Abstract
Recent studies have established SIRT1 as an important regulator of lipid metabolism, although the mechanism of its action at the molecular level has not been revealed. Here, we show that knockdown of SIRT1 with the help of small hairpin RNA decreases basal and isoproterenol-stimulated lipolysis in cultured adipocytes. This effect is attributed, at least in part, to the suppression of the rate-limiting lipolytic enzyme, adipose triglyceride lipase (ATGL), at the level of transcription. Mechanistically, SIRT1 controls acetylation status and functional activity of FoxO1 that directly binds to the ATGL promoter and regulates ATGL gene transcription. We have also found that depletion of SIRT1 decreases AMP-dependent protein kinase (AMPK) activity in adipocytes. To determine the input of AMPK in regulation of lipolysis, we have established a stable adipose cell line that expresses a dominant-negative α1 catalytic subunit of AMPK under the control of the inducible TET-OFF lentiviral expression vector. Reduction of AMPK activity does not have a significant effect on the rates of lipolysis in this cell model. We conclude, therefore, that SIRT1 controls ATGL transcription primarily by deacetylating FoxO1.
Keywords: adipose triglyceride lipase, adipose tissue; dyslipidemias; fatty acid/metabolism
In the mammalian organism, most energy is stored in adipose tissue in the form of triglycerides (TGs). Upon TG hydrolysis, free fatty acids (FFAs) are delivered by blood to starving cells and tissues. Meanwhile, it has long been known that elevated levels of circulating FFAs cause insulin resistance and diabetes mellitus (1–3) via mechanisms that are currently under intense investigation [as reviewed in (4–6)]. Clearly, the fine balance between healthy and unhealthy levels of circulating FFAs requires the tight control of lipolysis.
It is now appreciated that lipolysis is a complicated multi-step process. The complete hydrolysis of TG to glycerol and FFA is performed jointly by tri- di- and monoacylglyceride lipases (7–9). The recently discovered enzyme adipose triglyceride lipase, or ATGL (a.k.a. desnutrin, PNPLA2, TTS2.2, iPLA2ζ) (10–12) is responsible for the bulk of triacylglycerol hydrolase activity in various cells but has low affinity to di- and monoacylglycerides (7, 8). The major diacylglyceride lipase in adipocytes is hormone-sensitive lipase, or HSL. Monoacylglyceride products of HSL are hydrolyzed by monoacylglyceride lipase (7, 8).
According to current views, lipolysis is regulated primarily at the posttranslational level, with the cAMP/cGMP-mediated signaling pathways playing the key role in this process. Briefly, phosphorylation of the peripheral lipid droplet protein, perilipin, and HSL by cAMP-dependent protein kinase and/or cGMP-dependent protein kinase leads to the recruitment of HSL to the lipid droplet and activation of the enzyme. At the same time, a protein cofactor of ATGL, CGI-58 dissociates from phosphorylated perilipin and activates ATGL (9). Jointly, both processes rapidly and significantly stimulate lipolysis.
It has also been established that the rates of lipolysis are directly proportional to the levels of the ATGL protein, which is therefore considered the rate-limiting lipolytic enzyme. Basically, in every experimental model tested thus far, elevated ATGL expression increases while attenuated ATGL expression decreases both basal and isoproterenol-stimulated lipolysis (10–21). In particular, activation of lipolysis by fasting may be mediated by upregulation of ATGL expression (11, 17, 22, 23). Thus, not only posttranslational regulation of the enzymatic activity but also tight control of ATGL expression is necessary for normal lipolysis and FFA homeostasis. However, unlike the posttranslational regulation of lipolysis that has been studied in much detail (as reviewed in Refs. 7–9), relatively little is known about regulation of ATGL expression.
Mammalian sirtuins represent a group of seven proteins (SIRT1 through SIRT7) with NAD+-dependent deacetylase activity that controls various aspects of cell growth, division, and metabolism by deacetylating histones, transcription factors, corepressors, coactivators, cytoplasmic enzymes, and mitochondrial proteins (24, 25). In particular, SIRT1 plays an important role in the regulation of lipid metabolism both in vitro and in vivo. In cultured adipocytes, SIRT1 inhibits adipogenesis and accumulation of triglycerides (26). Interestingly, SIRT2 has an analogous activity in 3T3-L1 adipocytes (27, 28), suggesting some degree of redundancy in SIRT1 and SIRT2 action. In vivo, pharmacological activation (29) or overexpression (30) of SIRT1 inhibits diet-induced accumulation of fat and, in fact, mimics caloric restriction (31). In contrast, heterozygous SIRT1+/− animals show diminished release of FFA upon fasting (26). These data suggest that SIRT1 controls fat storage and mobilization, at least in part by regulating lipolysis. Therefore, in this study, we decided to explore the effect of SIRT1 on lipolysis and its potential mechanism(s). Our results demonstrate that SIRT1 regulates the expression of the rate-limiting lipolytic enzyme, ATGL, and hence lipolysis in cultured adipocytes. We also show that deacetylation of FoxO1 is essential for this process.
MATERIALS AND METHODS
Antibodies
Polyclonal antibodies against ATGL, SIRT1, perilipin, acetylated lysine, peroxisome proliferator-activated receptor γ (PPARγ), AMPK, and all the phospho-specific antibodies were from Cell Signaling (Beverly, MA). Polyclonal antibody against FoxO1 and 4E-BP1 was from Santa Cruz Biotechnology (Santa Cruz, CA). Monoclonal antibody against adiponectin was from Abcam (Cambridge, MA). Monoclonal anti-Flag tag antibody and monoclonal anti-β-actin antibody were from Sigma (St. Louis, MO). Polyclonal antibody against HSL was a gift from Dr. A. Greenberg (Tufts University). A rabbit polyclonal antibody against cellugyrin was described previously (32). Monoclonal anti-V5 tag antibody was from Invitrogen.
Cell culture
Stable 3T3-L1 cell line with decreased expression of SIRT1 was prepared using pSUPER-SIRT1 shRNA as described previously (33). Cells stably infected with the pSUPER empty vector (EV) were used for control experiments. 3T3-L1 cells stably expressing Flag-epitope-tagged wild-type SIRT1 or an acetylase-deficient dominant-negative mutant SIRT1 (H363Y) were prepared by retroviral infection. Briefly, HEK-293T cells were grown to 70% confluence in 100 mm-diameter dishes, at which stage they were transfected with a DNA-FUGENE cocktail consisting of 36 μl Fugene 6, 6 μg retrovirus plasmid pBABE-SIRT1, 6 μg pVPack-VSV-G vector, 6 μg pVPack-GAG-POL vector, and 164 μl DMEM without FBS. After 24 h, the medium was replaced with 6 ml fresh DMEM containing 10% FBS. On the following day, the culture medium containing high-titer retrovirus was harvested and filtered through a 0.45 μm filter. The viral filtrate was used to infect 3T3-L1 preadipocytes.
