Abstract
Culex flavivirus (CxFV) is an insect-specific flavivirus globally distributed in mosquitoes of the genus Culex. CxFV was positively associated with West Nile virus (WNV) infection in a case–control study of 268 mosquito pools from an endemic focus of WNV transmission in Chicago, United States. Specifically, WNV-positive Culex mosquito pools were four times more likely also to be infected with CxFV than were spatiotemporally matched WNV-negative pools. In addition, mosquito pools from residential sites characterized by dense housing and impermeable surfaces were more likely to be infected with CxFV than were pools from nearby urban green spaces. Further, 6/15 (40%) WNV-positive individual mosquitoes were also CxFV positive, demonstrating that both viruses can coinfect mosquitoes in nature. Phylogenetic analysis of CxFV from Chicago demonstrated a pattern similar to WNV, consisting of low global viral diversity and lack of geographic clustering. These results illustrate a positive ecological association between CxFV and WNV, and that coinfection of individual mosquitoes can occur naturally in areas of high flaviviral transmission. These conclusions represent a challenge to the hypothesis of super-infection exclusion in the CxFV/WNV system, whereby an established infection with one virus may interfere with secondary viral infection with a similar virus. This study suggests that infection with insect-specific flaviviruses such as CxFV may not exclude secondary infection with genetically distinct flaviviruses such as WNV, and that both viruses can naturally coinfect mosquitoes that are epidemic bridge vectors of WNV to humans.
Key Words: Arboviruses, Epidemiology, Flavivirus, Mosquito-only Flavivirus, West Nile
Introduction
Culex flavivirus (CxFV) is an insect-specific member of the family Flaviviridae, which includes a number of viruses of human health significance, including West Nile virus (WNV). CxFV was first isolated from Culex pipiens Linnaeus mosquitoes in Japan (Hoshino et al. 2007) but appears to have a global distribution, with additional variants identified in other Culex species and subspecies from Mexico, Uganda, Trinidad, and the United States (Morales-Betoulle et al. 2008, Blitvich et al. 2009, Cook et al. 2009, Farfan-Ale et al. 2009, Kim et al. 2009). The insect-specific designation of CxFV derives from the observation that it has been cultured in the laboratory only in mosquito cell lines and, additionally, that it has been identified in many natural populations of Culex mosquitoes (Hoshino et al. 2007). Its isolation from both female and male mosquitoes suggests vertical transmission (Hoshino et al. 2007, Farfan-Ale et al. 2009), although very little is known about its natural history.
The phenomenon of super-infection exclusion (Randolph and Hardy 1988, Tscherne et al. 2007) has been proposed in the case of CxFV, whereby a mosquito infected with CxFV may be refractory to coinfection with another related virus (Farfan-Ale et al. 2009). If so, then viruses such as CxFV, despite being restricted to insects, could provide indirect protection against the transmission of related viruses of human health importance, such as WNV, Dengue virus, or Yellow fever virus. We therefore sought to test the hypothesis that such negative virus–virus interactions might occur between CxFV and WNV in the field. We focused on an area of high WNV transmission in suburban Chicago, United States (Ruiz et al. 2004), where our previous studies have demonstrated predictable seasonal WNV amplification in Culex mosquitoes coincident with annual peaks in human cases (Hamer et al. 2008b). In particular, WNV transmission in this area is driven by Cx. pipiens, the dominant WNV vector in the Eastern and Midwestern United States and a bridge vector between avian and human hosts (Hamer et al. 2008a).
The availability of 1076 banked Culex spp. RNA samples from WNV-positive and WNV-negative Culex pools allowed us to conduct a case–control study comparing CxFV infection in spatiotemporally matched WNV-positive and WNV-negative samples. A smaller number of banked WNV-positive individual Culex spp. mosquitoes also allowed us to examine whether CxFV and WNV can coinfect mosquitoes in nature.
Methods
Mosquitoes were collected from our study area in the southwestern Chicago suburbs (∼150 km2 in Cook County; 87°44′W, 41°42′N), from 4 urban green space sites (cemeteries and parks) and 11 residential sites (characterized by dense housing and impermeable surfaces) (Bertolotti et al. 2008, Hamer et al. 2008a). A total of 1076 mosquito pools were available from 2006, a year of particularly high WNV transmission (Ruiz et al. 2010). Mosquitoes were captured using CO2-baited Centers for Disease Control and Prevention miniature light traps, Centers for Disease Control and Prevention gravid traps baited with rabbit pellet infusion, and battery-powered backpack aspirators. Mosquitoes were identified (Andreadis et al. 2005); pooled into groups of 31 or fewer; grouped by species, sex, collection site, and date; and processed for extraction of RNA, which was then frozen at −80°C for archiving after WNV testing by real-time reverse-transcription polymerase chain reaction (RT-PCR) (Hamer et al. 2008a).
