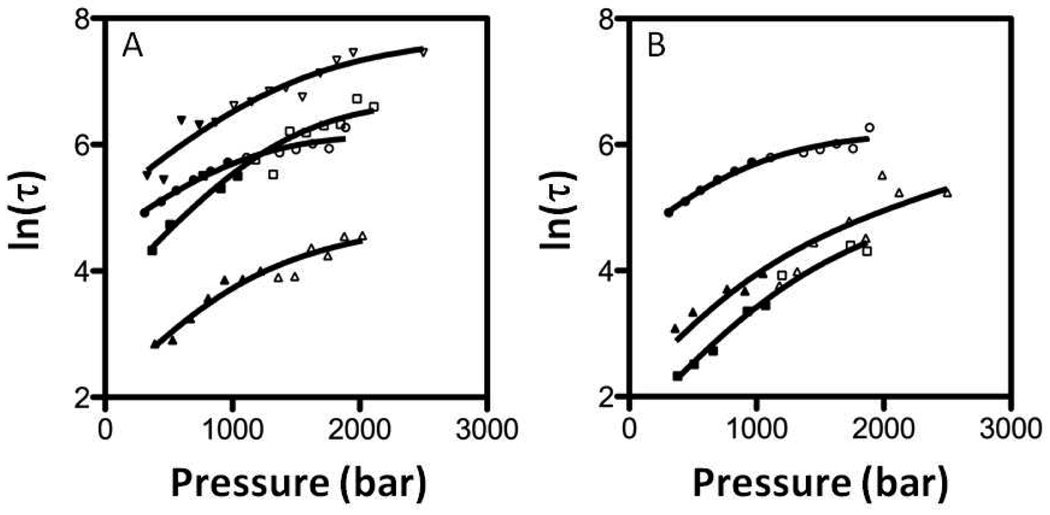
Pressure-jump relaxation kinetic data were acquired for the N and C terminal deletion constructs concomitant with the acquisition of the steady state data. The relaxation profiles were mono-exponential, as previously observed for Nank1–7, and were fit for the relaxation time, τ. The plots of lnτ vs pressure (Figure 4B), revealed a significant difference between the results obtained for the deletion mutants and those previously observed for Nank1–7 (also plotted in Figures 4A and B for comparison)23. In contrast to Nank1–7, activation volumes for unfolding were positive for all deletion constructs, placing the transition state at a higher molar volume than the folded state, and significantly larger (relative to the unfolded state) than the transition state of the full-length construct (Figures 5B and 6 and Table S2). The transition state of Nank1–7 lies between the folded and unfolded state at 20°C (Table S3), and only becomes larger than that of the folded state at higher temperature23. We note that the tryptophan residue that provides our observable is located in the central repeats, and is quite sensitive to the effects of terminal repeat deletions on the values of the activation volumes, indicating that the lack of effect of these deletions on the equilibrium volume change cannot be ascribed to the insensitivity of the observable.
Figure 4.