3T3-F442a AMPK DNα1 cells that express the dominant-negative mutant of human AMPK α1 catalytic subunit (D157A) were established using a TET-OFF lentiviral-inducible expression system. The cDNA for AMPKα1 (D157A) was kindly provided by Dr. Carling, and a Flag epitope was added to its N terminus by PCR. 3T3-F442a preadipocytes were first infected with the lentivirus that expresses tetracycline transactivator (tTA) and were selected with puromycin (2 µg/ml). The TET preadipocytes were then infected with the lentivirus containing a tTA-responsive vector with Flag-tagged AMPKα1 (D157A), and the cells were selected with blasticidin (2 µg/ml) to establish the stable cell line.
Mouse embryonic fibroblasts (MEFs) stably expressing V5-tagged human FoxO1 were established by lentiviral infection and blasticidine selection as described previously (21).
3T3-L1 preadipocytes were cultured, differentiated, and maintained as described previously (34). HEK 293T cells and MEFs were cultured in DMEM supplemented with 10% FBS in 2 mM l-glutamine, 100 U/ml penicillin, and 100 µg/ml streptomycin.
Transient transfections and reporter gene assays
Transient transfections with cDNA were performed using Lipofectamine 2000 (Invitrogen Life Technologies; Grand Island, NY) according to the manufacturer's instructions. Briefly, ∼80% confluent HEK293T cells and ∼50% confluent MEFs were transfected with 500 ng of luciferase and 100 ng of eGFP cDNA in a 6-well plate format. HEK293T cells were cotransfected with 500 ng of FoxO1 and SIRT1 cDNA, and MEFs were cotransfected with 1 µg of SIRT1 cDNA. All experiments were performed in triplicate. After 48 h of transfection, cells were harvested in Reporter Lysis Buffer (Promega). Luciferase activity was determined in whole-cell lysates using the Promega luciferase assay kit and expressed as relative light units. Expression of eGFP was measured fluorometrically. Firefly luciferase was normalized by eGFP fluorescence to correct for transfection efficiency.
RNA interference with siRNA
siRNA against the sequence TGAGGGAGTTAGAAGGTTCTTCATG of mouse PPARγ and scrambled siRNA were obtained from Integrated DNA Technologies (Coralville, IA). Transfection of siRNAs was performed with the help of the DeliverX Plus delivery kit (Panomics) according to the instructions of the manufacturer.
RNA extraction and quantitative PCR
Total RNA was extracted from differentiated 3T3-L1 cells using TRIzol reagent (Invitrogen). Reverse transcription of 500 ng total RNA was performed using random decamers (RETROscript kit; Ambion, Austin, TX), and the gene expression was determined by quantitative PCR (MX4000 Multiplex qPCR system; Stratagene, La Jolla, CA). Reactions were performed in triplicate in the total volume of 25 µl containing 2.5 µl 1:10-diluted cDNA, 1× SYBR green master mix (Brilliant II SYBR Green qPCR Master Mix; Stratagene), and gene-specific primers. Gene expression was normalized by GAPDH or 36B4 expression by the ΔΔCt method. DNase-treated samples and no-template controls were analyzed in parallel experiments to confirm specificity. Primer sequences are available upon request.
Lipolysis assay
Differentiated 3T3-L1 adipocytes were incubated in Phenol Red-free DMEM with 2% FA-free BSA for 1 h at 37°C in the presence or absence of 10 μM isoproterenol. Glycerol content in the media was measured colorimetrically at 540 nm using the Triglyceride (GPO) Reagent Set (Pointe Scientific; Canton, MI) against a set of glycerol standards. Nonesterified fatty acids (NEFAs) were measured colorimetrically at 560 nm using the HR Series NEFA-HR (2) kit (Wako Chemicals; Richmond, VA) against a set of palmitate standards. Cells were then washed with cold PBS and lysed in 1% Triton X-100 buffer, and the protein concentration was determined and used to normalize glycerol release. All experiments were carried out in triplicate.
Immunoprecipitation
Precleared cell lysates (500 µg/sample) were incubated with protein A-agarose beads (30 µl/sample) and primary antibody or IgG (2 µg/sample) overnight at 4°C with constant rotation. The lysates were then centrifuged at 4,000 g for 1 min, the pelleted beads were then washed three times with lysis buffer, and immunoprecipitated proteins were eluted from the beads by adding 2× sample loading buffer (40 µl/sample). Beads with immunoprecipitated proteins were incubated at 95°C for 5 min. After a brief centrifugation, the supernatants (immunoprecipitation samples) were analyzed by Western blotting.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) studies were carried out in 3T3-L1 adipocytes using the EZ-chIP kit (Millipore; Bedford, MA) according to the manufacturer's instructions. Briefly, proteins were cross-linked to DNA by 18.5% formaldehyde, lysed in SDS lysis buffer, and sonicated seven times for 15 s. FoxO1 proteins were then immunoprecipitated from the precleared lysates. Protein–DNA complexes were eluted, and cross-links were reversed. Purified DNA was subjected to PCR using the following primers: 5′-ATCTTTAAAAGGCAATTAAGCTG-3′ and 5′-TAAGTCCAGGTCTTAGAAATGT-3′. Purified DNA was also analyzed by quantitative PCR using SYBR Green reaction (Brilliant II SYBR Green qPCR Master Mix; Stratagene). For all PCR reactions, 10% input was used.
Gel electrophoresis and Western blotting
Proteins were separated in SDS-polyacrylamide gels and transferred to Immobilon-P membranes (Millipore) in 25 mM Tris, 192 mM glycine. Following transfer, the membrane was blocked with 10% nonfat milk in PBS with 0.5% Tween-20 for 2 h. Blots were probed overnight with specific primary antibodies at 4°C followed by 1 h incubation at room temperature with HRP-conjugated secondary antibodies (Sigma). Protein bands were detected with the enhanced chemiluminescence substrate kit (PerkinElmer Life Sciences; Boston, MA) using a Kodak Image Station 440CF (Eastman Kodak; Rochester, NY).