Using these samples, we designed a case–control study in which we selected 268 banked Culex spp. mosquito RNA pools consisting of 134 pools that were WNV-positive (cases) and 134 WNV-negative pools (controls). Mosquito pools selected were randomly chosen for each site and each time period (early vs. late in the season). Each WNV-positive sample was paired with a WNV-negative sample collected from the same site as close in time as possible (usually on the same day). In addition, we were able to examine RNA extracted from 15 individual blood-fed WNV-positive Culex spp. mosquitoes collected between 2005 and 2007, available as a result of a previous study of WNV transmission and mosquito feeding preferences (Hamer et al. 2009).
To detect CxFV directly from mosquito-derived RNA, we designed a novel, nested RT-PCR (Table 1). RT-PCR was performed using the Qiagen OneStep RT-PCR Kit (Qiagen, Valencia, CA) and pan-flavivirus primers FU2 and cFD3 (Table 1), which span 845 nucleotides of the viral nonstructural protein 5 (NS5) gene of all flaviviruses (Kuno et al. 1998). Cycling conditions consisted of initial RT at 48°C for 60 min, denaturation at 95°C for 15 min, followed by 35 cycles of PCR (94°C for 30 s, 50°C for 30 s, 68° for 2.5 min), a terminal extension at 68°C for 10 min, and an indefinite soak at 4°C. PCR product from the initial RT-PCR was then used as a template in a secondary reaction using the FailSafe™ PCR System (Epicentre Biotechnologies, Madison, WI) and primers CxFV_9131F and CxFV_9337R (Table 1), which we designed to be specific for a 206-nucleotide region of the CxFV NS5 gene. Cycling conditions for the internal PCR consisted of initial denaturation 94°C for 4 min, followed by 35 cycles of PCR (94°C for 30 s, 57°C for 30 s, and 72°C for 45 s), a terminal extension at 72°C for 7 min, and an indefinite soak at 4°C.
Table 1.
Polymerase Chain Reaction Primers Used for Detection and Sequencing of Culex Flavivirus from RNA of Culex Mosquito Pools
| Primer namea | Sequence (5′ to 3′) | |
|---|---|---|
| Diagnostic PCR for detection of CxFVb | ||
| External 1-step RT-PCR | FU2 | GCTGATGACACCGCCGGCTGGGACAC |
| cFD3 | AGCATGTCTTCCGTGGTCATCCA | |
| Internal PCR | CxFV_9131F | TTGTGGTTCTTGCTGGACCAAGTG |
| CxFV_9337R | ATTCTCCCAACCTGGTTCTTCCCA | |
| PCRs for sequencing of CxFV NS5 gene segment for phylogenetic analysisb | ||
| External 1-step RT-PCR | CxFV_8880F | GGAGAAGAAGCCGTCCTCTTTCGG |
| CxFV_10814R | AGACGTGAACAAAAGCTTGCCCAC | |
| Internal PCR 1 | CxFV_8880F | GGAGAAGAAGCCGTCCTCTTTCGG |
| CxFV_9538R | CTCGGTCGGTTGCAAGTTCTTG | |
| Internal PCR 2 | CxFV_9516F | CCACACCAGTCTAAGGTACATC |
| CxFV_10188R | GTTGTTCTCTACGAGTCGCGTG | |
| Internal PCR 3 | CxFV_10182F | CAACCGACGRCGTGTTCTGGTG |
| CxFV_10814R | AGACGTGAACAAAAGCTTGCCCAC | |
Primer names indicate the nucleotide coordinate of the 3′ base of the primer with respect to the CxFV reference sequence (GenBank accession number NC_008604) and the forward (F) or reverse (R) direction. Primers FU2 and cFD3 are from Kuno et al. (1998).
CxFV, Culex flavivirus; NS5, nonstructural protein 5; RT-PCR, reverse-transcription polymerase chain reaction.