Natural logarithm of the pressure-jump relaxation time as a function of pressure obtained at 20°C for A) N-terminal deletions (■,□) Nank 2–7 at 1.9 M urea, (▲, △) Nank 3–7 at 1.3 M urea, (▼, ▽) Nank4– 7 at 1.75 M urea and 750 mM TMAO and (●, ○) previous results from Nank1–7 at 2.3 M urea. These data were recalculated for the spectral span 320–400 nm from our previous experiments23, B) The C-terminal deletions (▲, △) Nank1–6 at 1.6 M urea and (■, □) Nank1–5 at 1.1 M urea, and (●, ○) previous results from Nank1–7 at 2.3 M) urea. Closed symbols represents jumps at to pressure below the unfolding midpoint, while open symbols represent results obtained for jumps to pressure above the unfolding midpoint
The temperature dependence of the equilibrium and activation volume changes corresponds to Δα, the difference in thermal expansivity between the two states of the transition. We investigated the temperature dependence of the equilibrium and activation volumes of two of the Nank N-terminal deletion mutants, Nank2–7 and Nank3–7 and compared them to the Nank1–7 construct23. These experiments yield the difference in thermal expansivity between the folded and unfolded state (dΔVu/dT = Δαu), between the transition state and the unfolded state (dΔV*f/dT = Δ α *f), and the transition state and the folded state (dΔV*u/dT = Δ α*u) (Table S3). The thermal expansivity of the transition state of the full-length construct was previously shown to be similar to that of the unfolded state and much larger than that of the folded state23; i.e., the values of Δαu and Δ α*u were equivalent within experimental uncertainty. The Nank2–7 construct exhibited a temperature dependence equivalent to that of Nank1–7, with the transition state expansivity equivalent to that of the unfolded state. In contrast, the Nank3–7 construct exhibited a transition state expansivity equivalent to that of the folded state. The very large expansivity of the transition state of Nank2–7 is consistent with the large increase in its molar volume upon deletion of the N-terminal repeat. These results indicate that the basis for the large expansivity of the transition state in the full-length Nank1–7 resides in repeat 2 or at the interface between repeats 2 and 3.
Finally, we examined the pressure unfolding behavior of two variants of the full-length construct bearing alanine to glycine mutations at equivalent positions in the second and fifth repeat (NAG2 and NAG5)41,42. These mutations were shown to be strongly destabilizing, and in particular, in the case of repeat 2, disrupt to some extent the folded structure. Equilibrium pressure unfolding and p-jump relaxation kinetics studies on these mutants (Figure S1 and Table S4) reveal that the absolute value of the equilibrium volume change increases slightly relative to that of Nank1–7 for the NAG2 variant, while it remains the same for NAG5. Interestingly, the activation volume for folding the NAG2 construct is found to be very large, indeed the largest of all of the variants tested (Figure 6). These observations indicate that this destabilizing mutation in the second repeat leads to some expansion of the folded state, and a very large expansion of the transition state, reinforcing the notion that the region responsible for the large expansivity of the transition state in the full-length Nank1–7 involves repeat 2 or the 2–3 interface.
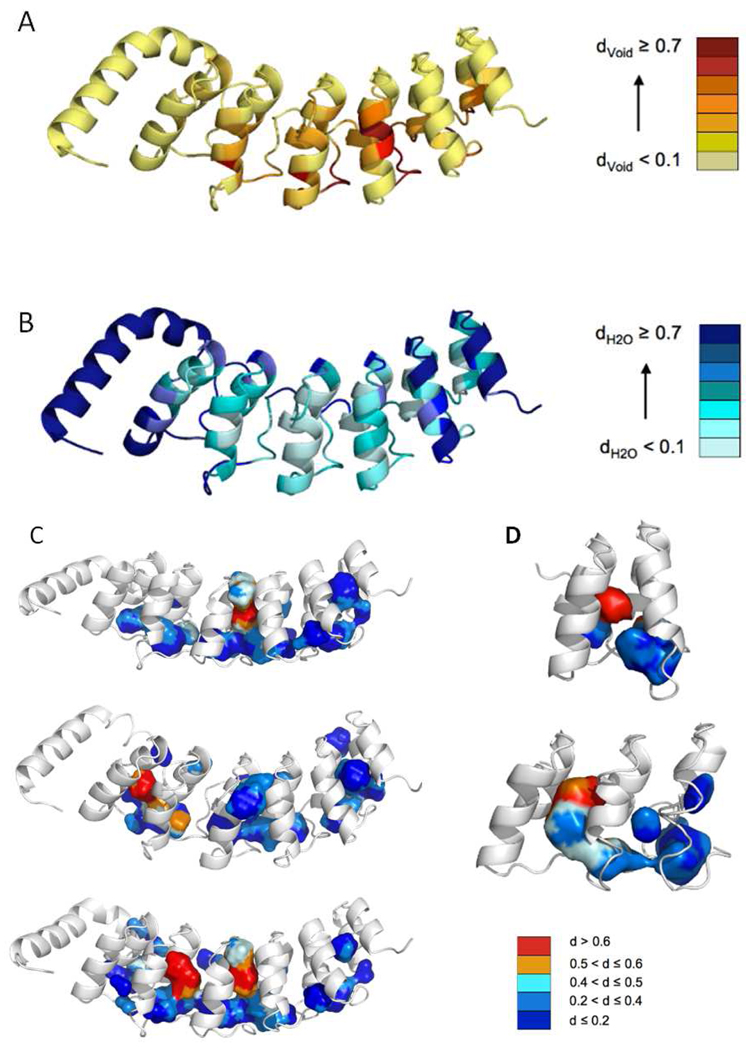