Statistics
Student's paired two-tailed t-test was used to evaluate the statistical significance of the results.
RESULTS AND DISCUSSION
Knockdown of SIRT1 decreases lipolysis and ATGL expression in adipocytes
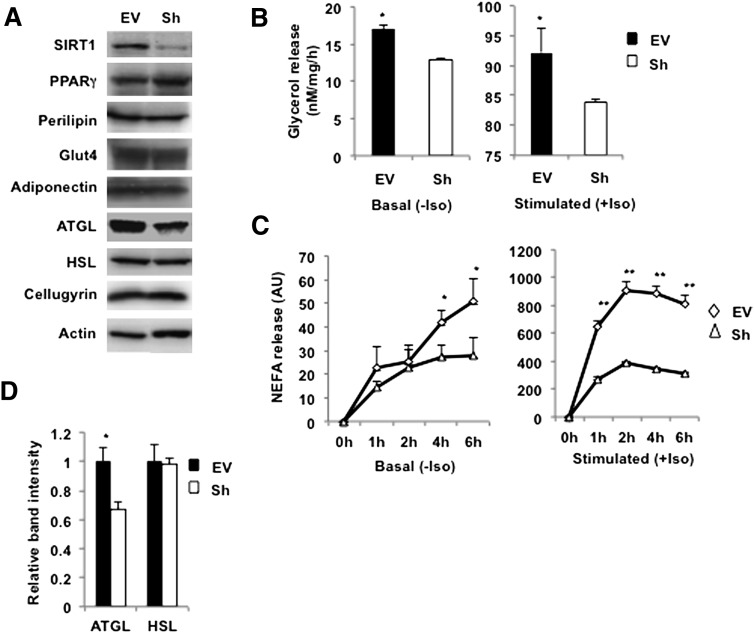
To determine the role of SIRT1 in lipolysis, its expression was significantly attenuated by constitutive production of a SIRT1-shRNA in 3T3-L1 adipocytes employing a retroviral vector as described previously (33). It has been reported that overexpression of SIRT1 attenuates differentiation of 3T3-L1 adipocytes (26). In agreement with this study, we have found that knockdown of SIRT1 somewhat accelerates differentiation, so that the cells acquire fat droplets 1–1.5 days earlier than control empty vector-infected cells (not shown). Nonetheless, by day 8–10 of differentiation, control cells “catch up” with SIRT1-depleted cells, as evidenced by equal levels of expression of adipogenic markers such as PPARγ, perilipin, Glut4, and adiponectin (Fig. 1A) (33).
Fig. 1.
Knockdown of SIRT1 decreases lipolysis and ATGL expression in 3T3-L1 adipocytes. A: Control (EV) and SIRT1-ablated (Sh) 3T3-L1 adipocytes were homogenized on day 10 of differentiation, and total lysates (50 µg) were analyzed by Western blotting for the indicated proteins. Cellugyrin and actin were used as loading controls. B: Empty vector (EV)-infected and stably depleted SIRT1 (Sh) 3T3-L1 adipocytes on day 10 of differentiation were incubated in Phenol Red-free DMEM with 2% FA BSA without (basal) or with (stimulated) 10 μM isoproterenol (Iso) for 1 h. Glycerol was measured in media aliquots in triplicate and normalized by protein concentration in whole-cell lysates. A representative result of three independent experiments is shown. Data are presented as nM glycerol/mg of protein/h and expressed as mean ± SD; * P < 0.05. C: Release of NEFA was measured in triplicate samples from the EV and Sh cells for the indicated time periods and normalized by protein concentration in whole-cell lysates. Data are expressed as mean ± SD; *P < 0.05; ** P < 0.001. D: Relative band intensities for ATGL and HSL in EV (black bars) and Sh (white bars) cells were determined in three independent experiments and normalized by cellugyrin. Data are presented relative to nonstimulated EV cells and are expressed as mean ± SD. * P < 0.05.
In agreement with results obtained in vivo (26), we have found that knockdown of SIRT1 decreases rates of basal and isoproterenol-stimulated lipolysis in cultured adipocytes (Fig. 1B,C). To get insight into this effect, we have measured the levels of expression of HSL and ATGL. It turns out that knockdown of SIRT1 does not affect HSL, whereas expression of ATGL is significantly diminished (Fig. 1A, D). This result is consistent with the recent report of Shan et al. (35), who have determined that knockdown of Sirt1 in primary porcine adipocytes decreases ATGL mRNA. Given the large body of literature showing that the rates of lipolysis tightly correlate with ATGL levels, we suggest that SIRT1 controls lipolysis by regulating expression of ATGL. In the following experiments, we decided to explore the potential mechanisms of this effect.
The role of PPARγ in SIRT1-regulated expression of ATGL
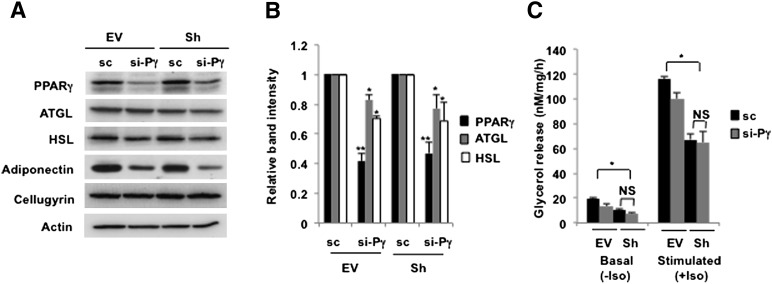
One possible connection between SIRT1 and ATGL may be PPARγ, which has been shown to stimulate ATGL expression (22, 36–38) and lipolysis (37). It was reported, however, that SIRT1 represses PPARγ by docking with its corepressors, NCoR and SMRT (26, 33), and the inhibitory effect of SIRT1 on PPARγ activity in vitro may reach 2-fold (26). Thus, we decreased levels of PPARγ in control and in SIRT1-ablated adipocytes to ∼50% using siRNA technology. As shown in Fig. 2A, this had a significant effect on the expression of the known PPARγ target, adiponectin (39). Partial knockdown of PPARγ also had a small negative effect on the expression of the two major lipases in adipocytes, ATGL and HSL, regardless of the levels of SIRT1 expression (Fig. 2A, B). Lipolysis in adipocytes with partial knockdown of PPARγ showed a tendency to decrease, although the numbers did not always reach statistical significance (Fig. 2C). These results confirm that PPARγ has a positive effect on ATGL expression. Thus, inhibition of PPARγ can explain the inhibitory role of SIRT1 in adipogenesis (26) but cannot account for stimulation of lipolysis in differentiated cells.