Negative and positive controls were carried through all steps. Amplicons were observed using gel electrophoresis on 1% agarose gels stained with ethidium bromide. Ten amplicons (five from WNV-positive pools and five from WNV-negative pools) were gel-excised and purified using the Zymoclean™ Gel DNA Recovery Kit (Zymo Research, Orange, CA) and sequenced directly for confirmation on ABI 3730xl DNA Analyzers (Applied Biosystems, Foster City, CA) at the University of Wisconsin–Madison Biotechnology Center.
We calculated infection rates (expressed per 1000 mosquitoes tested) using the maximum likelihood estimation method available in the Microsoft Excel (Microsoft Inc., Redmond, WA) add-in, Pooled Infection Rate version 3.0 (Biggerstaff 2006). We examined the association between WNV and CxFV infection using McNemar's test for case–control data with continuity correction, which considers only discordant pairs (pairs in which CxFV infection status differed between cases and controls) in a matched case–control study design (Fleiss 1981). To examine additional spatial and temporal factors influencing CxFV infection, we encoded variables to represent study site type (residential vs. urban green space) and time of sample collection (early vs. late season, defined as June 30–July 31 vs. August 1–September 7 based on dates from which positive mosquito pools were recovered in 2006). We then used logistic regression to examine associations between these factors and CxFV infection status. All statistical analyses were performed using SPSS v. 18 (SPSS, Inc., Chicago, IL).
To determine the phylogenetic position of CxFV circulating in our study area, we sequenced a 1239-nucleotide region of the viral NS5 gene corresponding to nucleotide positions 8899-10104 of the Japan 2003 CxFV prototype sequence (GenBank accession NC_008604) in five CxFV-positive individual mosquito samples. An initial long RT-PCR was performed with primers CxFV_8880F and CxFV_10814R (Table 1), and PCR product from this reaction was then used as template for internal PCR-amplification of three overlapping fragments (Table 1). Cycling conditions for the external PCR were identical to those described above for the diagnostic CxFV PCR external PCR, and cycling conditions for the three internal sequencing PCRs were identical, consisting of initial denaturation at 94°C for 4 min, followed by 35 cycles of PCR (94°C for 30 s, 55°C for 30 s, and 72°C for 1 min), a terminal extension at 72°C for 7 min, and an indefinite soak at 4°C.
PCR products were observed and purified from gels as described above and were sequenced directly using both forward and reverse PCR primers to resolve ambiguous bases. Sequences were assembled and hand-edited using the computer programs 4Peaks Version 1.7.2 (Mekentosj Inc., Amsterdam, The Netherlands) and Mesquite v. 2.72 (Maddison and Maddison 2009) and were aligned with respect to published Flavivirus sequences using ClustalW (Larkin et al. 2007), with manual adjustment. Phylogenetic analyses were conducted using Bayesian methods implemented in the computer program MrBayes v. 3.1 (Ronquist and Huelsenbeck 2003).
Results
The infection rate for WNV among the 1076 Culex pools tested in 2006 was 11.44 (95% confidence interval [CI]: 9.94–13.33). Within WNV-positive pools the infection rate for CxFV was 112.62 (95% CI: 91.25–142.89, based on 126/134 CxFV-positive pools), and within WNV-negative pools the infection rate for CxFV was 100.18 (95% CI: 82.59–123.22, based on 114/134 CxFV-positive pools). Sequencing of 10 CxFV amplicons from our diagnostic PCR revealed only CxFV-specific sequences, even in samples known also to be WNV positive, demonstrating the specificity of this PCR for CxFV.
Among the 134 spatiotemporally matched pairs of mosquito pools, 20 discordant pairs were identified in which WNV-positive and WNV-negative pools differed in their CxFV infection status (Table 2). Of these, we identified 16 (80%) pairs in which the WNV-positive pool was CxFV positive, whereas the WNV-negative pool was CxFV negative, but only 4 (20%) pairs in which the WNV-positive pool was CxFV negative, whereas the WNV-negative pool was CxFV positive. This difference was statistically significant based on McNemar's test for paired case–control data with continuity correction (χ2 = 6.050; 1 df; p = 0.014; odds ratio = 4.00; 95% CI: 1.29–16.44), indicating a four—fold increased odds of CxFV infection in WNV-positive mosquito pools relative to spatiotemporally matched WNV-negative mosquito pools. Among the 15 individual WNV-positive Culex mosquitoes tested, 6 (40%) were also CxFV positive, demonstrating clearly that coinfection of individual mosquitoes with both WNV and CxFV can occur in nature.
Table 2.