To gain insight into the possible contribution of internal solvent excluded voids (packing defects) to the observed volume changes and expansivity values, we carried out molecular dynamic simulations in explicit solvent on the Nank1–7 full-length protein and on molecular homology models of the two consensus repeat constructs, NW and NWC. The simulations ran for 5 ns and configurations were saved every 5 ps for a total of 1000 configurations. We determined the magnitude of internal voids by measuring the number of Monte Carlo points that could be inserted into the each of the configurations (the presence of a protein atom at any coordinate value precluded insertion of a Monte Carlo point). The average density of Monte Carlo points for all configurations found at distances < 0.4 nm from the C-α atom of each residue was then calculated. We refer to this as void density. To gain a sense of the degree of hydration across the structure we also calculated for each configuration, the number of water molecules within 0.4 nm of the C-α atom of each residue. We refer to the average over all configurations as the hydration density.
The central repeats exhibit the largest average void density, and the lowest average hydration density (Figure 7A, B), consistent with their nearly exclusive contribution to the volume change of unfolding. To better visualize the internal voids, Figure 7C and D shows three representative configurations of the Nank1–7 construct and the two consensus repeat constructs, NW and NWC, highlighting the internal voids (by linking the Monte Carlo points by proximity and calculating the surface of the void). The largest and “driest” voids in Nank1–7 are found at the interfaces between repeats 2 and 3, 3 and 4 and 4 and 5, consistent with the preponderant contribution of the central region of the protein to the magnitude of the volume change of unfolding. The N and C terminal repeats 1, 7 and 6 do not appear to contribute significantly to the solvent excluded void, in accord with the observation that their deletion does not diminish the volume change of unfolding. MD simulations of a construct containing repeats 3, 4, and 5 (taken directly from the crystal structure of the full-length protein) revealed very similar void density and hydration properties for these repeats as in the context of the full-length protein (Figure S3). These three “snapshots” of the protein illustrate the fact that the measured volume changes are representative of a large conformational ensemble that would not be represented by calculations on the pdb coordinates only. The average void and hydration densities depicted in Figures 7 A and B provide a qualitative appreciation of solvent excluded voids in this ensemble. Because other thermodynamic factors, such as the difference in expansivity between unfolded and folded states (Δα), are known to contribute significantly to the magnitude of ΔVu, the MD simulations present limitations to the quantitative evaluation of cavity size and its contribution to the volumetric change. However, they serve to identify qualitatively the void propensity and hydration probability throughout the 3D structure and the degree of variation among conformations of the folded state ensemble generated by the simulation. The smaller voids found for the NW and NWC constructs are consistent with their small volume changes of unfolding.
Figure 7.
Results of the molecular dynamics simulations and subsequent calculations for the Nank1–7 full-length construct and the NW and NWC consensus repeats. A) Ribbon diagram of Nank1–7 colored according to the average internal void density around the Cα atom of each residue from the 1000 configuration of the 10 ns simulation as described in the text, B) Average distance from the nearest water molecule of the Cα atom of each residue calculated for the 1000 configurations of the 10 ns simulation. C–D) Representations of the internal cavities detected using a 0.11 nm probe for a representative configuration from simulations of Nank1–7, and the homology models of the NW and NWC consensus repeats, respectively, colored blue to red for the distance from their center to the nearest water molecule.
