Abstract
Many human cancer cells are sensitive to killing by the pro-apoptotic ligand TRAIL which is under study for cancer treatment in clinical trials. The TRAIL receptor TRAIL-R1 (also known as DR4) is a transmembrane receptor that mediates TRAIL-induced apoptosis in cancer cells. In this study, we show that retinoids sensitize cancer cells to TRAIL-induced apoptosis by upregulating expression of TRAIL-R1. All-trans retinoic acid (ATRA) upregulated TRAIL-R1 expression in human cancer cells at the transcriptional level. The ability of ATRA to activate TRAIL-R1 expression was inhibited by retinoic acid receptor (RAR) antagonists or small interfering RNAs, but augmented by several RAR agonists. In analyzing how ATRA induces RAR-dependent transcriptional upregulation of TRAIL-R1 we identified two putative RA response elements (RARE) termed Pal-17 (a palindrome separated by 17 bases) and DR-11 (a direct repeat separated by 11 bases) in the 5′ flanking region of TRAIL-R1 gene. Deletion of DR-11 but not Pal-17 abrogated the ability of ATRA to stimulate TRAIL-R1 promoter activity. Consistent with this observation, RAR binding to DR-11 but not to Pal-17 was detected by ChIP assay in ATRA-treated cells, arguing that DR-11 was responsible for ATRA-mediated activation of the TRAIL-R1 gene. ATRA augmented TRAIL-induced apoptosis of cancer cells and this activity was attenuated by a blockade to upregulation of TRAIL-R1 expression. Taken together, our findings establish that ATRA accentuates TRAIL-induced apoptosis, reveal a novel mechanism by which retinoids modulate apoptosis, and suggest a novel strategy to augment the anti-cancer activity of TRAIL.
Keywords: Retinoic acid, retinoic acid receptors (RARs), TRAIL receptor 1, apoptosis
INTRODUCTION
Retinoic acid (RA), a natural metabolite of vitamin A, plays an essential role in regulation of differentiation, growthand development of normal and malignant epithelial cells in various tissues and is therefore an important regulator of cancer formation and progression (1, 2). RA and its derivatives (retinoids) have long been recognized as a group of cancer chemopreventive and therapeutic agents. Currently RA is an approved drug for the treatment of patients with acute promyelocytic leukemia (1). One underlying mechanism by which retinoids exert their anticancer effects is induction of apoptosis (3). However, the mechanisms by which retinoids including RA induce apoptosis have not been fully elucidated even though extensive studies have been done in this regard (2, 3).
Retinoid carries out most of its function by regulation of gene expression through the activation of its nuclear receptors known as retinoic acid receptors (RARs) and retinoid X receptors (RXRs). The activated and dimerized receptors (e.g., RXR/RAR) bind to retinoic acid response element (RARE) present in the untranslated regions of target genes to execute the trans-activation (4). A RARE sequence is a hexamer repeat, spaced by N numbers of nucleotides 5′-PuGGTCA (N) PuGGTCA-3′ (Pu = A or G). The repeat can be direct, everted or even palindromic (4, 5).
Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL, also called APO-2L), a member of the tumor necrosis factor family, has attracted much attention recently because it induces apoptosis in a wide variety of transformed cells but does not seem to be cytotoxic to normal cells in vitro and in vivo. TRAIL is therefore considered to be a tumor-selective, apoptosis-inducing cytokine and is being tested in clinical trials as a potential anticancer agent (6–8). TRAIL induces apoptosis in human cells by interacting with two death domain-containing receptors: TRAIL receptor 1 (TRAIL-R1; also called death receptor 4) and TRAIL receptor 2 (TRAIL-R2; also known as DR5) (7).
It has been shown that RA induces interferon regulatory factor-1 (IRF-1)-dependent TRAIL expression and subsequently triggers apoptosis in leukemia cells through a paracrine action of TRAIL (9, 10). In this study, we show that RA and other retinoids upregulate TRAIL-R1 expression through a RAR-dependent mechanism in addition to induction of TRAIL. Accordingly, RA cooperates with TRAIL to augment apoptosis. Therefore, the current study has revealed a novel mechanism by which RA modulates apoptosis, extending our understanding on RA regulation of the extrinsic apoptotic pathway.
MATERIALS AND METHODS
Reagents
All-trans RA (ATRA), actinomycin D (Act D) and cycloheximide (CHX) were purchased from Sigma Chemicals (St. Louis, MO). Am580, Ro415253 and AC55649 were purchased from Enzo Life Sciences (Plymouth Meeting, PA). CH55, CD271 and LE135 were purchased from Tocris Bioscience (Ellisville, MO). These agents were dissolved in DMSO as stock solutions and stored at −80 °C. The soluble recombinant human TRAIL was purchased from PeproTech, Inc. (Rocky Hill, NJ).
Cell Lines and Cell Culture
Except for PLA-801C and Tu686, which were provided by Dr. Z. Chen (Emory University, Atlanta, GA), all other human non-small cell lung cancer (NSCLC) and head and neck squamous cell carcinoma (HNSCC) cell lines used in this study were generously provided by Dr. R. Lotan (M.D. Anderson Cancer Center, Houston, TX). These cell lines were cultured as described previously (11, 12). Among them, H157, A549, 22B and 183A were authenticated by Genetica DNA Laboratories, Inc. (Cincinnati, OH) and PLA-801C and Tu686 were authenticated by RADIL (Columbia MO) recentlyby analyzing STR DNA profile. To avoid potential interference from retinoids in serum, all the treatments with ATRA and other retinoids were done in serum-free culture medium.
Western Blot Analysis
Preparation of whole cell protein lysates and the procedures for the Western blotting were described previously (13). Mouse monoclonal anti-TRAIL-R1 antibody (B-N28) was purchased from Diaclone (Stamford, CT). Rabbit polyclonal anti-TRAIL-R2 antibody was purchased from ProSci Inc. (Poway, CA). Mouse monoclonal anti-caspase-3 and anti-TRAIL antibody were purchased from Imgenex (San Diego, CA). Rabbit polyclonal anti-caspase-9 and anti-PARP antibodies and mouse anti-caspase-8 antibody were purchased from Cell Signaling Technology, Inc. (Beverly, MA). Rabbit polyclonal anti-β-actin antibody was purchased from Sigma Chemical Co. Rabbit polyclonal anti-human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody was purchased from Trevigen (Gaithersburg, MD).