Fig. 2.
PPARγ upregulates ATGL independently of SIRT1 expression. A: Control (EV) and SIRT1-ablated (Sh) 3T3-L1 adipocytes were transfected with 30 nM of scrambled (sc) RNA or siRNA directed against mouse PPARγ (si-Pγ) on day 8 of differentiation. After 48 h, cells were homogenized and total lysates (50 µg) were analyzed by Western blotting. B: Relative band intensities for ATGL (gray bars), HSL (white bars), and PPARγ (black bars) were determined in three independent experiments and normalized by cellugyrin. C: Control (EV) and SIRT1-ablated (Sh) 3T3-L1 adipocytes were transfected with scrambled (sc, black bars) RNA or siRNA directed against mouse PPARγ (si-Pγ, gray bars). Glycerol was measured in media aliquots without (basal) or with (stimulated) 10 μM isoproterenol (Iso) for 2 h. A representative result of three independent experiments is shown. Data are presented as nM glycerol/mg of protein/h and expressed as mean ± SD. NS, not significant. * P < 0.05; ** P < 0.001.
Downregulation of AMPK activity does not play a major role in regulation of lipolysis by SIRT1
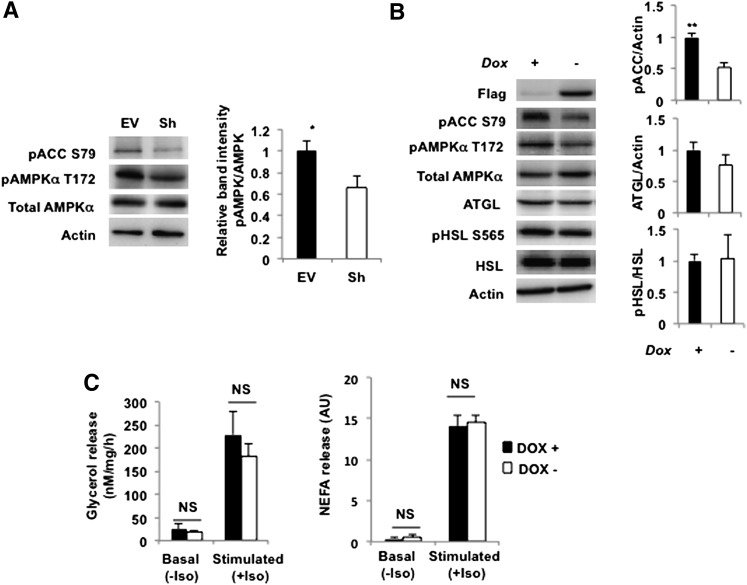
It has been shown previously that SIRT1 activates AMPK in HEK293T cells and hepatocytes, presumably by deacetylating and promoting the cytoplasmic localization of LKB1 (40–42). On the basis of these studies, we suggested that in SIRT1-depleted cells, the activity of AMPK would be decreased. Indeed, we have found that knockdown of SIRT1 decreases phosphorylation of AMPK and its substrate, acetyl-CoA carboxilase (Fig. 3A). Because AMPK may participate in the regulation of lipolysis in adipocytes (43), we decided to explore this connection further.
Fig. 3.
Decrease in AMPK activity in SIRT1-depleted adipocytes does not affect lipolysis. A: Left panels, control (EV) and SIRT1-ablated (Sh) 3T3-L1 adipocytes on day 10 of differentiation were serum starved for 2 h, and total cell lysates (50 µg) were analyzed by Western blotting. Actin was used as a loading control. A representative result of four independent experiments is shown. Right panel, relative band intensities for phosphorylated AMPK in EV (black bar) and Sh (white bar) cells were determined in four independent experiments and normalized by total AMPK. * P < 0.05. B: Left panels, 3T3-F442a cells stably expressing AMPK DNα1 under the control of tetracycline responsive element were differentiated in the presence of 2 µg/ml doxycycline (Dox+) for 6 days. Doxycycline was then withdrawn for 2 days (Dox−). On day 8, cells were serum starved for 2 h, and the total cell lysates were analyzed by Western blotting. A representative result of three independent experiments is shown. Right panels, relative band intensities for phosphorylated ACC, phosphorylated HSL and total ATGL in Dox+ (black bars) and Dox− (white bars) cells were determined in three independent experiments and normalized by actin, HSL, and actin, respectively. ** P < 0.001. C: Glycerol (left panel) and NEFA (right panel) released from 3T3-F442a cells stably expressing AMPK DNα1 incubated with (Dox+, black bars) or without (Dox−, white bars) doxycycline were measured in the media on day 8 of differentiation. For experimental details, see legend to Fig. 1. Data are presented relative to the nonstimulated Dox+ cells and are expressed as mean ± SD. NS, not significant. A representative result of three to five independent experiments is shown.
To this end, we established a line of cultured adipocytes that express the Flag-tagged dominant negative (DN) α1 catalytic subunit (D157A) of AMPK under the control of an inducible TET-OFF lentiviral expression system. As is shown in supplementary Fig. I, withdrawal of doxycycline from the cell medium leads to the induction of the Flag-tagged DN α subunit, increasing total AMPKα levels in the cell by ∼50–80%. Correspondingly, phosphorylation of AMPK and the AMPK substrate, ACC, is decreased by ∼50% (Fig. 3B). However, reduction of AMPK activity in our cell model has virtually no effect on either expression of ATGL and HSL or on phosphorylation of HSL on the critical residue, Ser565 (Fig. 3B). The rates of basal and isoproterenol-stimulated lipolysis also do not change significantly (Fig. 3C).
The role of AMPK in the regulation of lipolysis has been controversial, inasmuch as different groups have reported both lipolytic and anti-lipolytic activity of AMPK (43–48). The reasons for this controversy are not yet clear. Because AMPK may regulate various aspects of lipid metabolism and, in particular, biosynthesis and oxidation of FAs (43), its effect on lipolysis may be indirect and multifactorial. For example, Gaidhu et al. (46) have recently shown that activation of AMPK has antagonistic effects on HSL and ATGL. This mechanism should minimize the overall effect of AMPK on glycerol release from adipocytes. It is also clear that lipolysis affects AMPK activity as well (44), adding another level of complexity to the potential role of AMPK in lipolysis. In any case, and regardless of the potential mechanism or lack thereof, our results suggest that inhibition of AMPK activity by ∼50% may not represent a major factor in regulation of lipolysis in adipocytes.