Discussion
The first clear conclusion that can be drawn from the present work is that the determinants of the volume change of unfolding are located nearly exclusively in the central repeats of Nank1–7. How can we explain the non-uniformity of the contributions of the ankyrin domains to pressure effects on Nank1–7? Obviously, the volume of the atoms does not contribute to the volume change of unfolding, such that the explanation must lie in the volumetric consequences of differential hydration or the elimination of solvent excluded voids upon unfolding. Differential hydration contributions involve volumetric effects on the unfolded state, since they arise from changes in the density of water molecules that move from the bulk to interaction with the protein moieties upon their exposure to solvent upon unfolding. We make the implicit assumption that the contribution of solvent excluded voids, on the other hand, is defined by the structural and dynamic properties of the folded state, since any cavities in the unfolded state should be minimized and homogeneously distributed throughout the protein. Indeed, a recent analysis of packing density in proteins revealed that the hierarchy of sizes due to bond constraints in the polymer leads to inhomogeneous packing and a fractal packing dimension between the ideal crystalline and Apollonian limit that scales with size43. These conclusions are globally consistent with our present results, although the weaker correlation of ΔVu with size is a consequence of hydration of certain packing defects, typically those found closer to the surface. For the Nank system, it is likely that its repetitive nature puts it close to the ideal crystalline packing limit and could explain the small ΔVu of Nank1–7 despite its large size.
The thermal expansivity of amino acids in water, which arises from the temperature dependent loss of hydrating water molecules to the bulk, has provided estimates of the volume effects due to hydration44–46. The expansivity of polar and charged amino-acid side chains is large and positive at low to moderate temperatures because the density of the bulk solvent is lower than that of the electrostricted waters hydrating the polar amino acids. Since dehydration (upon increasing temperature) results in a large and positive expansion of volume, we can deduce that hydrating newly exposed polar residues upon unfolding should cause an increase in density (decrease in volume) for the newly interacting water molecules compared to their volume in bulk. Therefore, deletion of polar moieties should lead to an overall decrease in the magnitude of the ΔVu. The opposite situation holds for the volumetric properties of hydration of hydrophobic moieties which exhibit a negative thermal expansivity at low to moderate temperature. Dehydration upon increasing temperature leads to a decrease in volume, indicating that the density of water hydrating polar residues is lower than in bulk44–46. Therefore, hydration of exposed hydrophobic residues upon unfolding should lead to an increase in the volume of the hydrating water molecules relative to their volume in bulk solvent. This in turn should diminish the magnitude of the volume change of unfolding. Hence, deletion of hydrophobic moieties should lead to a smaller positive contribution to the molar volume of the unfolded state, and hence a larger magnitude for ΔVu.
Arguing against a major role for differential hydration as the basis for pressure effects in the Nank system studied here are the sequence differences between the repeats (Figure 1C and D). Indeed, the central repeats, those that contribute almost exclusively to the negative value of ΔVu, contain the largest number of buried hydrophobic residues (that should make a positive contribution to ΔVu) and the smallest number of buried polar and charged residues (those that should contribute negatively to ΔVu). Based on this lack of correlation between the sequence differences between the repeats and their predicted effects on ΔVu, and the measured values we conclude that the volume changes of the Nank1–7 protein and its deletion constructs do not arise primarily from differential hydration effects of exposed amino acid side chains. Moreover the repeats are of nearly identical 3D structure and size, and their unfolding should involve the exposure of a similar number of peptide bonds. Hence any contribution of the main chain to the volume change should be uniformly distributed across the structure. If the hydration of peptide bonds upon unfolding were the major contributing factor to the volume change, then, given that these are polar, removing up to 3/7 of the protein by deletion should have resulted in a significantly smaller magnitude for the volume change of unfolding. Instead, we observe either no change or even a small increase in the magnitude of ΔVu upon deletion of repeat units. We note that in contrast to the volume change, the m-values for urea induced unfolding are strongly dependent upon the number of repeats22, which is expected since the larger the protein, the larger the exposed surface area. Thus, since the amplitude of ΔVu is not correlated with the magnitude of the change in exposed surface area upon unfolding, we conclude that differential hydration is not a major contributing factor to its value.