Analysis of Cell Surface TRAIL-R1
The procedure for direct antibody staining and subsequent flow cytometric analysis of cell surface TRAIL-R1 was described previously (14). The mean fluorescence intensity (MFI) that represents antigenicdensity on a per cell basis was used to represent cell surface TRAIL-R1 expression level. Phycoerythrin (PE)-conjugated mouse anti-human TRAIL-R1 (DJR1) monoclonal antibody and PE mouse IgG1 isotype control (MOPC-21/P3) were purchased from eBioscience (San Diego, CA).
Isolation of Total RNA and Reverse Transcription PCR (RT-PCR)
Total RNA was extracted from cells using TRI Reagent (Sigma Chemical Co). Complementary DNA was synthesized from 1 μg of total RNA using iScript cDNA synthesis kit (Bio-Rad Laboratories; Hercules, CA) and was subsequently used for the amplification of TRAIL-R1. The following primers were used for the amplification of 170 bp fragment from TRAIL-R1 gene: Forward 5′-CAGAGGGATGGTCAAGGTCAAGG-3′ and reverse 5′-CCACAACCTGAGCCGATGC-3′. GAPDH (239 bp) was also amplified as an internal control with the primers, forward 5′-CACAGTCAAGGCTGAGAATGGGAA-3′ and reverse 5′-GTGGTTCACACCCATCACAAACATG-3′.
Generationof Deletion Mutants of 5′ Flanking Region of TRAIL-R1 Gene
The reporter plasmids in pGL3-basic-luc carrying the 5′ flanking regions of TRAIL-R1 gene (−1773/+63, −586/+63 and −208/+63)were constructed previously in our laboratory (15). The following primers, forward (−1289/+63) 5′-CGACGCGTGGGTTCAAGCGATTCTCCT-3′ and forward (−174/+63) 5′-CGACGCGTTGGGACCAAGTGGCAAAA-3′ were used along with the reverse primer, 5′-CTAGCTAGCCATCCTGCCAGGTCAATCC-3′ for the amplification of −1289/+63 and −174/+63 fragments. The amplified fragments were then cloned into pGL3-basic-luc reporter plasmid through Mlu I and Nhe I restriction sites. The authenticities of the constructs were confirmed by sequencing.
Transient Transfection and Luciferase Reporter Assay
The given cells plated in 24-well plates were transfected with a TRAIL-R1 reporter plasmid together with pCH110 plasmid harboring β-galactosidase gene using Fugene 6 reagent (Roche Applied Science, Indianapolis, IN) following the manufacturer’s protocol. Twenty-four hours later, the cells were treated with DMSO or RA in serum-free medium for 16 h and then harvested in cell-lysis buffer for measurement of the luciferase activity with the Luciferase Assay kit (Promega; Madison, WI) using a Sirius Luminometer (Berthold Detection Systems; Huntsville, AL). The luciferase activity was normalized with the β-galactosidase activity, which was measured as described previously (16).
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assay was carried out using EZ ChIP ™ Chromatin Immunoprecipitation kit (Millipore; Billerica, MA) with the manufacturer’s instructions. The immunoprecipitation was carried out with 2 μg of rabbit anti-RAR antibody (M-454; Santa Cruz Biotechnology Inc., Santa Cruz, CA) or rabbit IgG (as the negative control). The amplification of DNA fragment containing RARE Pal-17 or RARE DR-11 was conducted using PCR with the following primers: forward 5′-TTAGTAGAGGCTGCGTTTCTCC-3′ and reverse 5′GCCTGTAATCCCAGCACTTT-3′ for 102 bp fragment containing RARE Pal-17 and forward 5′-CCGTAAAAGCCTCTTAGAGGCC-3′ and reverse 5′-TCGTTTTGCCACTTGGTCCC-3′ for 166 bp region having the RARE DR-11.
Silencing of TRAIL-R1 with Short Hairpin RNA (shRNA)
TRAIL-R1 shRNA in pLKO1 lentiviral vector (TRCN0000005934) was purchased from Thermo Scientific Open Biosystems (Huntsville, AL). The lentivirus preparation and infection were carried out following the manufacturer’s instructions. To assure silencing effect, the same infection was conducted twice in a 24 h interval. Twenty four hours after the second infection, the given cells were split and treated next day with DMSO or ATRA. After additional 48 h, the efficiency of the TRAIL-R1 silencing was checked with the Western blotting. The infected cells were also re-seeded in 96-well plates for cell survival and DNA fragmentation assays as described below.
Silencing of RARs with Small Interfering RNA (siRNA)
Non-silencing control siRNA was described previously (13). siRNA against both RARα and γ was used as reported (17). RARβ siRNA (sc-29466) was purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Gene silencing effects were confirmed by real time quantitative RT-PCR with the primers for RARα (18), RARβ (forward: 5′ GAATTCCCCCATGCGAGCTGTT 3′ and reverse: 5′ TCCAGGCTTGCTCGGCCAAT 3′) and RARγ (forward: 5′ AGAGTCAGTGTGCGGTTTGGGAGA 3′ and reverse: 5′ TGGGCCTCCAAAAGTCCCGCT 3′), respectively, as described below.
Real Time Quantitative RT-PCR
Quantitative RT-PCR was performed using power SYBR green PCR master mix (Applied Biosystems; Carlsbad, CA) with respective primers described above in a 7500 Fast Real Time PCR system from Applied Biosystems. The following reaction parameters were used for amplification: 50°C for 2 min and 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. GAPDH was used for normalization. The formula, 2−ΔΔCT, was applied to calculate the fold change of gene expression.
Cell Survival Assay
The cell survival was measured by sulforhodamine B (SRB) assay as described previously (11).
DNA Fragmentation Assay
The amounts of cytoplasmic histone-associated DNA fragments (mononucleosome and oligonucleosomes) formed during apoptosis were measured using a Cell Death Detection ELISA kit (Roche Applied Science) according to the manufacturer’s instructions.
