Abstract
Triacylglycerols (TG) are the major storage form of energy in eukaryotic organisms and are synthesized primarily by acyl CoA:1,2-diacylglycerol acyltransferase (DGAT) enzymes. In vitro DGAT activity has previously been quantified by measuring the incorporation of either radiolabeled fatty acyl CoA or diacylglycerol (DG) into TG. We developed a modified acyltransferase assay using a fluorescent fatty acyl CoA substrate to accurately quantify in vitro DGAT activity. In the modified assay, radioactive fatty acyl CoA is replaced with fluorescent NBD-palmitoyl CoA, which is used as a substrate by DGAT with DG to produce NBD-TG. After extraction with organic solvents and separation by thin layer chromatography, NBD-TG formation can be detected and accurately quantified using a fluorescent imaging system. We demonstrate that this method can be adapted to detect other acyltransferase activities. Because NBD-palmitoyl CoA is commercially available at a much lower cost compared with radioactive acyl CoA substrates, it is a more economical alternative to radioactive tracers. In addition, the exposure of laboratory personnel to radioactivity is greatly reduced.
Keywords: triacylglycerol, lipid synthesis, radioactivity
Triacylglycerols (TG) are a class of neutral lipid and are the major storage form of energy in eukaryotic organisms. TG are composed of a glycerol backbone and three fatty acids attached by ester bonds. Although abundant in the liver, muscle, small intestine, and mammary gland, TG is primarily stored in cytosolic lipid droplets in adipose tissue. When needed, stored TG in adipocytes undergo lipolysis to release fatty acids from TG that are transported to tissues, such as muscle, where they are oxidized to generate ATP. However, excessive accumulation of TG in adipose tissue leads to obesity. In nonadipose tissue, such as the pancreas, skeletal muscle, and liver, metabolites derived from excessive TG can be lipotoxic, leading to insulin resistance and type 2 diabetes (1, 2). Elevated TG in the circulation is a risk factor for cardiovascular disease (3).
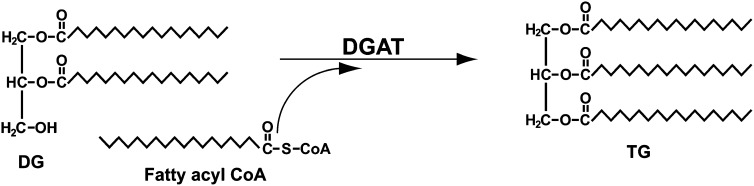
TG synthesis occurs via the multistep Kennedy (glycerol-3-phosphate) pathway with the final reaction of the pathway catalyzed by acyl CoA:1,2-diacylglycerol acyltransferase (DGAT, EC 2.3.1.20) (4). DGAT catalyzes the formation of an ester bond between an activated fatty acid (fatty acyl CoA) and the free hydroxyl group of 1,2-diacylglycerol (DG) (Fig. 1). Although never purified to homogeneity, two unrelated DGAT enzymes, DGAT1 and DGAT2, have now been identified (5, 6). These two enzymes share no sequence homology, and although they catalyze the same biochemical reaction in vitro, they have distinct physiological roles in vivo in mice (7, 8). DGAT1 also possesses other acyltransferase activities and can catalyze the formation of diacylglycerol, retinol, and wax esters (9).
Fig. 1.
TG synthesis by DGAT. DGAT catalyzes the formation of an ester bond between a fatty acyl CoA and the free hydroxyl group of 1,2-diacylglycerol.
The standard method to measure DGAT activity in vitro involves incubating cell or tissue extracts with radiolabeled substrate, either fatty acyl CoA or DG, which is incorporated into TG (5, 10, 11). The radiolabeled TG products are then separated from substrates by extraction with organic solvents and thin layer chromatography (TLC). TG is then scraped off of the TLC plate, and the radioactivity incorporated into TG is quantified by scintillation counting. Although this is a sensitive method, the use of radioactivity requires special precautions and dedicated equipment that will be contaminated with radioactivity. Other challenges include minimizing the exposure of laboratory personnel to radioactivity and the disposal of radioactive waste. Additionally, the availability of radioactive substrates is very limited, and the associated cost of this assay is becoming more prohibitive.