Culex Flavivirus Infection in 268 Culex Mosquito Pools Consisting of 134 Spatiotemporally Matched Pairs of West Nile Virus–Positive and West Nile Virus–Negative Pools from Suburban Chicago
|
Mosquito poolsa | |||
|---|---|---|---|
| CxFV positive | CxFV negative | Total | |
| WNV positive | 126 (95, 31) | 8 (4, 4) | 134 (99, 35) |
| WNV negative | 114 (79, 35) | 20 (13, 7) | 134 (92, 42) |
| Total | 240 (174, 66) | 28 (17, 11) | 268 (191, 77) |
|
Spatiotemporally matched pairs of WNV-positive and WNV-negative mosquito poolsb | |||
|---|---|---|---|
| Type of pair | Case (WNV positive) | Control (WNV negative) | No. of pairs |
| Discordant | CxFV positive | CxFV negative | 16 (10, 6) |
| Discordant | CxFV negative | CxFV positive | 4 (1, 3) |
| Concordant | CxFV positive | CxFV positive | 110 (74, 36) |
| Concordant | CxFV negative | CxFV negative | 4 (1, 3) |
| Total | — | — | 134 (86, 48) |
Numbers in parentheses indicate numbers of pools/pairs collected between June 30 and September 7, 2006, from 11 residential sites characterized by dense housing and 4 urban green space sites characterized by more diverse landscapes, respectively.
Results are significant by McNemar's test with continuity correction (χ2 = 6.050; p = 0.014).
WNV, West Nile virus.
Logistic regression confirmed a statistically significant positive association between CxFV infection and WNV infection in mosquito pools (χ2 = 7.97; p = 0.005; odds ratio = 3.411; 95% CI: 1.402–8.317). In addition, we observed a weak association between CxFV infection and site type, with mosquito pools from residential sites more likely to test positive for CxFV than mosquito pools from urban green space sites (χ2 = 6.85; p = 0.033; odds ratio = 2.51; 95% CI: 1.05–6.02). CxFV infection was also slightly likelier in mosquito pools collected early in the season (June–July) than later in the season (August–September), although this trend was not significant (χ2 = 2.91; p = 0.088; odds ratio = 2.05; 95% CI: 0.88–4.78). These results were not influenced by variation in pool size; median pool sizes for WNV-positive and WNV-negative pools, CxFV-positive and CxFV-negative pools, and pools from residential and urban green space sites were 25 in all cases.
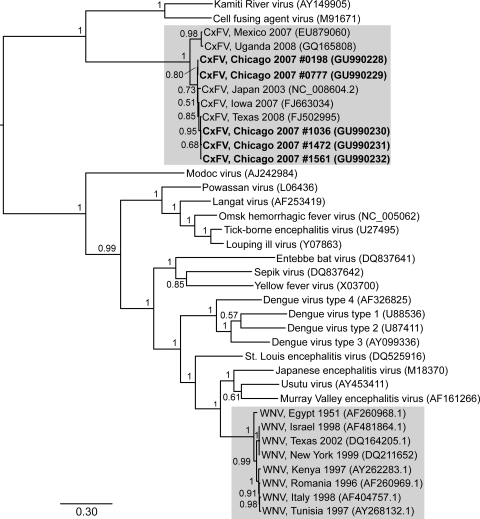
Our phylogenetic analyses produced a tree very similar to other recently published Flavivirus phylogenies, in which CxFV is most closely related to other insect-specific flaviviruses (Kamiti River virus and cell fusing agent virus) and more distantly related to flaviviruses that infect humans (Fig. 1). Our five new CxFV sequences from Chicago (GenBank accession numbers GU990228-GU990232) cluster with other CxFV sequences from Japan, Iowa, and Texas, forming a subclade that is closely related to CxFV from Mexico and Uganda. Overall, CxFV global phylogenetic diversity appears low within NS5 and is comparable to the global diversity of WNV at the same locus, although CxFV has to date been less thoroughly sampled.
FIG. 1.