RESULTS
Identification of Two Putative RAREs in the 5′ Flanking Region of TRAIL-R1 Gene
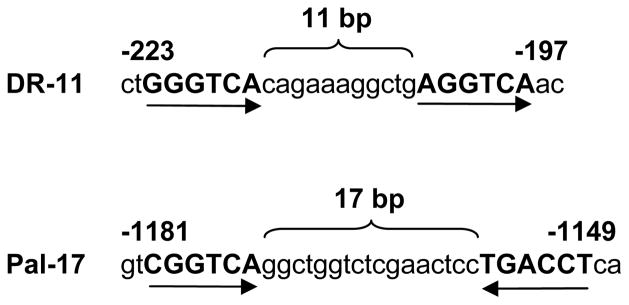
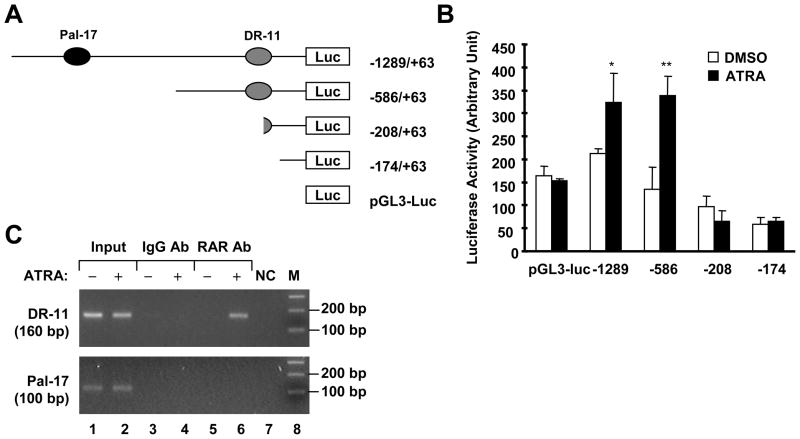
In an effort to understand the mechanisms by which TRAIL-R1 is regulated at the transcriptional level, we identified two putative RAREs in the 5′ flanking region of TRAIL-R1 gene. The first RARE had direct repeat motifs of GGGTCA and AGGTCA separated by 11 nucleotides and thus termed as DR-11. The second RARE had palindromic motifs of CGGTCA and TGACCT separated by 17 nucleotides and hence named as Pal-17 (Fig. 1).
Fig. 1.
Two putative RAREs, DR-11 and Pal-17, in the 5′ flanking region of TRAIL-R1 gene.
ATRA Induces TRAIL-R1 Expression in Human Cancer Cells
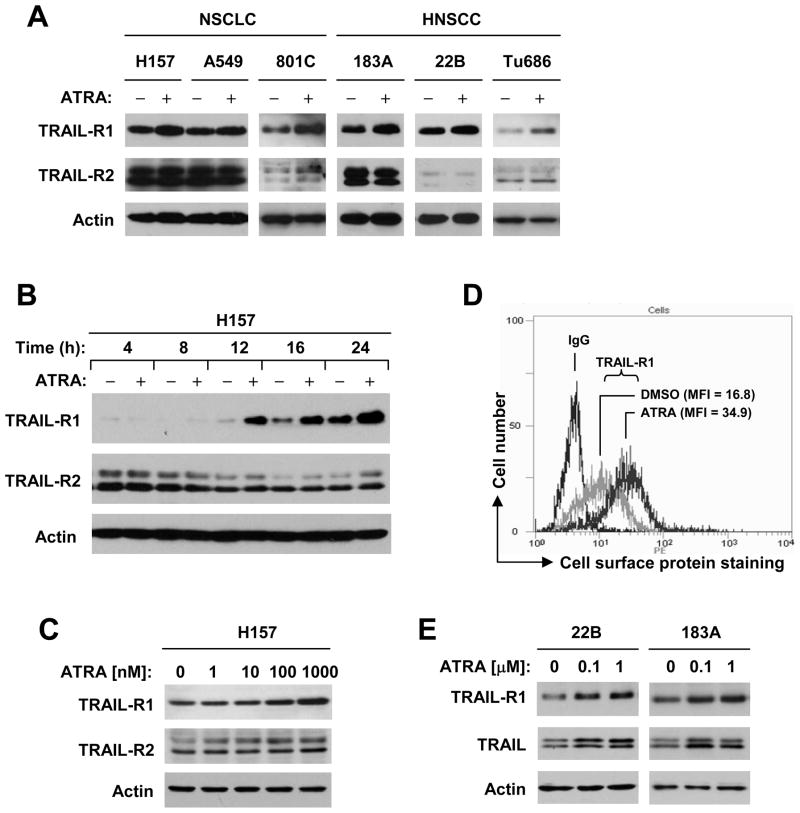
If the aforementioned RAREs in the 5′ flanking region of TRAIL-R1 gene are functional, it is plausible to speculate that ATRA, a natural ligand of RAR, would induce TRAIL-R1 expression. Thus, we examined the effects of ATRA on TRAIL-R1 expression in a panel of NSCLC and HNSCC cell lines. By Western blotting, we found that ATRA increased the levels of TRAIL-R1 protein in H157, A549, PLA-801C, 22B, 183A and Tu686 cells (Fig. 2A) as well as other NSCLC and HNSCC cell lines (supplemental Fig. S1). The induction of TRAIL-R1 occurred after 12 h post treatment with ATRA and was sustained up to 24 h (Fig. 2B). ATRA even at 100 nM could increase TRAIL-R1 protein levels (Fig. 2C). Moreover, we detected cell surface TRAIL-R1 levels in ATRA-treated cells and found that the MFI was increased from 16.8 in DMSO-treated cells to 34.9 in ATRA-treated cells, indicating that ATRA also increases cell surface TRAIL-R1 expression (Fig. 2D). Together, these results indicate that ATRA induces TRAIL-R1 expression.
Fig. 2. ATRA increases the expression of TRAIL-R1 (A–C), cell surface TRAIL-R1 (D) and TRAIL (E) in various cancer cell lines.