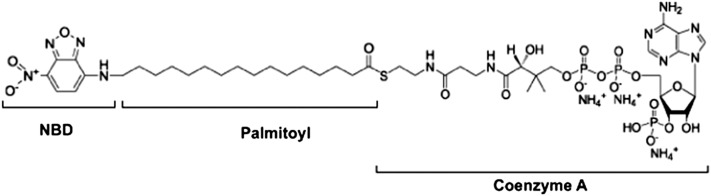
Here we demonstrate a modified DGAT assay where we use a fluorescent acyl CoA substrate instead of a radioactive one without compromising sensitivity. The modified protocol involves substituting the radiolabeled acyl CoA substrate with the fluorescent fatty acyl CoA {N-[(7-nitro-2-1,3-benzoxadiazol-4-yl)-methyl]amino} (NBD)-palmitoyl CoA (Fig. 2). This NBD substrate is commercially available and is approximately 25% of the cost of its radiolabeled counterpart usually used for DGAT and other acyl CoA-dependent assays. The formation of the reaction product NBD-TG is then separated by TLC and quantified using a molecular imager capable of fluorescence detection. We also show that this method can be modified to assay other acyltransferase enzymes.
Fig. 2.
{N-[(7-nitro-2-1,3-benzoxadiazol-4-yl)-methyl]amino} (NBD)-palmitoyl CoA substrate.
EXPERIMENTAL PROCEDURES
Materials
NBD-palmitoyl CoA (catalog number 810705P) was from Avanti Polar Lipids. Tris-HCl, MgCl2, sucrose, bovine serum albumen (BSA), hexane, ethyl ether, CHCl3, methanol, acetone, acetic acid, 1,2 dioleoyl-sn-glycerol (DOG) (catalog number D0138), and 1-hexadecanol (catalog number H6800) were from Sigma. Partisil K6 60Å channeled TLC plates (250 µm thickness) with a pre-adsorbent zone (catalog number 05-713-329) were from Fisher Scientific.
Cell culture and transfection
HEK-293T cells (American Type Tissue Culture Collection) were cultured in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum in a 37°C incubator with 5% CO2. HEK293T cells were transfected with the mammalian expression vector, pcDNA3.1 (Invitrogen) containing FLAG-tagged DGAT1 using 0.1% polyethylenimine (pH 7.0) (12).
3T3-L1 cells were induced to differentiate into adipocytes by incubating ∼90% confluent pre-adipocytes in DMEM supplemented with 10% FBS, 10 μM dexamethasone, 0.5 mM isobutylmethylxanthine, and 10 μg/ml insulin for 72 h (13). Medium was changed every 24 h. 72 h postinduction, the differentiation medium was removed and cells were maintained in DMEM containing 10% FBS.
Total membrane isolation
Cells were washed twice with ice-cold PBS, scraped from the tissue culture dish and placed in a 1.5 ml tube. Cells were pelleted by centrifugation at 1,000 g for 2 min and the cell pellet was resuspended in 500 μl of 50 mM Tris-HCl (pH 7.6)/250 mM sucrose. Cells were disrupted by 15 passages through a 27-gauge needle. Cell debris and nuclei were pelleted by centrifugation at 600 g for 5 min. To obtain total cellular membranes, the supernatant was centrifuged at 100,000 g for 30 min at 4°C. The supernatant was removed and the membrane pellet was resuspended in 50 mM Tris-HCl (pH 7.6)/250 mM sucrose and used for DGAT assays at the indicated concentrations.
Fluorescent DGAT assay
The method is a modification of that described by Coleman and Bell (10, 11). The stock solutions used for the assay were 1 M Tris-HCl (pH 7.6), 1 M MgCl2, 4 mM DOG in acetone, 12.5 mg/ml BSA, 500 μM NBD-palmitoyl CoA in 20 mM Tris-HCl (pH 7.6), and 20-100 μg protein sample (cell lysate or total membranes) diluted in 50 μl of 50 mM Tris-HCl (pH 7.6)/250 mM sucrose. Assays were performed in 16 × 100 mm glass test tubes in a final reaction volume of 200 μl.