If differential hydration cannot be invoked to explain the magnitude of pressure effects, can the contribution of internal solvent excluded void explain the totality of our results on the volumetric properties of these Nank constructs? Removing repeats should result in a decrease in the magnitude of the volume change of unfolding to the extent that significant solvent excluded voids exist in each repeat and its interface with its neighboring repeat. The lack of effect of deletion of repeats 1, 2, 6, and 7 is consistent with the notion, supported by the results of our MD simulations, that these repeats present very little solvent excluded void volume. We note that our probe, the tryptophan residue, is located at the center of the protein, and could conceivably be insensitive to volume effects of deletion of the terminal domains. However, the observation of large increases in the activation volumes for all of the constructs indicates that the tryptophan is indeed sensitive to effects of terminal repeat deletions and to the global unfolding. The agreement between the stabilities measured in the present the high pressure experiments and those obtained from previous urea melts further supports this assumption.
How can repeat deletions result in an increase in the molar volume of the transition state relative to the unfolded state? The actual size of solvent excluded void volumes in proteins at any temperature will depend on the three dimensional structure, as ascertained by coordinates from structural studies. In addition, the capacity of the protein to expand should contribute to the magnitude of internal void volume at any given temperature, and this will depend upon the strength of the interactions. If removal of the N and C terminal repeats leads to a decrease in the stabilizing interactions of the folded and/or transition states, then internal solvent-excluded voids could conceivably expand in volume, relative to their size at a standard temperature or their size at the same temperature in the context of the full length construct. This expansion could occur either globally across the structure, or locally around the affected regions, leading to larger solvent excluded voids, and hence larger equilibrium or activation volume changes. An expansion of protein structure has been proposed recently to constitute the first step in their unfolding47. Our results suggest that this expansion is local in nature in the case of the Nank constructs, and is particularly apparent at the unfolding barrier. Indeed, the small decreases in the magnitude of ΔVu observed for Nank3–7 and Nank4–7 relative to Nank2–7 are consistent with loss of cavity volume in the interfaces that occurs upon repeat deletion.
Conclusion
This systematic investigation of the role of size and sequence in determining the magnitude of pressure effects on protein structure and stability provides strong evidence that differential hydration does not significantly contribute to the pressure-induced unfolding of proteins. In contrast, these results implicate the imperfect internal packing of the folded state as the major contributing factor to the volume change of unfolding. These packing defects are specifically localized in central region of the Nank structure, rendering pressure effects local in character, in contrast to temperature or denaturant effects that depend upon properties that are uniformly distributed across the proteins sequence and structure. As a consequence of the local nature of the pressure effect, the degree to which pressure globally affects protein structure depends upon the cooperative interactions within the folded state. Pressure therefore represents a unique probe for the long-range coupling in proteins and their intrinsic cooperativity of unfolding. Furthermore, the present results demonstrate that the energetics of the protein’s conformations can significantly modulate their volumetric properties. First, the unique configuration corresponding to the crystal structure does not provide a comprehensive view of internal solvent excluded voids, as the multiple conformations in the native state manifold can exhibit different amounts and distributions of internal void depending upon side-chain and even main chain orientation or incursions by solvent. Moreover, loss of constraining interactions upon mutation can lead to expansion of internal voids. Given this more profound understanding of the factors contributing to the volumetric properties of, and hence pressure effects on proteins, we are confident that further pressure effect studies will prove extremely useful tool to understand protein conformations and energetics.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by a grant from the Agence National pour la Recherche PiriBio number 09-455024 to CAR, from the National Institutes of Health grant GM060842 to DB and National Science Foundation MCB-0543769 to AEG.
Footnotes
SUPPORTING INFORMATION
Supplementary Methods. Analysis of the NW and NWC unfolding profiles.