A, The indicated cell lines were treated with 1 μM ATRA for 16 h. B, H157 cells were treated with 1 μM ATRA for the given times as indicated. C, H157 cells were treated with the indicated concentrations of ATRA for 16 h. E, The indicated cell lines were treated with the given concentrations of ATRA for 48 h. After the aforementioned treatments, the whole cell protein lysates were prepared from these cells and used for the detection of indicated proteins with Western blot analysis. D, H157 cells were exposed to DMSO or 1 μM ATRA for 24 h and then harvested for detection of cell surface TRAIL-R1 with a flow cytometer.
TRAIL-R2 is another TRAIL death receptor with similar function to TRAIL-R1. We also examined the effects of ATRA on TRAIL-R2 and TRAIL expression. ATRA had almost no effect on TRAIL-R2 expression in the tested cell lines (Figs. 2A-2C). However, it increased TRAIL levels while inducing TRAIL-R1 expression (Fig. 2E).
ATRA Induces TRAIL-R1 Expression at the Transcriptional Level
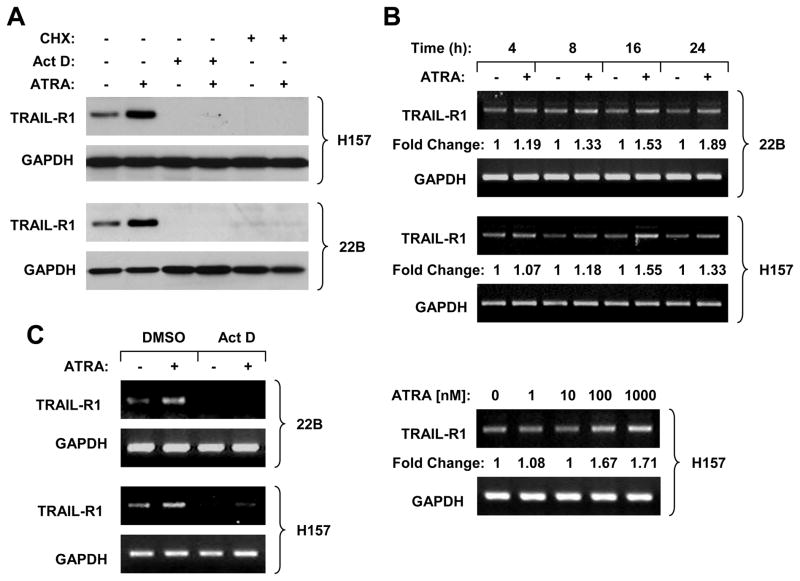
To demonstrate whether ATRA increases TRAIL-R1 expression through RAR-dependent transactivation of TRAIL-R1 gene, we began to test whether ATRA increases TRAIL-R1 transcription, leading to upregulation of TRAIL-R1 expression. As presented in Fig. 3A, ATRA increased TRAIL-R1 protein levels in the absence of the transcriptional inhibitor, Act D or the protein synthesis inhibitor, CHX, but not in the presence of Act D or CHX, indicating that inhibition of RNA transcription blocks ATRA-induced TRAIL-R1 expression. Subsequently, we studied whether ATRA increases TRAIL-R1 mRNA levels. Indeed, increased levels of TRAIL-R1 mRNA were detected in cells exposed to ATRA with the RT-PCR (Fig. 3B); the increase of TRAIL-R1 mRNA occurred at 8 h and was sustained up to 24 h post ATRA treatment. In agreement with increase in TRAIL-R1 protein level, ATRA at 100 nM elevated TRAIL-R1 mRNA levels (Fig. 3B). Consistently, ATRA-induced increase in TRAIL-R1 mRNA could be abolished by the presence of Act D (Fig. 3C). We further used quantitative real time RT-PCR to validate the induction of TRAIL-R1 mRNA by ATRA. Again we observed that ATRA increased TRAIL-R1 mRNA levels in the absence of Act D, but not in the presence of Act D (supplemental Fig. S2). Collectively, it is clear that ATRA increases TRAIL-R1 transcription.
Fig. 3. ATRA increases TRAIL-R1 expression at the transcriptional level.
A, Act D and CHX block ATRA-induced TRAIL-R1 upregulation. The indicated cell lines were pre-treated with 2.5 μg/ml Act D or 5 μg/ml CHX for 30 min and then co-treated with 1 μM ATRA for an additional 16 h. The cells were then harvested for preparation of whole cell protein lysates and subsequent Western blotting. B, ATRA increases TRAIL-R1 mRNA levels. The indicated cells lines were treated with 1 μM ATRA for the given times or treated with the indicated concentrations of ATRA for 16 h. C, The indicated cell lines were pre-treated with 2.5 μg/ml Act D for 30 min before the co-treatment with 1 μM ATRA for 16 h. After the aforementioned treatments in B and C, the cells were subjected to preparation of cellular total RNA and subsequent RT-PCR for the indicated genes.
ATRA Induces TRAIL-R1 through a RAR-dependent Mechanism
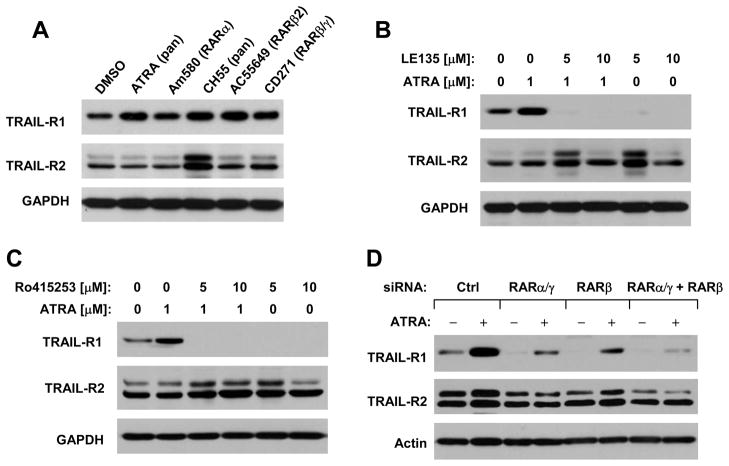
We next determined whether ATRA increases TRAIL-R1 expression through a RAR-mediated mechanism. To this end, we first examined the effects of other RAR agonists on TRAIL-R1 expression. As shown in Fig. 4A, the tested RAR agonists, Am580 (RARα), CH55 (RARα/β/γ), AC55649 (RARβ), and CD271 (RARβ/γ), increased TRAIL-R1 level as ATRA did. The presence of the RAR antagonist, LE135 (RARβ) or Ro415253 (RARα), not only substantially decreased basal levels of TRAIL-R1, but also abolished TRAIL-R1 upregulation by ATRA (Figs. 4B and 4C). We also examined the effects of RAR knockdown on ATRA-induced TRAIL-R1 expression. With real time RT-PCR, we could show that the tested siRNAs effectively reduced the expression of RARα, β and γ, respectively, in our cell system (supplemental Fig. S3), indicating good knockdown efficiencies of these siRNAs against their corresponding genes. With these siRNAs, we found that the transfection of either RARβ or RARα/γ siRNA substantially attenuated the ability of ATRA to increase TRAIL-R1 expression. Co-transfection of RARβ and RARα/γ siRNAs together was more potent than either siRNA alone in blocking TRAIL-R1 upregulation by ATRA (Fig. 4D). These data taken together clearly indicate that ATRA induces a RAR-dependent TRAIL-R1 expression.