The procedure was as follows: 1) Prepare a master mix containing 20 μl of 1 M Tris-HCl (pH 7.6), 4 μl of 1 M MgCl2, 10 μl of 4 mM DOG, 10 μl of 12.5 mg/ml BSA, 10 μl of 500 μM NBD-palmitoyl CoA, and 96 μl of water per reaction. Scale up the volumes proportionally to accommodate the desired number of reactions. Protect the master mix from direct light. 2) Aliquot 150 μl master mix per test tube. Preincubate tubes in a 37°C water bath for 2 min. Start the reaction by adding 50 μl of protein sample (50 μg protein per assay is routinely used). Incubate at 37°C for 10 min with occasional shaking. 3) Terminate the reaction by adding 4 ml CHCl3/methanol (2:1, v/v). Mix by vortexing. Add 800 μl of water and mix by vortexing. Allow samples to sit at room temperature for 1 h. 4) Vortex and centrifuge at 3,000 rpm for 5 min to separate aqueous and organic phases. 5) Aspirate the upper aqueous phase and dry the organic phase under stream of nitrogen. Solvents can be evaporated faster by placing test tubes in a warm water bath. 6) Resuspend lipids in 50 μl CHCl3/methanol (2:1) and spot on a channeled 20 × 20 cm TLC plate. 7) Develop the TLC plate in the solvent system hexane/ethyl ether/acetic acid (80:20:1, v/v/v). Allow the plate to air dry for 1 h before quantification of reaction products.
The newly synthesized NBD-TG was analyzed with a VersaDoc 4000 molecular imaging system (Bio-Rad Laboratories, Inc.), and fluorescence was quantified with Quantity One software (Bio-Rad Laboratories, Inc.). The excitation and emission wavelengths of NBD were 465 nm and 535 nm, respectively. A blue LED laser light source and 530BP emission filter were used. Data are presented as units (fluorescence intensity) of NBD-TG formed per minute per mg protein.
Statistical analyses
Where appropriate, data are presented as means ± SD. Means were compared by Student's t-test.
RESULTS AND DISCUSSION
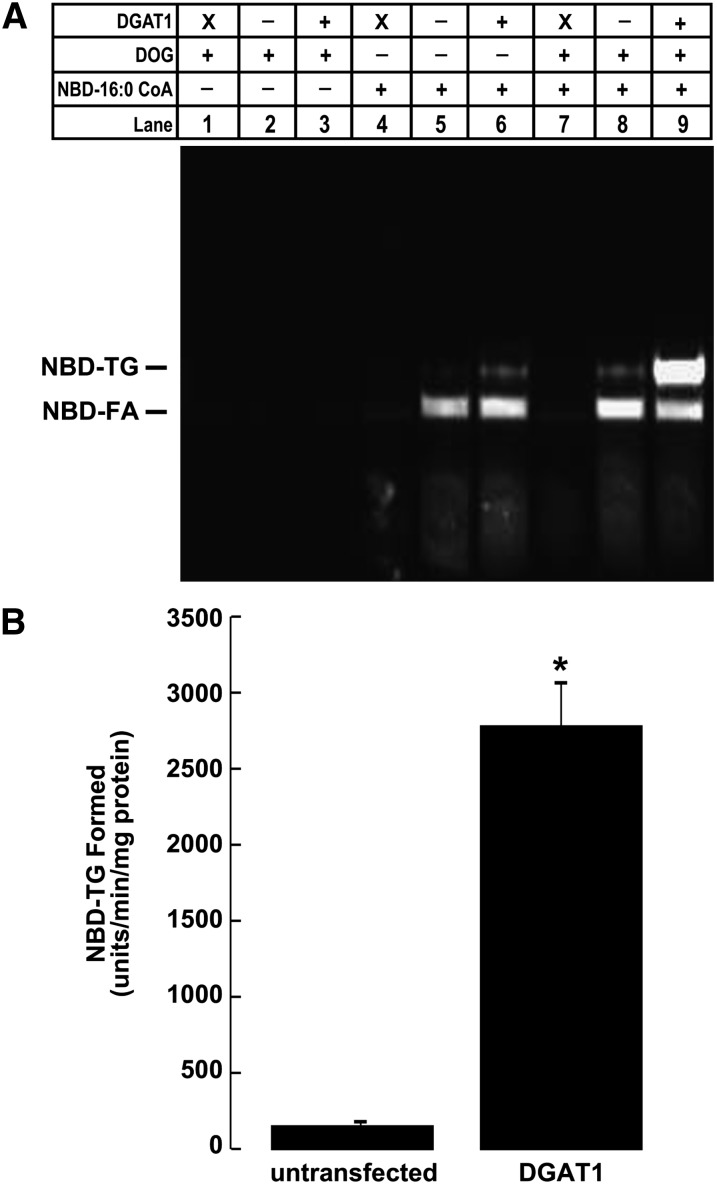
To validate our modified fluorescent DGAT activity assay, we performed a series of assays on samples where DGAT activity has been reliably quantified using the traditional radioactive method. Total membranes isolated from untransfected HEK293T cells and HEK293T cells expressing DGAT1 were incubated in the DGAT reaction mixture as described in Experimental Procedures. In some cases, reactions were carried out in the absence of protein, DOG, or NBD-16:0 CoA. After reactions were terminated, reaction products were extracted with organic solvents and separated by TLC. The TLC plate was then scanned with a fluorescence imager to visualize and quantify NBD-TG formed during the assay (excitation = 465 nm; emission = 535 nm).