The role of FoxO1 in SIRT1-regulated expression of ATGL
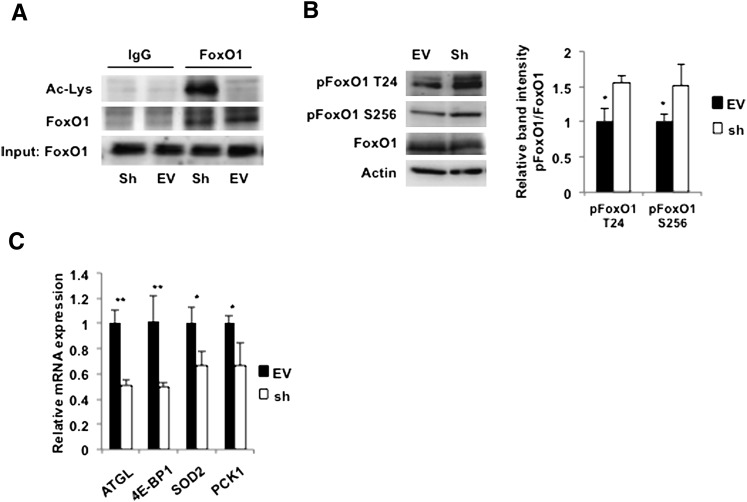
One of the well-known direct targets of SIRT1 is FoxO1, which is activated by deacetylation (49–52). Because we have recently demonstrated that FoxO1 directly stimulates expression of ATGL and lipolysis in adipocytes (21), we decided to test the hypothesis that SIRT1 controls lipolysis via FoxO1-mediated stimulation of ATGL expression. We immunoprecipitated FoxO1 from control and SIRT1-depleted cells and showed, by Western blotting with anti-acetylated lysine antibodies, that acetylation of FoxO1 is dramatically increased in the latter (Fig. 4A). Phosphorylation of FoxO1 on T24 and S253 is also increased in SIRT1-ablated cells (Fig. 4B). Phosphorylation of these sites inhibits the functional activity of FoxO1 (52); correspondingly, we have found that expression of ATGL mRNA as well as mRNAs for several other FoxO1 target genes, such as 4E-BP1, SOD2, and PCK1, are suppressed by knockdown of SIRT1 (Fig. 4C).
Fig. 4.
Knockdown of SIRT1 leads to acetylation and inhibition of FoxO1 in adipocytes. A: Control (EV) and SIRT1-ablated (Sh) 3T3-L1 adipocytes on day 10 of differentiation were serum starved for 4 h, and FoxO1 was immunoprecipitated from the whole-cell lysates (500 µg). Proteins were eluted from the beads, and acetylated FoxO1 was analyzed by Western blotting with an antibody against acetylated lysine (Ac-Lys). Input lane shows 50 µg of the total cell lysates. B: Left panel, control (EV) and SIRT1-ablated (Sh) 3T3-L1 adipocytes on day 10 of differentiation were serum starved for 2 h, and total cell lysates (50 µg) were analyzed by Western blotting with the indicated antibodies. Right panel, relative band intensities for phosphorylated FoxO1 in EV (black bar) and Sh (white bar) cells were determined in three independent experiments and normalized by total FoxO1. Data are expressed as mean ± SD relative to the EV cells. * P < 0.05. C: Levels of mRNA in control (EV, black bars) and SIRT1-ablated (Sh, white bars) adipocytes were determined in triplicate by quantitative PCR and normalized by GAPDH mRNA levels. Data are expressed as mean ± SD relative to the expression levels in EV cells. 4E-BP1, eIF4E binding protein 1; SOD2, superoxide dismutase 2; PCK1, phosphoenolpyruvate carboxykinase 1. * P < 0.05; ** P < 0.001.
To confirm the role of SIRT1 in regulating ATGL expression, we stably expressed wild-type SIRT1, as well as a deacetylase-defective dominant-negative mutant, each tagged with the Flag epitope in 3T3-L1 adipocytes (see supplementary Fig. IIA). Using quantitative PCR, we found that expression of ATGL mRNA is increased in response to overexpression of SIRT1 and decreased by the mutant protein (see supplementary Fig. IIB). In parallel, we determined that overexpression of wild-type SIRT1 decreases specific acetylation (see supplementary Fig. IIC) and negative phosphorylation (see supplementary Fig. IID) of FoxO1. In these experiments, however, we were unable to detect statistically significant changes in the ATGL protein (not shown). We attribute this to the fact that quantitative PCR is a more sensitive and accurate technique in comparison to semi-quantitative Western blotting.
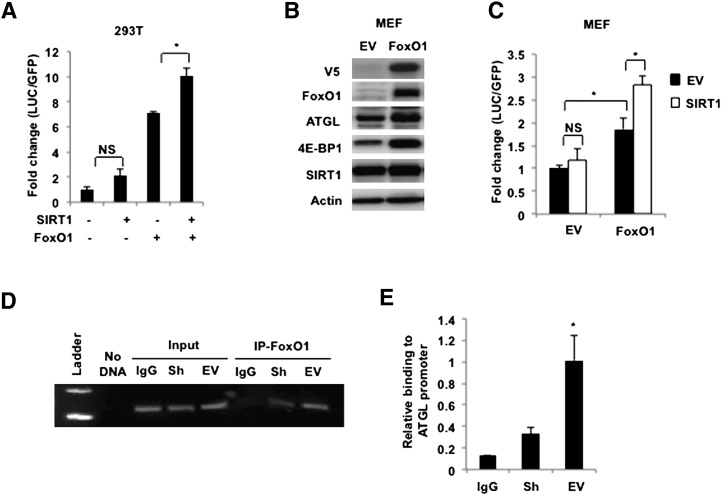
In any case, our results show that SIRT1 controls the acetylation status and functional activity of FoxO1, as well as ATGL expression in adipocytes. Given that FoxO1 binds to and regulates the activity of the ATGL promoter (21), we suggest that FoxO1 mediates the effect of SIRT1 on ATGL gene transcription. To confirm this regulatory connection, we carried out several experiments. First, we cotransfected HEK293T cells with a luciferase reporter construct containing the ATGL promoter (22) together with FoxO1 and SIRT1 cDNA in different combinations. In agreement with previously published results (21), FoxO1 increases expression of luciferase ∼7-fold, whereas cotransfection with SIRT1 increases the stimulatory effect of FoxO1 on the ATGL promoter even further (Fig. 5A). We have confirmed this result in mouse embryonic fibroblasts (MEFs) that do not express FoxO1 endogenously (Fig. 5B). In the absence of FoxO1, transfection of SIRT1 into wild-type MEFs has no effect on the activity of the ATGL promoter (Fig. 5C). Stable transfection of FoxO1 into MEFs (21) not only increases levels of endogenous ATGL (Fig. 5B) but also confers SIRT1 responsiveness to the ATGL promoter (Fig. 5C).