Fig. 4. ATRA and other RAR agonists induce TRAIL-R1 expression (A), which can be abolished by RAR antagonists (B and C) or by the knockdown of RARs (D).
A, H157 cells were treated with 1 μM ATRA or other indicated retinoids for 16 h. B and C, H157 cells were pre-treated with the indicated concentrations of LE135 or Ro415253 for 30 min and then co-treated with 1 μM ATRA for 16 h. D, H157 cells were transfected with control (Ctrl), RARα/γ, RARβ, or RARα/γ plus RARβ siRNAs for 72 h. The cells were then treated with DMSO or 1μM ATRA for additional 16 h. After the aforementioned treatments, the whole cell protein lysates were prepared from these cells and used for the detection of indicated proteins with Western blot analysis.
Besides, we detected TRAIL-R2 expression in the above experiments. We noted that CH55, but not other RAR agonists, increased TRAIL-R2 expression. Moreover, the RAR antagonists, LE135 (at 5 μM) and Ro415253, substantially reduced basal levels of TRAIL-R1, but increased TRAIL-R2 expression (Figs. 4A–4C). Since knockdown of RARs did not increase TRAIL-R2 levels even though they decreased basal levels of TRAIL-R1 (Fig. 4D), it is likely that TRAIL-R2 induction by CH55, LE530 and Ro415253 is independent of RAR-mediated mechanism.
DR-11 is a Functional RARE in the Mediation of ATRA-induced TRAIL-R1 Transactivation
As stated earlier, there are two putative RAREs in the 5′ flanking region of TRAIL-R1 gene (Fig. 1). The unaddressed question is whether they are functional RAREs that mediate RAR-dependent TRAIL-R1 transactivation by ATRA. To answer this question, we made several reporter constructs with different deletions of TRAIL-R1 5′ flanking region, which contain both DR-11 and Pal-17 (-1289/+63), DR-11 only (-586/+63) or none of DR-11 and Pal-17 (−208/+63 and −174/+63) (Fig. 5A). Luciferase assay from the cells transfected with these reporter constructs followed by ATRA treatment showed that ATRA significantly increased luciferase activity in cells transfected with pGL3 (−1289/+63)-luc or pGL3 (−586/+63)-luc, but failed to do so in the cells transfected with pGL3 (−208/+63)-luc or pGL3 (−174/+63)-luc. Moreover, ATRA induced comparable luciferase activities between cells transfected with pGL3 (−1289/+63)-luc and with pGL3 (−586/+63)-luc (Fig. 5B). Thus, it appears that the RARE, DR-11, is more important than Pal-17 in mediating ATRA-induced TRAIL-R1 transactivation. In support of this finding, we further carried out a ChIP assay to test which RARE in the TRAIL-R1 gene binds to RARs in response to ATRA treatment. As shown in Fig. 5C, the ChIP assay detected a DNA fragment containing DR-11, but not a DNA fragment with Pal-17, from DNA precipitated with an anti-RAR antibody, indicating that RARs bind to DR-11, but not to Pal-17, in response to ATRA treatment. Taken together, it is clear that DR-11 is the functional RARE responsible for ATRA-induced TRAIL-R1 expression.
Fig. 5. Deletion analysis (A and B) and ChIP assay (C) demonstrate that DR-11 is the functional RARE responsible for ATRA-induced TRAIL-R1 transactivation.
A, Schematic illustration for generating deletion mutant reporter constructs carrying different lengths of TRAIL-R1 5′ flanking regions. B, H157 cells were co-transfected with the indicated reporter constructs and pCH110 plasmid for 24 h and then treated with DMSO or 1 μM ATRA for additional 16 h. The cells were harvested for luciferase assay. Columns, means of triplicate determinations; bars, ± SDs. C. H157 cells treated without and with 1 μM ATRA for 24 h and were subjected to ChIP assay for detection of RARE-containing DNA fragments as described in “Materials and Methods”.
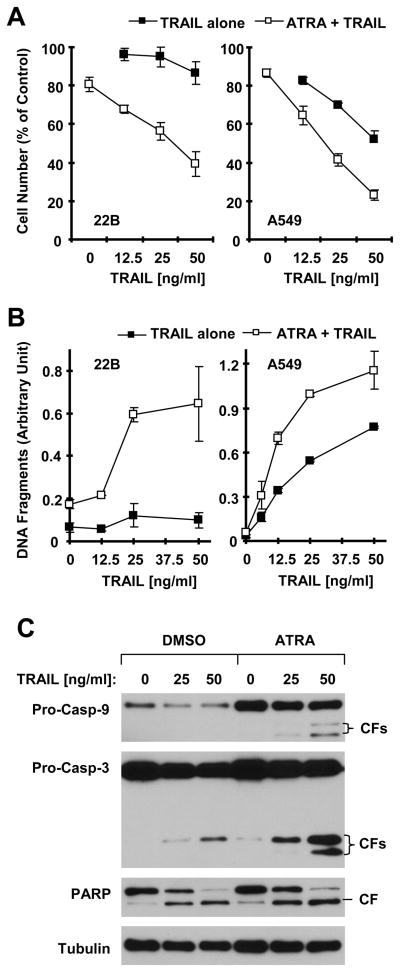
ATRA Enhances TRAIL-induced Apoptosis
It has been documented in many studies that upregulation of TRAIL death receptors can sensitize cancer cells to TRAIL-induced apoptosis (19, 20). Thus, we wanted to know whether ATRA augments TRAIL-mediated apoptosis. Indeed, ATRA and TRAIL combination was much more potent than each single agent in decreasing cell survival (Fig. 6A) and in increasing DNA fragmentation (Fig. 6B) in both 22B and A549 cells. Moreover, the combination of ATRA and TRAIL was also more effective than ATRA or TRAIL alone in inducing cleavage of caspase-3 and PARP (Fig. 6C). Thus, it is apparent that ATRA augments induction of apoptosis mediated by TRAIL.