Table S1. Comparison of the Free Energies and m-values for the urea denaturation of Nank constructs from the pressure unfolding studies with previously reported values from urea melts.
Table S2. Equilibrium and Activation volume changes for the Nank deletion constructs
Table S3. Temperature dependence of the equilibrium and activation volume changes for the Nank1–7, 2–7 and 3–7 constructs.
Table S4. Equilibrium and Activation volume changes for the NAG2 and NAG5 variants
Figure S1 Correlation between the measured volume change upon unfolding ΔVu and protein size.
Figure S2. Pressure unfolding equilibrium and kinetic profiles of the Nag2 (A and C) and Nag5 (B and D) mutants.
Figure S3. Results of MD simulations and void density and hydration calculations on a Nank 345 construct.
Reference List
- 1.Bridgman PW. The coagulation of albumin by pressure. J. Biol. Chem. 1914;19:511–512. [Google Scholar]
- 2.Brandts JF, Oliveira RJ, Westort C. Thermodynamics of protein denaturation. Effect of pressu on the denaturation of ribonuclease A. Biochemistry. 1970;9(4):1038–1047. doi: 10.1021/bi00806a045. [DOI] [PubMed] [Google Scholar]
- 3.Hawley SA. Reversible pressure--temperature denaturation of chymotrypsinogen. Biochemistry. 1971;10(13):2436–2442. doi: 10.1021/bi00789a002. [DOI] [PubMed] [Google Scholar]
- 4.Smeller L, Meersman F, Heremans K. Refolding studies using pressure: the folding landscape of lysozyme in the pressure-temperature plane. Biochim. Biophys. Acta. 2006;1764(3):497–505. doi: 10.1016/j.bbapap.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 5.Zipp A, Kauzmann W. Pressure denaturation of metmyoglobin. Biochemistry. 1973;12(21):4217–4228. doi: 10.1021/bi00745a028. [DOI] [PubMed] [Google Scholar]
- 6.Royer CA. Revisiting volume changes in pressure-induced protein unfolding. Biochim. Biophys. Acta. 2002;1595(1–2):201–209. doi: 10.1016/s0167-4838(01)00344-2. [DOI] [PubMed] [Google Scholar]
- 7.Chalikian TV, Macgregor RB., Jr Origins of pressure-induced protein transitions. J. Mol. Biol. 2009;394(5):834–842. doi: 10.1016/j.jmb.2009.10.020. [DOI] [PubMed] [Google Scholar]
- 8.Ando N, Barstow B, Baase WA, Fields A, Matthews BW, Gruner SM. Structural and thermodynamic characterization of T4 lysozyme mutants and the contribution of internal cavities to pressure denaturation. Biochemistry. 2008;47(42):11097–11109. doi: 10.1021/bi801287m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins MD, Hummer G, Quillin ML, Matthews BW, Gruner SM. Cooperative water filling of a nonpolar protein cavity observed by high-pressure crystallography and simulation. Proc. Natl. Acad. Sci. U. S. A. 2005;102(46):16668–16671. doi: 10.1073/pnas.0508224102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frye KJ, Royer CA. Probing the contribution of internal cavities to the volume change of protein unfolding under pressure. Protein Sci. 1998;7(10):2217–2222. doi: 10.1002/pro.5560071020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lassalle MW, Yamada H, Morii H, Ogata K, Sarai A, Akasaka K. Filling a cavity dramatically increases pressure stability of the c-Myb R2 subdomain. Proteins. 2001;45(1):96–101. doi: 10.1002/prot.1128. [DOI] [PubMed] [Google Scholar]
- 12.Brun L, Isom DG, Velu P, Garcia-Moreno B, Royer CA. Hydration of the folding transition state ensemble of a protein. Biochemistry. 2006;45(11):3473–3480. doi: 10.1021/bi052638z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herberhold H, Winter R. Temperature- and pressure-induced unfolding and refolding of ubiquitin: a static and kinetic Fourier transform infrared spectroscopy study. Biochemistry. 2002;41(7):2396–2401. doi: 10.1021/bi012023b. [DOI] [PubMed] [Google Scholar]