Fig. 5.
SIRT1 increases ATGL promoter activity and regulates binding of FoxO1 to the ATGL promoter. A: HEK 293T cells were transiently transfected with −2,979/+21 luciferase ATGL promoter construct together with enhanced green fluorescence protein. Cells were cotransfected with either empty vector, SIRT1 or FoxO1, or with both SIRT1 and FoxO1 cDNA. After 48 h, cells were washed three times in cold PBS and harvested in the reporter lysis buffer. Luciferase activity in cell lysates was assayed as described in MATERIALS AND METHODS and normalized by GFP fluorescence. Data are presented in triplicate as mean ± SD. * P < 0.05; NS, not significant. A representative result of three independent experiments is shown. B: Control (EV) and FoxO1-infected MEFs were homogenized, and total cell lysates (50 µg) were analyzed by Western blotting. Actin was used as loading control. C: Control (EV) and FoxO1-infected MEFs were transiently transfected with −2,979/+21 LUC ATGL promoter construct together with eGFP. Cells were cotransfected with either empty vector or SIRT1 cDNA. After 48 h, luciferase activity in cell lysates was assayed and normalized by GFP fluorescence. A representative result of two independent experiments is shown. Data are presented in triplicate as mean ± SD. * P < 0.05; NS, not significant. D, E: Chromatin immunoprecipitation assay was performed in control (EV) and SIRT1-ablated (Sh) 3T3-L1 adipocytes after 4 h serum withdrawal. Following cross-linking and sonication, genomic fragments were immunoprecipitated with antibody against FoxO1 or rabbit IgG and amplified by PCR (D) or by SYBR green reaction (E) as described in MATERIALS AND METHODS. Data are representative of three independent experiments and are expressed as mean ± SD relative to the promoter binding in EV cells. * P < 0.05.
In addition, we have performed ChIP analysis on control and SIRT1-depleted adipocytes and demonstrated that knockdown of SIRT1 decreases the interaction of endogenous FoxO1 with the endogenous ATGL promoter (Fig. 5D, E). Taken together, these results suggest that SIRT1 controls ATGL transcription by deacetylating FoxO1.
It may be intriguing to discuss this result in the context of lifespan regulation in mammals. At present, a significant body of evidence shows the key role of adipose tissue in regulating longevity in different organisms (53–56). Multiple experimental findings in both lower eukaryotes and mammals have uncovered the following major pathways of the extension of healthy life span: caloric restriction, inhibition of mTORC1 signaling, and the activation of FoxO1 and SIRT1 (57–60). The key question is whether these pathways have a common downstream effector in adipocytes that may be responsible for their effect on longevity. Here, we put forward a hypothesis that all of these pathways converge on the activation of ATGL expression and lipolysis.
The effect of food restriction on ATGL expression has been documented earlier (11, 17, 22, 23). Our previous work showed that both inhibition of mTORC1 (20) and activation of FoxO1 (21) also increase ATGL expression and lipolysis and may thus mimic the effect of calorie restriction. Here, we demonstrate that the effect of SIRT1 on fat mobilization (26, 31) may be attributed, at least in part, to the deacetylation of FoxO1 and a corresponding increase in ATGL expression. Thus, all pathways presently known to extend life activate ATGL expression and lipolysis in adipocytes and reduce TG stores. Given that depletion of fat reserves by lipolysis may represent an evolutionarily conserved longevity signal (61, 62), we speculate that ATGL may represent the “life-extending lipase” postulated by Cynthia Kenyon (57) in a recent review.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. Carling, Greenberg, and Smas for their generous gifts of reagents.
Footnotes
Abbreviations:
- AMPK
- AMP-dependent protein kinase
- ATGL
- adipose triglyceride lipase
- ChIP
- chromatin immunoprecipitation
- DN
- dominant negative
- EV
- empty vector
- HSL
- hormone-sensitive lipase
- MEF
- mouse embryonic fibroblast
- NEFA
- nonesterified fatty acid
- TG
- triglyceride
- tTA
- tetracycline transactivator
This work was supported by research Grants DK-52057 and DK-56736 from the National Institutes of Health (K.V.K) and DK-51586 and DK-58825 (S.R.F.). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of two figures.