For DGAT assays performed with the complete reaction mixture, two fluorescent lipids were observed (Fig. 3A). The lower lipid was NBD-palmitate, and the upper was the reaction product NBD-TG. Consistent with the radioactive DGAT assay, NBD-TG formation was higher in samples from HEK293T cells expressing DGAT1 than in those from untransfected cells. This was the case when DGAT activity was determined using endogenous DG or when DG was added exogenously (Fig. 3A). No fluorescent products could be detected when either NBD-16:0 CoA or protein was omitted from the reaction. The fluorescence intensity of NBD-TG formed during the assay (Fig. 3A, lanes 8 and 9) was quantified using Quantity One software. Using 50 μg of membrane protein from untransfected HEK293T cells, which is the amount typically used in the radioactive assay, DGAT activity could be quantified and was significantly higher than background (Fig. 3B). For HEK293T cells expressing DGAT1, NBD-TG formation was ∼19-fold greater than in that of untransfected cells (Fig. 3B), which is consistent with values obtained using the radioactive method (12).
Fig. 3.
NBD-TG formation in HEK-293T cells expressing DGAT1. A: TLC plate showing the results of a DGAT assay using the fluorescent NBD-palmitoyl CoA (NBD-16:0 CoA) substrate. Assays were performed on total membranes from either untransfected HEK293T cells or HEK293T cells transiently transfected with DGAT1 (“X” indicates assays performed without any protein) in the presence or absence of either 200 μM DOG or 25 μM NBD-16:0 CoA. B: Quantification of NBD-TG formation in DGAT assays performed on membranes from untransfected HEK293T cells or HEK293T cells transiently transfected with DGAT1 (lanes 8 and 9). *P < 0.001 (n =3).
The NBD-palmitate observed on the TLC plate was likely formed by the hydrolysis of NBD-palmitoyl CoA during the DGAT assay. This phenomenon has also been observed in the radioactive DGAT assay and is caused by an acyl CoA hydrolase present in the samples (14). It is unlikely that DGAT1 or DGAT2 possesses this function, as this hydrolase activity is still apparent in both cells lacking both DGAT1 and DGAT2 (15). It has been proposed that acyl CoA hydrolase generates an acyl intermediate that can be utilized by acyltransferases, such as DGAT1 and DGAT2 (14). Unfortunately, because the NBD-palmitate formed during the DGAT assay partitions into the organic phase, reaction products must still be separated by TLC prior to quantification.