Phylogenetic tree of partial nonstructural protein 5 sequences from Culex flavivirus (CxFV), West Nile virus (WNV), and other viruses in the genus Flavivirus (CxFV and WNV clades highlighted). An initial neighbor-joining tree was constructed using FastTree version 2.0.1 (Price et al. 2009) and a Jukes-Cantor + CAT model, to serve as a guide tree for more robust Bayesian phylogenetic inference. The substitution model used in the analysis was estimated using jModelTest (Posada 2008) and was of the form GTR + Γ with the following parameters: nucleotide frequencies A = 0.3035; C = 0.2506; G = 0.2734; T = 0.1725; substitution rates AC = 1.7354, AG = 2.4902, AT = 1.6292, CG = 0.9655, CT = 7.2558, and GT = 1; and Γ (gamma distribution of among-site rate variation) = 0.4130. Phylogenetic analyses were performed using this model and the Bayesian method available in MrBayes v3.1.2 (Huelsenbeck and Ronquist 2001) with two parallel runs of four Markov Chain Monte Carlo (MCMC) chains each for 600,000 generations, with subsampling every 100th generation. Stationarity was assessed at effective sample sizes >400 (469 and 559, respectively, for each Markov Chain Monte Carlo (MCMC) chain), using Tracer v1.5 (Drummond and Rambaut 2007) and a burn-in value of 10%. Posterior probability support is shown next to the nodes of the tree. GenBank accession numbers are in parentheses following taxon names; newly generated CxFV sequences from the Chicago study area are in bold, with reference numbers. The scale bar indicates genetic divergence (nucleotide substitutions per site).
Discussion
Our results demonstrate a high infection rate of CxFV in Culex mosquitoes from suburban Chicago, and a four—fold increased likelihood of infection of WNV-positive mosquito pools with CxFV relative to spatiotemporally matched WNV-negative pools. In addition, 40% of individual Culex spp. mosquitoes positive for WNV were also positive for CxFV. These results indicate an unexpected positive statistical association between CxFV and WNV. Further, the results from individual mosquitoes appear to counter the hypothesis of super-infection exclusion, whereby infection with one virus is protective against subsequent infection with a related virus.
The mechanisms accounting for positive association between CxFV and WNV in our study are not clear. Such positive association could result from immunological processes within individual mosquitoes. For example, a recent study has shown that insect-specific Flavivirus genomes are subject to similar selective pressures as those operating on the genomes of their mosquito hosts, suggesting host–virus coevolution (Lobo et al. 2009). If so, then CxFV may have evolved mechanisms to reduce immune recognition by its mosquito host, which could facilitate secondary infections with similar agents such as WNV. In this case, CxFV may modulate or even suppress the immune response of the mosquito, making the mosquito more susceptible to infection with a broad range of secondary pathogens. In this light, a recent study by Kent and colleagues (2010) demonstrated enhanced transmission of WNV by Culex quinquefasciatus mosquitoes simultaneously infected with CxFV under laboratory conditions.
The super-infection exclusion hypothesis is based on the idea of homologous interference, which is the ability of an established infection with one virus to interfere with secondary viral infection. Specific examples of this phenomenon have been documented in cell culture not only with flaviviruses (Randolph and Hardy 1988, Sundin and Beaty 1988, Burivong et al. 2004, Pepin et al. 2008), but also with other arboviruses of the genera Alphavirus (Karpf et al. 1997), Orbivirus (Ramig et al. 1989), and Vesiculovirus (Legault et al. 1977, Whitaker-Dowling et al. 1983). Such observations have led to the proposition that super-infection exclusion may be a generalized phenomenon that occurs broadly across the genus Flavivirus (Farfan-Ale et al. 2009, Kim et al. 2009), but evidence from the laboratory has been equivocal. For example, Vero cells infected with St. Louis encephalitis virus can be super-infected with two related flaviviruses, Japanese encephalitis virus and Yellow fever virus (Randolph and Hardy 1988), although not with the same strain of St. Louis encephalitis virus. On the whole, laboratory studies seem to suggest that superinfection exclusion can occur between closely related viruses but that more distantly related viruses do not generally interfere with each other. Our results demonstrating natural coinfection of individual mosquitoes and positive statistical association between CxFV and WNV suggest that the homologous interference concept may not apply to highly divergent flaviviruses such as the insect-specific flaviviruses and their vertebrate pathogen relatives, and that superinfection exclusion may be of limited relevance in a field setting.
It is also possible, and not mutually exclusive, that positive association between CxFV and WNV results from ecological factors. Ecological conditions varying on very fine spatial and temporal scales within our study area could create localized conditions favorable for the infection of mosquito pools with both CxFV and WNV. For example, the high CxFV infection rate of mosquito pools in our study area could reflect a high local density of Culex mosquitoes, especially if CxFV transmission is density dependent. High Culex density could, in turn, also enhance WNV transmission. Resolving the extent to which immunological versus ecological processes account for positive association between CxFV and WNV will require experimental studies of coinfection as well as more detailed studies of individual mosquitoes collected from areas with endemic CxFV and other cocirculating flaviviruses. Our documentation of CxFV infection in 40% of WNV-positive individual mosquitoes demonstrates clearly, however, that coinfection can occur in nature.