Fig. 6. Combination of ATRA and TRAIL are more potent than ATRA or TRAIL alone in decreasing cell survival (A), and in increasing DNA fragmentation (B) and caspase cleavage (C).
A and B, The indicated cells in 96-well plates were treated with the given doses of TRAIL alone, 1 μM ATRA alone and their respective combinations. After 24 h, the cell numbers were estimated with the SRB assay (A) and DNA fragmentation was measured with the ELISA kit (B) described in “Materials and Methods”. Data are means ± SDs of four replicates (A) or triplicate determinations (B). C, 22B cells plated in 10-cm dishes were treated with the indicated doses of TRAIL alone, 1 μM ATRA alone and their combinations. After 24 h, the cells were harvested for preparation of whole cell protein lysates and subsequent Western blot analysis.
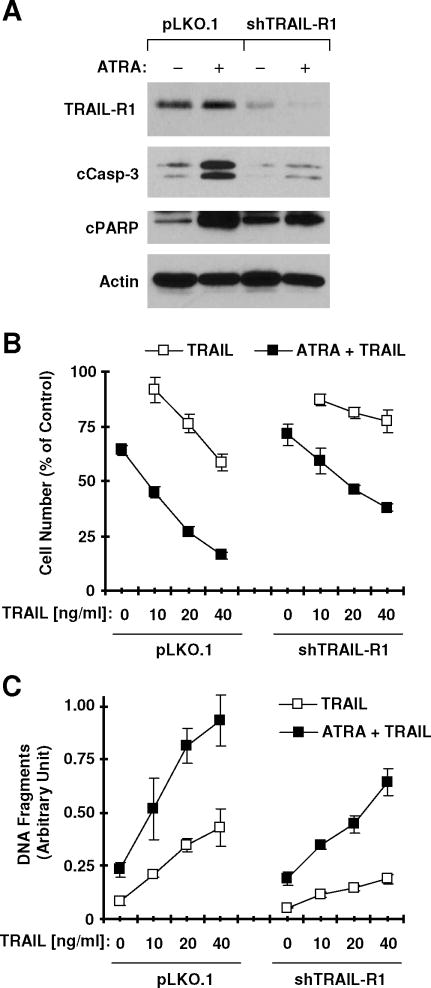
Involvement of TRAIL-R1 Upregulation in ATRA-induced Apoptosis and Enhancement of TRAIL-induced Apoptosis
Finally, we determined whether TRAIL-R1 induction plays a role in ATRA-induced apoptosis and enhancement of TRAIL-induced apoptosis. Hence, we knocked down TRAIL-R1 expression using TRAIL-R1 shRNA (shTRAIL-R1) and then examined its effects on ATRA-induced apoptosis and on ATRA/TRAIL-induced apoptosis. As presented in Fig. 7A, infection of cells with lentivirus carrying shTRAIL-R1 not only decreased basal levels of TRAIL-R1 but also blocked ATRA-induced TRAIL-R1 expression when compared to cells infected with control vector viruses (pLKO1), indicating a successful knockdown of TRAIL-R1. After prolonged treatment with ATRA, we detected increased levels of cleaved forms of caspase-3 and PARP in pLKO1-infected cells, but much less cleaved forms of caspase-3 and PARP in TRAIL-R1-silenced cells (compared to pLKO1 control cells) (Fig. 7A). This result suggests that TRAIL-R1 upregulation contributes to ATRA-induced apoptosis. Moreover, we examined the impact of TRAIL-R1 knockdown on induction of apoptosis by ATRA and TRAIL combination. As presented in Figs. 7B and 7C, combination of ATRA and TRAIL were still more effective than either agent alone in decreasing cell survival and in increasing DNA fragmentation in shTRAIL-R1-infected cells; however these effects in shTRAIL-R1 cells were substantially attenuated in comparison with their effects in pLKO1 control cells. These data suggest that TRAIL-R1 contributes to ATRA/TRAIL-induced apoptosis, but other mechanisms are involved in this process as well.
Fig. 7. shRNA-mediated knockdown of TRAIL-R1 attenuates ATRA-induced caspase activation (A) and enhancement of TRAIL-induced apoptosis (B and C).
22B cells were infected with lentiviruses carrying shTRAIL-R1 twice in a 24 h interval. Twenty four hours after the second infection, the cells were re-seeded in 6-well plates or 96-well plates. On the second day, the cells in 6-well plates were treated with 1 μM ATRA for 48 h and then harvested for Western blot analysis of the given proteins (A). The cells in 96-well plates were pretreated with 1 uM ATRA for 36 h, co-treated with TRAIL for additional 19 h and then used for the estimation of cell numbers (B) and for the measurement of DNA fragments (C). Data are means ± SDs of four replicates (B) or triplicate determinations (C)
DISCUSSION
Given the critical role of RA/RAR signaling in regulation of the growth of normal and malignant cells, understanding the underlying mechanisms, particularly by identifying RA/RAR-regulated genes has been a major focus of investigations (21). The previous discovery of TRAIL induction by RA and its contribution to RA-induced apoptosis via a paracrine mechanism (9) had facilitated our understanding of the mechanisms underlying RA-induced apoptosis. In the current study, we could detect increased TRAIL expression in our cell systems exposed to ATRA (Fig. 2E). We also detected cleavage of caspase-8 in cells exposed to ATRA (data not shown), suggesting that ATRA activates the extrinsic or death receptor-mediated apoptotic pathway. The novel finding in this study is that ATRA induces TRAIL-R1 expression as we demonstrated in a panel of NSCLC and HNSCC cell lines (Figs. 2A and S1). Although ATRA is in general a weaker inducer of apoptosis in solid tumor cells, we indeed detected apoptosis in cells after prolonged exposure to ATRA as supported by appearance of cleaved forms of caspase-3 and PARP (Fig. 7). shRNA-mediated blockage of TRAIL-R1 upregulation by ATRA did attenuate the ability of ATRA to induce apoptosis as evidenced by inhibition of caspase-3 and PARP cleavage in shTRAIL-R1 cells (Fig. 7A). These data collectively suggest that ATRA induces TRAIL-R1 expression that contributes to ATRA-induced apoptosis. Given that TRAIL-R1 is a known TRAIL receptor, it is plausible to speculate that TRAIL-R1 induction is involved in TRAIL-dependent paracrine model of RA-induced apoptosis as suggested previously (9). Thus, our current finding provides complementary evidence supporting the involvement of TRAIL/TRAIL-R1 signaling in RA-induced apoptosis.