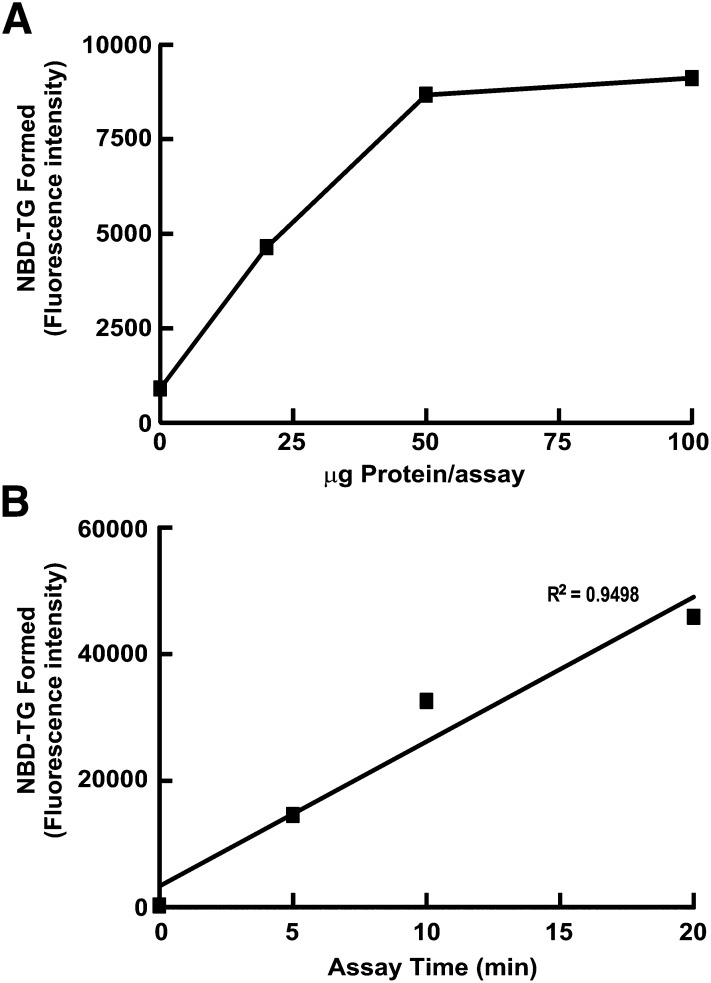
We also determined that NBD-TG formation was linear with respect to time and amount of protein in the modified assay. Fluorescent DGAT assays were performed using 0-100 μg of membrane protein isolated from HEK293T cells. The concentrations of other reaction components were at the concentrations described in Experimental Procedures. Without any protein in the assay, a weak fluorescent signal corresponding to NBD-TG was detected, which was likely due to autofluorescence of the TLC plate (Fig. 4A). NBD-TG formation was linear up to 50 μg of protein; it reached a plateau as the concentrations of substrates became limiting with higher amounts of protein. NBD-TG formation was also linear with time from 5-20 min (R2 = 0.9498) (Fig. 4B).
Fig. 4.
NBD-TG formation is linear with respect to time and protein. DGAT activity of membranes from HEK293T cells was measured with (A) 0-100 μg protein per assay and (B) 0-20 min using 20 μg protein per assay.
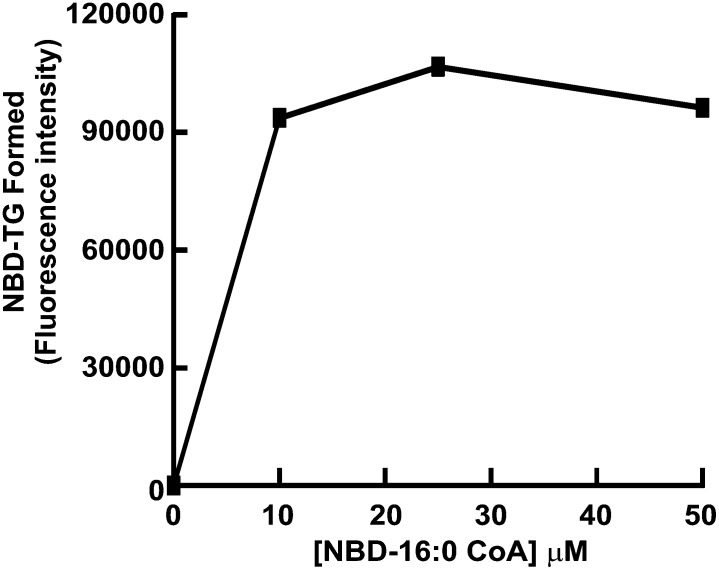
Because our modified DGAT assay replaces the traditional radioactive acyl CoA substrate with one containing the fluorescent NBD group at the methyl end of the molecule, it was important to determine that the amount of NBD-palmitoyl CoA in the assay was not limiting. DGAT assays were performed on samples from HEK293T cells using NBD-palmitoyl CoA at a concentration of 0-50 μM. When NBD-palmitoyl CoA was not included in the reaction, no NBD-TG formation could be detected (Fig. 5). Maximum NBD-TG formation (apparent Vmax) occurred at 10 μM NBD-palmitoyl CoA, indicating that the amount of substrate was not limiting in the assay and could be used at the same concentrations as radioactive acyl CoA (usually 25 μM) (Fig. 5).