Although little is known about the natural history of CxFV in mosquitoes, it is likely vertically transmitted (Hoshino et al. 2007, Farfan-Ale et al. 2009). This is based on the observation of CxFV-positive pools containing only male mosquitoes (Farfan-Ale et al. 2009) and the identification of insect-specific flaviviruses in mosquito pupae (Crabtree et al. 2003). Therefore, we suggest that any interaction between CxFV and WNV in individual mosquitoes would most likely involve prior infection with CxFV. In this light, Kent et al. (2010) recently demonstrated enhanced WNV transmission in the laboratory after simultaneous inoculation of Cx. quinquefasciatus with CxFV and WNV, but not after sequential inoculation with CxFV then by WNV, supporting the notion that the timing of coinfection may be critical determinant of the nature of virus–virus interaction. Regardless of the underlying mechanism, Culex spp. mosquitoes are the principal vectors of WNV in suburban Chicago, and our previous work using blood-meal analysis and analytical risk assessment shows that Cx. pipiens serves as both the primary enzootic vector for mosquito–bird transmission and a bridge vector for human WNV infection (Hamer et al. 2009). Given the importance of Culex mosquitoes to maintaining WNV in birds and transmitting it to humans, our results showing a positive association between CxFV and WNV suggest that an insect-specific flavivirus could influence animal and human disease risk, or at least serve as an ecological indicator of such risk.
Our results also reveal an association between CxFV infection status of mosquito pools and collection-site type; CxFV-positive mosquito pools were more likely to be recovered from residential sites characterized by dense housing than from urban green spaces. Culex mosquitoes are the most common mosquito species inhabiting urban areas in the northeastern United States and thrive in peridomestic habitats, where they feed on both humans and birds (including reservoir-competent hosts for WNV) (Vinagradova 2000, Spielman 2001). Higher CxFV infection in residential areas might therefore indicate higher Culex density or different Culex population dynamics in such locations. Our past studies of these same sites have shown a consistent trend across years of higher WNV genetic diversity in residential sites than in urban green space sites (Bertolotti et al. 2008, Amore et al. 2010). Fine-scale effects of the urban environment may therefore affect the transmission dynamics of both WNV and CxFV. The absence of a strong temporal trend in CxFV infection, despite pronounced seasonal amplification of WNV, again supports a difference in the mode of transmission of these two viruses, with vertical transmission in the case of CxFV and horizontal transmission in the case of WNV.
Our phylogenetic results (Fig. 1) confirm that CxFV from Chicago is similar to previously characterized CxFV and is a distant relative of WNV within the genus Flavivirus (Hoshino et al. 2007). Our new sequences from Chicago fall into a cluster of closely related CxFV sequences from both Japan and the United States, slightly divergent from CxFV from Mexico and Uganda. CxFV phylogenetic diversity will likely increase with additional sampling, but these preliminary data suggest that CxFV global diversity is low and not strongly geographically apportioned. Indeed, our phylogeny shows a degree of genetic divergence within CxFV that is comparable to that in WNV (our analysis included the most divergent published WNV sequences). Overall levels of CxFV and WNV diversity might imply similarity in the history of epidemic expansion of the two viruses. Confirmation of this hypothesis will have to await more extensive sampling of CxFV and further epidemiological and evolutionary analyses. Nevertheless, our results suggest that examining ecological and evolutionary associations between insect-specific viruses such as CxFV and other flaviviruses of human and animal health importance could provide significant new insights into both arbovirus biology and public health.
Acknowledgments
We thank the Archdiocese of Chicago, and the municipalities of Evergreen Park, Palos Hills, Burbank, Alsip, Blue Island, the City of Chicago, and private landowners in these municipalities for allowing us to conduct this research, as well as to the Village of Oak Lawn for providing field laboratory facilities and logistical support. We thank S. Dallmann, D. Gohde, S. Loss, and T. Thompson for assisting with mosquito collection; L. Abernathy and J. McClain for assistance in the laboratory; and L. Bartholomay and B. Blivitch for providing positive control CxFV samples and invaluable discussion. This material is based upon work supported by the National Science Foundation/National Institutes of Health Ecology of Infectious Diseases program under Award No. 0840403.