REFERENCES
- 1.Fraze E., Donner C. C., Swislocki A. L., Chiou Y. A., Chen Y. D., Reaven G. M. 1985. Ambient plasma free fatty acid concentrations in noninsulin-dependent diabetes mellitus: evidence for insulin resistance. J. Clin. Endocrinol. Metab. 61: 807–811. [DOI] [PubMed] [Google Scholar]
- 2.Abbasi F., McLaughlin T., Lamendola C., Reaven G. M. 2000. Insulin regulation of plasma free fatty acid concentrations is abnormal in healthy subjects with muscle insulin resistance. Metabolism. 49: 151–154. [DOI] [PubMed] [Google Scholar]
- 3.Reaven G. M., Chang H., Hoffman B. B. 1989. Impaired insulin-mediated inhibition of lipolysis and glucose transport with aging. Horm. Metab. Res. 21: 168–171. [DOI] [PubMed] [Google Scholar]
- 4.Guilherme A., Virbasius J. V., Puri V., Czech M. P. 2008. Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol. 9: 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kennedy A., Martinez K., Chuang C. C., LaPoint K., McIntosh M. 2009. Saturated fatty acid-mediated inflammation and insulin resistance in adipose tissue: mechanisms of action and implications. J. Nutr. 139: 1–4. [DOI] [PubMed] [Google Scholar]
- 6.Savage D. B., Petersen K. F., Shulman G. I. 2007. Disordered lipid metabolism and the pathogenesis of insulin resistance. Physiol. Rev. 87: 507–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zechner R., Kienesberger P. C., Haemmerle G., Zimmermann R., Lass A. 2009. Adipose triglyceride lipase and the lipolytic catabolism of cellular fat stores. J. Lipid Res. 50: 3–21. [DOI] [PubMed] [Google Scholar]
- 8.Duncan R. E., Ahmadian M., Jaworski K., Sarkadi-Nagy E., Sul H. S. 2007. Regulation of lipolysis in adipocytes. Annu. Rev. Nutr. 27: 79–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lafontan M., Langin D. 2009. Lipolysis and lipid mobilization in human adipose tissue. Prog. Lipid Res. 48: 275–297. [DOI] [PubMed] [Google Scholar]
- 10.Zimmermann R., Strauss J. G., Haemmerle G., Schoiswohl G., Birner-Gruenberger R., Riederer M., Lass A., Neuberger G., Eisenhaber F., Hermetter A., et al. 2004. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science. 306: 1383–1386. [DOI] [PubMed] [Google Scholar]
- 11.Villena J. A., Roy S., Sarkadi-Nagy E., Kim K. H., Sul H. S. 2004. Desnutrin, an adipocyte gene encoding a novel patatin domain-containing protein, is induced by fasting and glucocorticoids: ectopic expression of desnutrin increases triglyceride hydrolysis. J. Biol. Chem. 279: 47066–47075. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins C. M., Mancuso D. J., Yan W., Sims H. F., Gibson B., Gross R. W. 2004. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J. Biol. Chem. 279: 48968–48975. [DOI] [PubMed] [Google Scholar]
- 13.Smirnova E., Goldberg E. B., Makarova K. S., Lin L., Brown W. J., Jackson C. L. 2006. ATGL has a key role in lipid droplet/adiposome degradation in mammalian cells. EMBO Rep. 7: 106–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., et al. 2006. Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science. 312: 734–737. [DOI] [PubMed] [Google Scholar]
- 15.Bezaire V., Mairal A., Ribet C., Lefort C., Girousse A., Jocken J., Laurencikiene J., Anesia R., Rodriguez A. M., Ryden M., et al. 2009. Contribution of adipose triglyceride lipase and hormone-sensitive lipase to lipolysis in hMADS adipocytes. J. Biol. Chem. 284: 18282–18291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miyoshi H., Perfield J. W., II, Obin M. S., Greenberg A. S. 2008. Adipose triglyceride lipase regulates basal lipolysis and lipid droplet size in adipocytes. J. Cell. Biochem. 105: 1430–1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kershaw E. E., Hamm J. K., Verhagen L. A., Peroni O., Katic M., Flier J. S. 2006. Adipose triglyceride lipase: function, regulation by insulin, and comparison with adiponutrin. Diabetes. 55: 148–157. [PMC free article] [PubMed] [Google Scholar]
- 18.Gronke S., Mildner A., Fellert S., Tennagels N., Petry S., Muller G., Jackle H., Kuhnlein R. P. 2005. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 1: 323–330. [DOI] [PubMed] [Google Scholar]
- 19.Kurat C. F., Natter K., Petschnigg J., Wolinski H., Scheuringer K., Scholz H., Zimmermann R., Leber R., Zechner R., Kohlwein S. D. 2006. Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J. Biol. Chem. 281: 491–500. [DOI] [PubMed] [Google Scholar]
- 20.Chakrabarti P., English T., Shi J., Smas C. M., Kandror K. V. 2010. The mTOR complex 1 suppresses lipolysis, stimulates lipogenesis and promotes fat storage. Diabetes. 59: 775–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakrabarti P., Kandror K. V. 2009. FoxO1 controls insulin-dependent ATGL expression and lipolysis in adipocytes. J. Biol. Chem. 284: 13296–13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim J. Y., Tillison K., Lee J. H., Rearick D. A., Smas C. M. 2006. The adipose tissue triglyceride lipase ATGL/PNPLA2 is downregulated by insulin and TNF-alpha in 3T3–L1 adipocytes and is a target for transactivation by PPARgamma. Am. J. Physiol. Endocrinol. Metab. 291: E115–E127. [DOI] [PubMed] [Google Scholar]
- 23.Lake A. C., Sun Y., Li J. L., Kim J. E., Johnson J. W., Li D., Revett T., Shih H. H., Liu W., Paulsen J. E., et al. 2005. Expression, regulation, and triglyceride hydrolase activity of Adiponutrin family members. J. Lipid Res. 46: 2477–2487. [DOI] [PubMed] [Google Scholar]
- 24.Guarente L. 2006. Sirtuins as potential targets for metabolic syndrome. Nature. 444: 868–874. [DOI] [PubMed] [Google Scholar]
- 25.Finkel T., Deng C-X., Mostoslavsky R. 2009. Recent progress in the biology and physiology of sirtuins. Nature. 460: 587–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Picard F., Kurtev M., Chung N., Topark-Ngarm A., Senawong T., Machado De Oliveira R., Leid M., McBurney M. W., Guarente L. 2004. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 429: 771–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jing E., Gesta S., Kahn C. R. 2007. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 6: 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang F., Tong Q. 2009. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARgamma. Mol. Biol. Cell. 20: 801–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Feige J. N., Lagouge M., Canto C., Strehle A., Houten S. M., Milne J. C., Lambert P. D., Mataki C., Elliott P. J., Auwerx J. 2008. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 8: 347–358. [DOI] [PubMed] [Google Scholar]
- 30.Pfluger P. T., Herranz D., Velasco-Miguel S., Serrano M., Tschop M. H. 2008. Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl. Acad. Sci. USA. 105: 9793–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bordone L., Cohen D., Robinson A., Motta M. C., van Veen E., Czopik A., Steele A. D., Crowe H., Marmor S., Luo J., et al. 2007. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 6: 759–767. [DOI] [PubMed] [Google Scholar]