In addition to protein change, ATRA increased TRAIL-R1 mRNA levels. Moreover, ATRA-induced TRAIL-R1 expression at both the protein and mRNA levels was abolished by the transcriptional inhibitor Act D (Fig. 3). Thus, it is apparent that ATRA increases TRAIL-R1 levels at the transcriptional level. We noted that the fold changes of TRAIL-R1 induction at the mRNA levels were generally more modest than the changes at the protein level. Whether this discrepancy is caused by differential stabilities of TRAIL-R1 mRNA and protein or suggests additional regulatory mechanism at the protein level is unclear.
ATRA typically regulates expression of its target genes through RAR-dependent transactivation via RAREs in their promoter regions. The identification of two putative RAREs in the 5′ flanking region of TRAIL-R1 gene led us to speculate that TRAIL-R1 should be a RAR target gene. Indeed, other synthetic retinoids or RAR agonists with different RAR selectivity increased TRAIL-R1 expression as ATRA did (Fig. 4A). Besides, the presence of RAR antagonists, LE135 or Ro415253, not only reduced basal levels of TRAIL-R1 protein expression, but also abolished ATRA-induced TRAIL-R1 expression (Figs. 4B and 4C). In agreement, knockdown of RARs with siRNA also reduced basal levels of TRAIL-R1 and blocked TRAIL-R1 induction by ATRA (Fig. 4D). Hence, it is apparent that ATRA induces TRAIL-R1 through a RAR-dependent mechanism. We noted that both RAR antagonists and siRNAs substantially reduced basal levels of TRAIL-R1 expression. It is likely that the presence of endogenous RA in cells is involved in maintaining basal levels of TRAIL-R1 expression. RAR antagonists or siRNAs block this effect, leading to downregulation of basal levels of TRAIL-R1.
The two putative RAREs in the 5′ flanking region of TRAIL-R1 are not typical RARE. Deletion experiments showed that the deletion of the putative RARE, Pal-17, did not impair ATRA’s ability to increase TRAIL-R1 promoter activity; however, further deletion of the putative RARE, DR-11, abolished ATRA-induced TRAIL-R1 promoter activity. Moreover, the ChIP assay showed that RAR antibody could pull down DNA fragment containing the DR-11 RARE, but not the DNA fragment harboring the Pal-17 RARE from cells exposed to ATRA, indicating that ATRA treatment induces binding of RARs to DR-11, but not Pal-17. Collectively, these results clearly demonstrate that DR-11 is a functional RARE in the TRAIL-R1 gene that mediates TRAIL-R1 transactivation upon ATRA treatment. Taken all the data discussed above together, we conclude that TRAIL-R1 is a novel RAR target gene.
TRAIL-R1 is known to be a death receptor for the death ligand, TRAIL (22). Upregulation of TRAIL death receptors often results in sensitization of cancer cells to TRAIL-induced apoptosis (19, 20). Thus, it is reasonable to assume that ATRA can sensitize cancer cells to TRAIL-induced apoptosis since it upregulates TRAIL-R1 expression. Indeed, the combination of ATRA and TRAIL was much more potent than either agent alone in decreasing cell survival and in inducing apoptosis in the tested cancer cell lines (Fig. 6), indicating that ATRA does sensitize cancer cells to TRAIL-induced apoptosis. Moreover, we found that blockage of ATRA-induced TRAIL-R1 expression through shRNA-mediated gene silencing substantially attenuated apoptosis induced by the combination of ATRA and TRAIL (Fig. 7), indicating that TRAIL-R1 upregulation contributes to enhancement of TRAIL-induced apoptosis by ATRA. We noted that blockage of TRAIL-R1 induction failed to abolish ATRA’s ability to enhance TRAIL-induced apoptosis although it attenuated this process (Fig. 7). Thus, the data argue that other mechanisms might also be involved in mediating the enhancement of TRAIL-induced apoptosis by ATRA. Given that ATRA is an approved drug for treatment of patients with acute promyelocytic leukemia (1) and that TRAIL and TRAIL-R1 agonistic antibodies are being tested in clinical trials (7, 8), our findings warrant further study to look at whether ATRA can enhance TRAIL- or TRAIL-R1 agonistic antibody-based cancer therapy in vivo and even in the clinic.
In summary, the current study has revealed for the first time that TRAIL-R1 is a RAR target gene. Accordingly, ATRA and other RAR agonists regulate its expression. TRAIL-R1 upregulation contributes to ATRA-induced apoptosis and enhancement of TRAIL-induced apoptosis, thus highlighting a novel mechanism by which ATRA or retinoids modulate apoptosis.
Supplementary Material
Acknowledgments
Grant Support: Georgia Cancer Coalition Distinguished Cancer Scholar award (to S-Y. Sun), Department of Defense grant W81XWH-04-1-0142-VITAL (to S-Y. Sun for Project 4) and NIH/NCI SPORE P50 grant CA128613 (to S-Y. Sun for Project 2).
We thank Drs. R. Lotan and Z. Chen for providing cell lines.
Footnotes
Note: S. S. Ramalingam, K. R. Khuri and S-Y. Sun are the Georgia Cancer Coalition Distinguished Cancer Scholars.