Disclosure Statement
No competing financial interests exist.
References
- Amore G. Bertolotti L. Hamer GL. Kitron UD, et al. Multi-year evolutionary dynamics of West Nile virus in suburban Chicago, USA, 2005–2007. Philos Trans R Soc Lond B Biol Sci. 2010;365:1871–1878. doi: 10.1098/rstb.2010.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreadis TG. Thomas MC. Shepard JJ. Identification Guide to the Mosquitoes of Connecticut. New Haven, CT: The Connecticut Agricultural Experiment Station; 2005. [Google Scholar]
- Bertolotti L. Kitron UD. Walker ED. Ruiz MO, et al. Fine-scale genetic variation and evolution of West Nile Virus in a transmission “hot spot” in suburban Chicago, USA. Virology. 2008;374:381–389. doi: 10.1016/j.virol.2007.12.040. [DOI] [PubMed] [Google Scholar]
- Biggerstaff BJ. Pooled Infection Rate, Version 3.0. Ft. Collins, CO: Center for Disease Control and Prevention; 2006. [Google Scholar]
- Blitvich BJ. Lin M. Dorman KS. Soto V, et al. Genomic sequence and phylogenetic analysis of Culex flavivirus, an insect-specific Flavivirus, isolated from Culex pipiens (Diptera: Culicidae) in Iowa. J Med Entomol. 2009;46:934–941. doi: 10.1603/033.046.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burivong P. Pattanakitsakul SN. Thongrungkiat S. Malasit P. Flegel TW. Markedly reduced severity of Dengue virus infection in mosquito cell cultures persistently infected with Aedes albopictus densovirus (AalDNV) Virology. 2004;329:261–269. doi: 10.1016/j.virol.2004.08.032. [DOI] [PubMed] [Google Scholar]
- Cook S. Moureau G. Harbach RE. Mukwaya L, et al. Isolation of a novel species of flavivirus and a new strain of Culex flavivirus (Flaviviridae) from a natural mosquito population in Uganda. J Gen Virol. 2009;90:2669–2678. doi: 10.1099/vir.0.014183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree MB. Sang RC. Stollar V. Dunster LM, et al. Genetic and phenotypic characterization of the newly described insect Flavivirus, Kamiti River virus. Arch Virol. 2003;148:1095–1118. doi: 10.1007/s00705-003-0019-7. [DOI] [PubMed] [Google Scholar]
- Drummond AJ. Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farfan-Ale JA. Lorono-Pino MA. Garcia-Rejon JE. Hovav E, et al. Detection of RNA from a novel West Nile-like virus and high prevalence of an insect-specific flavivirus in mosquitoes in the Yucatan Peninsula of Mexico. Am J Trop Med Hyg. 2009;80:85–95. [PMC free article] [PubMed] [Google Scholar]
- Fleiss JL. Statistical Methods for Rates and Proportions. New York: John Wiley; 1981. [Google Scholar]
- Hamer GL. Kitron UD. Brawn JD. Loss SR, et al. Culex pipiens (Diptera: Culicidae): a bridge vector of West Nile virus to humans. J Med Entomol. 2008a;45:125–128. doi: 10.1603/0022-2585(2008)45[125:cpdcab]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Hamer GL. Kitron UD. Goldberg TL. Brawn JD, et al. Host selection by Culex pipiens mosquitoes and West Nile virus amplification. Am J Trop Med Hyg. 2009;80:268–278. [PubMed] [Google Scholar]
- Hamer GL. Walker ED. Brawn JD. Loss SR, et al. Rapid amplification of West Nile virus: the role of hatch-year birds. Vector Borne Zoonot Dis. 2008b;8:57–67. doi: 10.1089/vbz.2007.0123. [DOI] [PubMed] [Google Scholar]
- Hoshino K. Isawa H. Tsuda Y. Yano K, et al. Genetic characterization of a new insect Flavivirus isolated from Culex pipiens mosquito in Japan. Virology. 2007;359:405–414. doi: 10.1016/j.virol.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP. Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Karpf AR. Lenches E. Strauss EG. Strauss JH. Brown DT. Superinfection exclusion of alphaviruses in three mosquito cell lines persistently infected with Sindbis virus. J Virol. 1997;71:7119–7123. doi: 10.1128/jvi.71.9.7119-7123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent RJ. Crabtree MB. Miller BR. Transmission of West Nile virus by Culex quinquefasciatus say infected with Culex Flavivirus Izabal. PLoS Negl Trop Dis. 2010;4:e671. doi: 10.1371/journal.pntd.0000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DY. Guzman H. Bueno R., Jr. Dennett JA, et al. Characterization of Culex Flavivirus (Flaviviridae) strains isolated from mosquitoes in the United States and Trinidad. Virology. 2009;386:154–159. doi: 10.1016/j.virol.2008.12.034. [DOI] [PubMed] [Google Scholar]