Fig. 5.
Effect of substrate concentration on NBD-TG formation. DGAT activity of membranes from HEK293T cells (50 μg protein) was measured using NBD-16:0 CoA at a final concentration of 0-50 μM per assay.
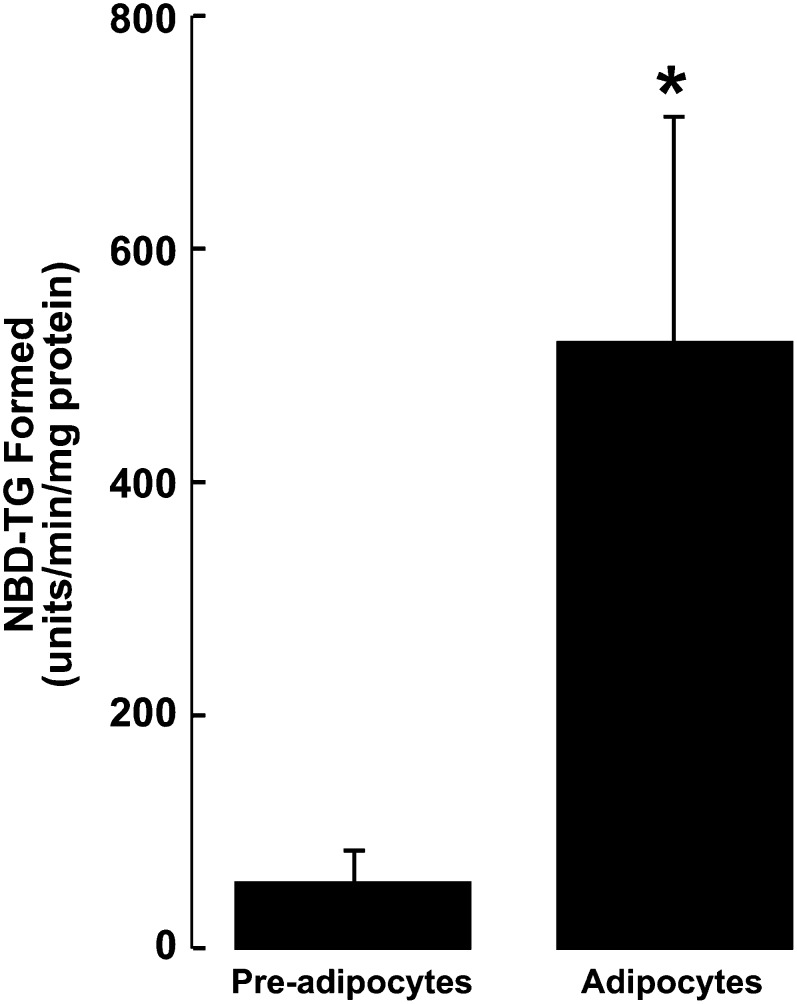
To further validate our modified fluorescent DGAT assay, we compared the DGAT activity of undifferentiated 3T3-L1 fibroblasts to that of differentiated 3T3-L1 adipocytes. Previous studies have demonstrated that during adipocyte differentiation both DGAT1 and DGAT2 gene expression are increased with a corresponding increase in DGAT activity in vitro (5, 6, 16). Consistent with these findings, our fluorescent assay showed that undifferentiated cells had a low level of DGAT activity (Fig. 6). However, after 10 days of differentiation, the fluorescent assay showed that DGAT activity of adipocytes was increased ∼9-fold compared with that of undifferentiated cells. This proportional increase was similar to what has been a reported elsewhere using a radioactive assay (5, 16).
Fig. 6.
Determination of NBD-TG formation in undifferentiated 3T3-L1 fibroblasts and differentiated adipocytes. Cell lysates (37.5 μg protein) were prepared from 3T3-L1 pre-adipocytes or adipocytes (10 days postdifferentiation), and DGAT activity was determined. *P < 0.001 (n =3).
Many other acyltransferase enzymes exist, and their activities are also usually determined by using assays with radioactive acyl CoA substrates. We sought to determine if the fluorescent NBD-palmitoyl CoA substrate could be a useful reagent for assaying other acyl CoA-dependent activities. To do this, we took advantage of the ability of DGAT1 to utilize acyl acceptors other than diacylglycerol. In addition to catalyzing TG synthesis, DGAT1 also possesses retinol acyltransferase, monoacylglycerol acyltransferase, and wax synthase activities (9, 17).