- 14.Jacob MH, Saudan C, Holtermann G, Martin A, Perl D, Merbach AE, Schmid FX. Water contributes actively to the rapid crossing of a protein unfolding barrier. J. Mol. Biol. 2002;318(3):837–845. doi: 10.1016/S0022-2836(02)00165-1. [DOI] [PubMed] [Google Scholar]
- 15.Kitahara R, Royer C, Yamada H, Boyer M, Saldana JL, Akasaka K, Roumestand C. Equilibrium and pressure-jump relaxation studies of the conformational transitions of P13MTCP1. J. Mol. Biol. 2002;320(3):609–628. doi: 10.1016/s0022-2836(02)00516-8. [DOI] [PubMed] [Google Scholar]
- 16.Mei G, Di VA, Campeggi FM, Gilardi G, Rosato N, De MF, Finazzi-Agro A. The effect of pressure and guanidine hydrochloride on azurins mutated in the hydrophobic core. Eur. J. Biochem. 1999;265(2):619–626. doi: 10.1046/j.1432-1327.1999.00751.x. [DOI] [PubMed] [Google Scholar]
- 17.Mohana-Borges R, Silva JL, Ruiz-Sanz J, de Prat-Gay G. Folding of a pressure-denatured model protein. Proc. Natl. Acad. Sci. U. S. A. 1999;96(14):7888–7893. doi: 10.1073/pnas.96.14.7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sasahara K, Nitta K. Pressure-induced unfolding of lysozyme in aqueous guanidinium chloride solution. Protein Sci. 1999;8(7):1469–1474. doi: 10.1110/ps.8.7.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Myers JK, Pace CN, Scholtz JM. Denaturant m values and heat capacity changes: relation to changes in accessible surface areas of protein unfolding. Protein Sci. 1995;4(10):2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damjanovic A, Garcia-Moreno B, Lattman EE, Garcia AE. Molecular dynamics study of water penetration in staphylococcal nuclease. Proteins. 2005;60(3):433–449. doi: 10.1002/prot.20486. [DOI] [PubMed] [Google Scholar]
- 21.Zweifel ME, Leahy DJ, Hughson FM, Barrick D. Structure and stability of the ankyrin domain of the Drosophila Notch receptor. Protein Sci. 2003;12(11):2622–2632. doi: 10.1110/ps.03279003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mello CC, Barrick D. An experimentally determined protein folding energy landscape. Proc. Natl. Acad. Sci. U. S. A. 2004;101(39):14102–14107. doi: 10.1073/pnas.0403386101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rouget JB, Schroer MA, Jeworrek C, Puhse M, Saldana JL, Bessin Y, Tolan M, Barrick D, Winter R, Royer CA. Unique features of the folding landscape of a repeat protein revealed by pressure perturbation. Biophys. J. 2010;98(11):2712–2721. doi: 10.1016/j.bpj.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van der Spoel S, Lindahl E, Hess B, Groenhof G, Mark A, Berendsen H. GROMACS: fast, flexible, and free. J Comput Chem. 2005;98:2712–2721. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 25.Kaminski G, Friesner R, Tirado-Rives J, Jorgensen W. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B. 2011;105:6474–6487. [Google Scholar]
- 26.Gnanakaran S, Garcia AE. Helix-coil transition of alanine peptides in water: force field dependence on the folded and unfolded structures. Proteins. 2005;59(4):773–782. doi: 10.1002/prot.20439. [DOI] [PubMed] [Google Scholar]
- 27.Sgourakis NG, Yan Y, McCallum SA, Wang C, Garcia AE. The Alzheimer's peptides Abeta40 and 42 adopt distinct conformations in water: a combined MD / NMR study. J Mol Biol. 2007;368(5):1448–1457. doi: 10.1016/j.jmb.2007.02.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 29.Berendsen H, Grigera J, Straatsma T. The missing term in effective pair potentials. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 30.Hess B, Bekker H, Berendsen H, Fraaije J. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 31.Miyamoto S, Kollman P. SETTLE - An analytical version of the SHAKE and RATTLE algorithm for rigid water models. J Comput Chem. 1992;13:952–962. [Google Scholar]