References
- 1.Sun SY, Lotan R. Retinoids and their receptors in cancer development and chemoprevention. Crit Rev Oncol Hematol. 2002;41:41–55. doi: 10.1016/s1040-8428(01)00144-5. [DOI] [PubMed] [Google Scholar]
- 2.Altucci L, Leibowitz MD, Ogilvie KM, de Lera AR, Gronemeyer H. RAR and RXR modulation in cancer and metabolic disease. Nat Rev Drug Discov. 2007;6:793–810. doi: 10.1038/nrd2397. [DOI] [PubMed] [Google Scholar]
- 3.Noy N. Between death and survival: retinoic acid in regulation of apoptosis. Annu Rev Nutr. 2010;30:201–17. doi: 10.1146/annurev.nutr.28.061807.155509. [DOI] [PubMed] [Google Scholar]
- 4.Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 5.Chambon P. A decade of molecular biology of retinoic acid receptors. Faseb J. 1996;10:940–54. [PubMed] [Google Scholar]
- 6.Newsom-Davis T, Prieske S, Walczak H. Is TRAIL the holy grail of cancer therapy? Apoptosis. 2009;14:607–23. doi: 10.1007/s10495-009-0321-2. [DOI] [PubMed] [Google Scholar]
- 7.Falschlehner C, Ganten TM, Koschny R, Schaefer U, Walczak H. TRAIL and other TRAIL receptor agonists as novel cancer therapeutics. Adv Exp Med Biol. 2009;647:195–206. doi: 10.1007/978-0-387-89520-8_14. [DOI] [PubMed] [Google Scholar]
- 8.Bellail AC, Qi L, Mulligan P, Chhabra V, Hao C. TRAIL agonists on clinical trials for cancer therapy: the promises and the challenges. Rev Recent Clin Trials. 2009;4:34–41. doi: 10.2174/157488709787047530. [DOI] [PubMed] [Google Scholar]
- 9.Altucci L, Rossin A, Raffelsberger W, Reitmair A, Chomienne C, Gronemeyer H. Retinoic acid-induced apoptosis in leukemia cells is mediated by paracrine action of tumor-selective death ligand TRAIL. Nat Med. 2001;7:680–6. doi: 10.1038/89050. [DOI] [PubMed] [Google Scholar]
- 10.Clarke N, Jimenez-Lara AM, Voltz E, Gronemeyer H. Tumor suppressor IRF-1 mediates retinoid and interferon anticancer signaling to death ligand TRAIL. Embo J. 2004;23:3051–60. doi: 10.1038/sj.emboj.7600302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun SY, Yue P, Dawson MI, Shroot B, Michel S, Lamph WW, et al. Differential effects of synthetic nuclear retinoid receptor-selective retinoids on the growth of human non-small cell lung carcinoma cells. Cancer Res. 1997;57:4931–9. [PubMed] [Google Scholar]
- 12.Sun SY, Yue P, Mao L, Dawson MI, Shroot B, Lamph WW, et al. Identification of receptor-selective retinoids that are potent inhibitors of the growth of human head and neck squamous cell carcinoma cells. Clin Cancer Res. 2000;6:1563–73. [PubMed] [Google Scholar]
- 13.Liu X, Yue P, Zhou Z, Khuri FR, Sun SY. Death receptor regulation and celecoxib-induced apoptosis in human lung cancer cells. J Natl Cancer Inst. 2004;96:1769–80. doi: 10.1093/jnci/djh322. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Yue P, Chen S, Hu L, Lonial S, Khuri FR, et al. The proteasome inhibitor PS-341 (bortezomib) up-regulates DR5 expression leading to induction of apoptosis and enhancement of TRAIL-induced apoptosis despite up-regulation of c-FLIP and survivin expression in human NSCLC cells. Cancer Res. 2007;67:4981–8. doi: 10.1158/0008-5472.CAN-06-4274. [DOI] [PubMed] [Google Scholar]
- 15.Guan B, Yue P, Lotan R, Sun SY. Evidence that the human death receptor 4 is regulated by activator protein 1. Oncogene. 2002;21:3121–9. doi: 10.1038/sj.onc.1205430. [DOI] [PubMed] [Google Scholar]
- 16.Pfahl M, Tzukerman M, Zhang XK, Lehmann JM, Hermann T, Wills KN, et al. Nuclear retinoic acid receptors: cloning, analysis, and function. Methods Enzymol. 1990;189:256–70. doi: 10.1016/0076-6879(90)89297-u. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka T, Suh KS, Lo AM, De Luca LM. p21WAF1/CIP1 is a common transcriptional target of retinoid receptors: pleiotropic regulatory mechanism through retinoic acid receptor (RAR)/retinoid X receptor (RXR) heterodimer and RXR/RXR homodimer. J Biol Chem. 2007;282:29987–97. doi: 10.1074/jbc.M701700200. [DOI] [PubMed] [Google Scholar]
- 18.Stromskaya TP, Rybalkina EY, Zabotina TN, Shishkin AA, Stavrovskaya AA. Influence of RARalpha gene on MDR1 expression and P-glycoprotein function in human leukemic cells. Cancer Cell Int. 2005;5:15. doi: 10.1186/1475-2867-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun SY. Chemopreventive agent-induced modulation of death receptors. Apoptosis. 2005;10:1203–10. doi: 10.1007/s10495-005-2274-4. [DOI] [PubMed] [Google Scholar]
- 20.Elrod HA, Sun SY. Modulation of death receptors by cancer therapeutic agents. Cancer Biol Ther. 2007;7:163–73. doi: 10.4161/cbt.7.2.5335. [DOI] [PubMed] [Google Scholar]
- 21.Sun SY. How much do we know about retinoid-regulated genes? Cancer Biol Ther. 2002;1:28–30. doi: 10.4161/cbt.1.1.36. [DOI] [PubMed] [Google Scholar]
- 22.Ammirante M, Di Giacomo R, De Martino L, Rosati A, Festa M, Gentilella A, et al. 1-Methoxy-canthin-6-one induces c-Jun NH2-terminal kinase-dependent apoptosis and synergizes with tumor necrosis factor-related apoptosis-inducing ligand activity in human neoplastic cells of hematopoietic or endodermal origin. Cancer Res. 2006;66:4385–93. doi: 10.1158/0008-5472.CAN-05-3895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.