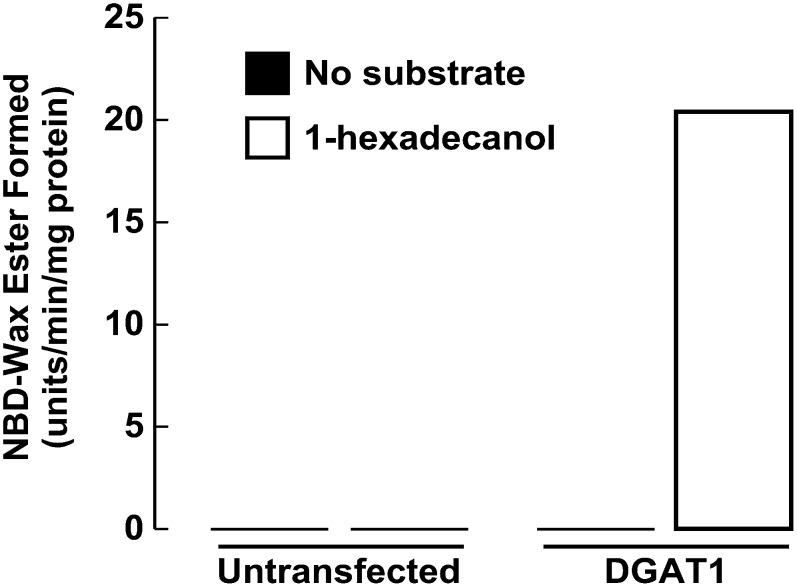
When membrane proteins from HEK293T cells expressing DGAT1 were incubated in assays with NBD-palmitoyl CoA and the acyl acceptor 1-hexadecanol, we detected robust wax synthase activity by monitoring NBD-wax ester formation (Fig. 7). No NBD-wax esters were formed in the absence of 1-hexadecanol in untransfected or transfected samples. NBD-wax ester formation also could not be detected in assays performed on untransfected HEK293T cells, which we previously found to have very little endogenous wax synthase activity as assessed by using the radioactive method (12).
Fig. 7.
NBD-16:0 CoA can be used to measure other acyltransferase activities. In vitro wax synthase acyltransferase assays were performed on total cellular membranes (50 μg protein) from untransfected HEK-293T cells or from HEK-293T cells expressing FL-DGAT1. NBD-16:0 CoA (25 μM) was the acyl donor and 1-hexadecanol (200 μM) was the acyl group acceptor. NBD-wax ester formation was determined as described for NBD-TG.
In summary, we have described a modified fluorescent assay that accurately measures in vitro DGAT activity. In our hands, this fluorescent method appears to be as sensitive and as reproducible as the traditional radioactive assay. Using the NBD-palmitoyl CoA substrate reduces the cost of the assay by ∼75% compared with the radioactive method, and it minimizes the exposure of lab personnel to radioactivity. Additionally, we have demonstrated that NBD-palmitoyl CoA could be used as a substrate to measure the in vitro activities of other acyltransferase enzymes. Specific assay conditions would have to be optimized for each enzyme.
Footnotes
Abbreviations:
- DG
- diacylglycerol
- DGAT
- acyl CoA:diacylglycerol acyltransferase
- DOG
- 1,2 dioleoyl-sn-glycerol
- NBD
- {N-[(7-nitro-2-1,3-benzoxadiazol-4-yl)-methyl]amino}
- TG
- triacylglycerol
This work was supported by Regional Partnership Program FRN# 88063 (S.S.) from the Canadian Institutes of Health Research, by the Canadian Foundation for Innovation Leaders Opportunity Fund, and by a grant-in-aid (S.S.) from the Heart and Stroke Foundation of Saskatchewan.