- 32.Xu Z., Kandror K. V. 2002. Translocation of small preformed vesicles is responsible for the insulin activation of glucose transport in adipose cells. Evidence from the in vitro reconstitution assay. J. Biol. Chem. 277: 47972–47975. [DOI] [PubMed] [Google Scholar]
- 33.Wang H., Qiang L., Farmer S. R. 2008. Identification of a domain within peroxisome proliferator-activated receptor gamma regulating expression of a group of genes containing fibroblast growth factor 21 that are selectively repressed by SIRT1 in adipocytes. Mol. Cell. Biol. 28: 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Jack A. K., Hamm J. K., Pilch P. F., Farmer S. R. 1999. Reconstitution of insulin-sensitive glucose transport in fibroblasts requires expression of both PPAR-gamma and C/EBP-alpha. J. Biol. Chem. 274: 7946–7951. [DOI] [PubMed] [Google Scholar]
- 35.Shan T., Ren Y., Liu Y., Zhu L., Wang Y. 2010. Breed difference and regulation of the porcine Sirtuin 1 by insulin. J. Anim. Sci. 88: 3909–3917. [DOI] [PubMed] [Google Scholar]
- 36.Kershaw E. E., Schupp M., Guan H. P., Gardner N. P., Lazar M. A., Flier J. S. 2007. PPARgamma regulates adipose triglyceride lipase in adipocytes in vitro and in vivo. Am. J. Physiol. Endocrinol. Metab. 293: E1736–E1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Festuccia W. T., Laplante M., Berthiaume M., Gelinas Y., Deshaies Y. 2006. PPARgamma agonism increases rat adipose tissue lipolysis, expression of glyceride lipases, and the response of lipolysis to hormonal control. Diabetologia. 49: 2427–2436. [DOI] [PubMed] [Google Scholar]
- 38.Nielsen R., Pedersen T. A., Hagenbeek D., Moulos P., Siersbaek R., Megens E., Denissov S., Borgesen M., Francoijs K. J., Mandrup S., et al. 2008. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 22: 2953–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu M., Liu F. 2010. Transcriptional and post-translational regulation of adiponectin. Biochem. J. 425: 41–52. [DOI] [PubMed] [Google Scholar]
- 40.Lan F., Cacicedo J. M., Ruderman N., Ido Y. 2008. SIRT1 modulation of the acetylation status, cytosolic localization, and activity of LKB1. Possible role in AMP-activated protein kinase activation. J. Biol. Chem. 283: 27628–27635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruderman N. B., Xu X. J., Nelson L., Cacicedo J. M., Saha A. K., Lan F., Ido Y. 2010. AMPK and SIRT1: a long-standing partnership? Am. J. Physiol. Endocrinol. Metab. 298: E751–E760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou X., Xu S., Maitland-Toolan K. A., Sato K., Jiang B., Ido Y., Lan F., Walsh K., Wierzbicki M., Verbeuren T. J., et al. 2008. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J. Biol. Chem. 283: 20015–20026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daval M., Foufelle F., Ferre P. 2006. Functions of AMP-activated protein kinase in adipose tissue. J. Physiol. 574: 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gauthier M. S., Miyoshi H., Souza S. C., Cacicedo J. M., Saha A. K., Greenberg A. S., Ruderman N. B. 2008. AMP-activated protein kinase is activated as a consequence of lipolysis in the adipocyte: potential mechanism and physiological relevance. J. Biol. Chem. 283: 16514–16524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Djouder N., Tuerk R. D., Suter M., Salvioni P., Thali R. F., Scholz R., Vaahtomeri K., Auchli Y., Rechsteiner H., Brunisholz R. A., et al. 2010. PKA phosphorylates and inactivates AMPKalpha to promote efficient lipolysis. EMBO J. 29: 469–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaidhu M. P., Fediuc S., Anthony N. M., So M., Mirpourian M., Perry R. L., Ceddia R. B. 2009. Prolonged AICAR-induced AMP-kinase activation promotes energy dissipation in white adipocytes: novel mechanisms integrating HSL and ATGL. J. Lipid Res. 50: 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yin W., Mu J., Birnbaum M. J. 2003. Role of AMP-activated protein kinase in cyclic AMP-dependent lipolysis In 3T3–L1 adipocytes. J. Biol. Chem. 278: 43074–43080. [DOI] [PubMed] [Google Scholar]
- 48.Ahmadian M., Abbott M. J., Tang T., Hudak C. S., Kim Y., Bruss M., Hellerstein M. K., Lee H. Y., Samuel V. T., Shulman G. I., et al. 2011. Desnutrin/ATGL is regulated by AMPK and is required for a brown adipose phenotype. Cell Metab. 13: 739–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daitoku H., Hatta M., Matsuzaki H., Aratani S., Ohshima T., Miyagishi M., Nakajima T., Fukamizu A. 2004. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl. Acad. Sci. USA. 101: 10042–10047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Motta M. C., Divecha N., Lemieux M., Kamel C., Chen D., Gu W., Bultsma Y., McBurney M., Guarente L. 2004. Mammalian SIRT1 represses forkhead transcription factors. Cell. 116: 551–563. [DOI] [PubMed] [Google Scholar]
- 51.Nakae J., Cao Y., Daitoku H., Fukamizu A., Ogawa W., Yano Y., Hayashi Y. 2006. The LXXLL motif of murine forkhead transcription factor FoxO1 mediates Sirt1-dependent transcriptional activity. J. Clin. Invest. 116: 2473–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Calnan D. R., Brunet A. 2008. The FoxO code. Oncogene. 27: 2276–2288. [DOI] [PubMed] [Google Scholar]
- 53.Bluher M., Kahn B. B., Kahn C. R. 2003. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 299: 572–574. [DOI] [PubMed] [Google Scholar]
- 54.Muzumdar R., Allison D. B., Huffman D. M., Ma X., Atzmon G., Einstein F. H., Fishman S., Poduval A. D., McVei T., Keith S. W., et al. 2008. Visceral adipose tissue modulates mammalian longevity. Aging Cell. 7: 438–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hwangbo D. S., Gershman B., Tu M. P., Palmer M., Tatar M. 2004. Drosophila dFOXO controls lifespan and regulates insulin signalling in brain and fat body. Nature. 429: 562–566. [DOI] [PubMed] [Google Scholar]
- 56.Giannakou M. E., Goss M., Junger M. A., Hafen E., Leevers S. J., Partridge L. 2004. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 305: 361. [DOI] [PubMed] [Google Scholar]
- 57.Kenyon C. J. 2010. The genetics of ageing. Nature. 464: 504–512. [DOI] [PubMed] [Google Scholar]
- 58.Fontana L., Partridge L., Longo V. D. 2010. Extending healthy life span–from yeast to humans. Science. 328: 321–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Canto C., Auwerx J. 2009. Caloric restriction, SIRT1 and longevity. Trends Endocrinol. Metab. 20: 325–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Donmez G., Guarente L. 2010. Aging and disease: connections to sirtuins. Aging Cell. 9: 285–290. [DOI] [PubMed] [Google Scholar]
- 61.Wang M. C., O'Rourke E. J., Ruvkun G. 2008. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 322: 957–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie T. 2008. Physiology. Burn fat, live longer. Science. 322: 865–866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.