- Kuno G. Chang GJ. Tsuchiya KR. Karabatsos N, et al. Phylogeny of the genus Flavivirus. J Virol. 1998;72:73–83. doi: 10.1128/jvi.72.1.73-83.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA. Blackshields G. Brown NP. Chenna R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Legault D. Takayesu D. Prevec L. Heterotypic exclusion between vesicular stomatitis viruses of the New Jersey and Indiana serotypes. J Gen Virol. 1977;35:53–65. doi: 10.1099/0022-1317-35-1-53. [DOI] [PubMed] [Google Scholar]
- Lobo FP. Mota BE. Pena SD. Azevedo V, et al. Virus-host coevolution: common patterns of nucleotide motif usage in Flaviviridae and their hosts. PLoS ONE. 2009;4:e6282. doi: 10.1371/journal.pone.0006282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP. Maddison DR. Mesquite: a modular system for evolutionary analysis, version 2.72. http://mesquiteprojectorg. 2009. http://mesquiteprojectorg
- Morales-Betoulle ME. Monzon Pineda ML. Sosa SM. Panella N, et al. Culex flavivirus isolates from mosquitoes in Guatemala. J Med Entomol. 2008;45:1187–1190. doi: 10.1603/0022-2585(2008)45[1187:cfifmi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Pepin KM. Lambeth K. Hanley KA. Asymmetric competitive suppression between strains of dengue virus. BMC Microbiol. 2008;8:28. doi: 10.1186/1471-2180-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Price MN. Dehal PS. Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramig RF. Garrison C. Chen D. Bell-Robinson D. Analysis of reassortment and superinfection during mixed infection of Vero cells with bluetongue virus serotypes 10 and 17. J Gen Virol. 1989;70:2595–2603. doi: 10.1099/0022-1317-70-10-2595. [DOI] [PubMed] [Google Scholar]
- Randolph VB. Hardy JL. Establishment and characterization of St Louis encephalitis virus persistent infections in Aedes and Culex mosquito cell lines. J Gen Virol. 1988;69:2189–2198. doi: 10.1099/0022-1317-69-9-2189. [DOI] [PubMed] [Google Scholar]
- Ronquist F. Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Ruiz MO. Chaves LF. Hamer GL. Sun T, et al. Local impact of temperature and precipitation on West Nile virus infection in Culex species mosquitoes in northeast Illinois, USA. Parasit Vectors. 2010;3:19. doi: 10.1186/1756-3305-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz MO. Tedesco C. McTighe TJ. Austin C, et al. Environmental and social determinants of human risk during a West Nile virus outbreak in the greater Chicago area, 2002. Int J Health Geogr. 2004;3:8. doi: 10.1186/1476-072X-3-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielman A. Structure and seasonality of nearctic Culex pipiens populations. Ann N Y Acad Sci. 2001;951:220–234. doi: 10.1111/j.1749-6632.2001.tb02699.x. [DOI] [PubMed] [Google Scholar]
- Sundin DR. Beaty BJ. Interference to oral superinfection of Aedes triseriatus infected with La Crosse virus. Am J Trop Med Hyg. 1988;38:428–432. doi: 10.4269/ajtmh.1988.38.428. [DOI] [PubMed] [Google Scholar]
- Tscherne DM. Evans MJ. von Hahn T. Jones CT, et al. Superinfection exclusion in cells infected with hepatitis C virus. J Virol. 2007;81:3693–3703. doi: 10.1128/JVI.01748-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinagradova EB. Culex pipiens pipiens Mosquitoes: Taxonomy, Distribution, Ecology, Physiology, Genetics, Applied Importance, and Control Sofia. Bulgaria: Pensoft Publishers; 2000. [Google Scholar]
- Whitaker-Dowling P. Youngner JS. Widnell CC. Wilcox DK. Superinfection exclusion by vesicular stomatitis virus. Virology. 1983;131:137–143. doi: 10.1016/0042-6822(83)90540-8. [DOI] [PubMed] [Google Scholar]