REFERENCES
- 1.Unger R. H., Zhou Y. T. 2001. Lipotoxicity of beta-cells in obesity and in other causes of fatty acid spillover. Diabetes. 50(Suppl. 1): S118–S121. [DOI] [PubMed] [Google Scholar]
- 2.Unger R. H., Orci L. 2002. Lipoapoptosis: its mechanism and its diseases. Biochim. Biophys. Acta. 1585: 202–212. [DOI] [PubMed] [Google Scholar]
- 3.Rosenbaum M., Leibel R. L., Hirsch J. 1997. Obesity. N. Engl. J. Med. 337: 396–406. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy E. P. 1957. Metabolism of lipides. Annu. Rev. Biochem. 26: 119–148. [DOI] [PubMed] [Google Scholar]
- 5.Cases S., Smith S. J., Zheng Y. W., Myers H. M., Lear S. R., Sande E., Novak S., Collins C., Welch C. B., Lusis A. J., et al. 1998. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 95: 13018–13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cases S., Stone S. J., Zhou P., Yen E., Tow B., Lardizabal K. D., Voelker T., Farese R. V., Jr 2001. Cloning of DGAT2, a second mammalian diacylglycerol acyltransferase, and related family members. J. Biol. Chem. 276: 38870–38876. [DOI] [PubMed] [Google Scholar]
- 7.Stone S. J., Myers H. M., Watkins S. M., Brown B. E., Feingold K. R., Elias P. M., Farese R. V., Jr 2004. Lipopenia and skin barrier abnormalities in DGAT2-deficient mice. J. Biol. Chem. 279: 11767–11776. [DOI] [PubMed] [Google Scholar]
- 8.Smith S. J., Cases S., Jensen D. R., Chen H. C., Sande E., Tow B., Sanan D. A., Raber J., Eckel R. H., Farese R. V., Jr 2000. Obesity resistance and multiple mechanisms of triglyceride synthesis in mice lacking DGAT. Nat. Genet. 25: 87–90. [DOI] [PubMed] [Google Scholar]
- 9.Yen C. L., Monetti M., Burri B. J., Farese R. V., Jr 2005. The triacylglycerol synthesis enzyme DGAT1 also catalyzes the synthesis of diacylglycerols, waxes, and retinyl esters. J. Lipid Res. 46: 1502–1511. [DOI] [PubMed] [Google Scholar]
- 10.Coleman R. A. 1992. Diacylglycerol acyltransferase and monoacylglycerol acyltransferase from liver and intestine. Methods Enzymol. 209: 98–104. [DOI] [PubMed] [Google Scholar]
- 11.Coleman R., Bell R. M. 1976. Triacylglycerol synthesis in isolated fat cells. Studies on the microsomal diacylglycerol acyltransferase activity using ethanol-dispersed diacylglycerols. J. Biol. Chem. 251: 4537–4543. [PubMed] [Google Scholar]
- 12.McFie P. J., Stone S. L., Banman S. L., Stone S. J. 2010. Topological orientation of acyl-CoA:diacylglycerol acyltransferase-1 (DGAT1) and identification of a putative active site histidine and the role of the N terminus in dimer/tetramer formation. J. Biol. Chem. 285: 37377–37387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brasaemle D. L., Barber T., Wolins N. E., Serrero G., Blanchette-Mackie E. J., Londos C. 1997. Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J. Lipid Res. 38: 2249–2263. [PubMed] [Google Scholar]
- 14.Lehner R., Kuksis A. 1993. Purification of an acyl-CoA hydrolase from rat intestinal microsomes. A candidate acyl-enzyme intermediate in glycerolipid acylation. J. Biol. Chem. 268: 24726–24733. [PubMed] [Google Scholar]
- 15.Harris C. A., Haas J. T., Streeper R. S., Stone S. J., Kumari M., Yang K., Han X., Brownell N., Gross R. W., Zechner R., et al. 2011. DGAT enzymes are required for triacylglycerol synthesis and lipid droplets in adipocytes. J. Lipid Res. 52: 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meegalla R. L., Billheimer J. T., Cheng D. 2002. Concerted elevation of acyl-coenzyme A:diacylglycerol acyltransferase (DGAT) activity through independent stimulation of mRNA expression of DGAT1 and DGAT2 by carbohydrate and insulin. Biochem. Biophys. Res. Commun. 298: 317–323. [DOI] [PubMed] [Google Scholar]
- 17.Shih M. Y., Kane M. A., Zhou P., Yen C. L., Streeper R. S., Napoli J. L., Farese R. V., Jr 2009. Retinol esterification by DGAT1 is essential for retinoid homeostasis in murine skin. J. Biol. Chem. 284: 4292–4299. [DOI] [PMC free article] [PubMed] [Google Scholar]