- 32.Essmann U, Perera L, Berkowitz M, Darden T, Lee H, Pedersen L. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577–8593. [Google Scholar]
- 33.Hoover W. Canonical Dynamics: Equilibrium Phase space distributions. Phys Rev. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 34.Nosé S. A molecular dynamics method for simulating the canonical ensemble. Mol Phys. 1984;52:255–268. [Google Scholar]
- 35.Nosé S, Klein M. Constant pressure moleculer dynamics for molecular systems. Mol Phys. 1983;50:1055–1076. [Google Scholar]
- 36.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 37.Till MS, Ullmann GM. McVol - a program for calculating protein volumes and identifying cavities by a Monte Carlo algorithm. J Mol Model. 2010;16(3):419–429. doi: 10.1007/s00894-009-0541-y. [DOI] [PubMed] [Google Scholar]
- 38.Ahmad S, Gromiha M, Fawareh H, Sarai A. ASA View: database and tool for solvent accessibility representation in proteins. BMC. Bioinformatics. 2004;5:51. doi: 10.1186/1471-2105-5-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mello CC, Bradley CM, Tripp KW, Barrick D. Experimental characterization of the folding kinetics of the notch ankyrin domain. J. Mol. Biol. 2005;352(2):266–281. doi: 10.1016/j.jmb.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 40.Pappenberger G, Saudan C, Becker M, Merbach AE, Kiefhaber T. Denaturant-induced movement of the transition state of protein folding revealed by high-pressure stopped-flow measurements. Proc. Natl. Acad. Sci. U. S. A. 2000;97(1):17–22. doi: 10.1073/pnas.97.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradley CM, Barrick D. The notch ankyrin domain folds via a discrete, centralized pathway. Structure. 2006;14(8):1303–1312. doi: 10.1016/j.str.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 42.Bradley CM, Barrick D. Limits of cooperativity in a structurally modular protein: response of the Notch ankyrin domain to analogous alanine substitutions in each repeat. J. Mol. Biol. 2002;324(2):373–386. doi: 10.1016/s0022-2836(02)00945-2. [DOI] [PubMed] [Google Scholar]
- 43.Lin LN, Brandts JF, Brandts JM, Plotnikov V. Determination of the volumetric properties of proteins and other solutes using pressure perturbation calorimetry. Anal. Biochem. 2002;302(1):144–160. doi: 10.1006/abio.2001.5524. [DOI] [PubMed] [Google Scholar]
- 44.Mitra L, Smolin N, Ravindra R, Royer C, Winter R. Pressure perturbation calorimetric studies of the solvation properties and the thermal unfolding of proteins in solution-- experiments and theoretical interpretation. Phys. Chem. Chem. Phys. 2006;8(11):1249–1265. doi: 10.1039/b516608j. [DOI] [PubMed] [Google Scholar]
- 45.Mitra L, Rouget JB, Garcia-Moreno B, Royer CA, Winter R. Towards a quantitative understanding of protein hydration and volumetric properties. Chemphyschem. 2008;9(18):2715–2721. doi: 10.1002/cphc.200800405. [DOI] [PubMed] [Google Scholar]
- 46.Chowdury PD, Gruebele M. Molecules: What kind of a bag of atoms. J. Phys. Chem. 2011;113:13139–13143. doi: 10.1021/jp903104p. [DOI] [PubMed] [Google Scholar]
- 47.Baldwin RL, Frieden C, Rose GD. Dry molten globule intermediates and the mechanism of protein unfolding. Proteins. 2010;78:2725–2737. doi: 10.1002/prot.22803. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.