Abstract
The conflict theory of genomic imprinting argues that parent-of-origin effects on allelic expression evolve as a consequence of conflict between maternally and paternally derived genomes. I derive explicit population-genetic models of this theory when individuals in a cohort with an arbitrary and variable number of sires and dams interact. I show that the evolution of imprinting is governed by the reciprocal of the harmonic mean number of fathers but the reciprocal of the arithmetic mean number of mothers per cohort. Thus, a few monandrous females in a polyandrous population decrease the strength of the genetic conflict and the opportunity for conflict-driven paternal imprinting. In contrast, in populations in which few males control large harems, rare males with small harems do not have such a disproportionate effect on genetic conflicts and maternal imprinting. Additionally, I demonstrate that under the conflict theory, selection for imprinted expression on paternally derived X chromosomes is much weaker than it is on maternally derived X chromosomes or autosomes.
Keywords: genomic imprinting, conflict theory, harem, polyandry, mating system, kin selection
Background
Genomic imprinting is an epigenetic phenomenon in which the phenotypic effect of an allele depends on its parental origin. Imprinting is achieved by differential allelic expression, and it is generally under the control of complex regulatory machinery such as methylation and other forms of chromatin remodeling (Bartolomei 2009). Although well documented in plants (e.g., Berger and Chaudhury 2009), imprinting is best understood in mammals, where imprinted genes are clustered in “imprinting centers,” and imprinted expression most often occurs in the placenta and the brain (Morison et al. 2005).
The conflict (or kinship) theory argues that genomic imprinting is an evolutionary outcome of conflict within families (Moore and Haig 1991). The canonical version of the conflict theory is based on conflict between maternally and paternally derived genes over maternal resource allocation. This conflict arises because relatedness between maternal siblings (matrisibs) at maternally and paternally derived alleles is asymmetric: when mothers mate multiply, maternally inherited alleles in a maternal family have higher probabilities of being identical by descent than do paternally inherited alleles. Thus, mothers favor a more uniform distribution of resources across developing progeny than do fathers, who favor skewing resources toward their own offspring and away from those of other patrilines within the brood. This classic version of the conflict theory successfully accounts for many observed patterns of parent-of-origin gene expression, especially in the mammalian placenta and the plant endosperm (Wilkins and Haig 2003b).
Some empirical findings have cast doubt on the generality of the classic version of the conflict theory (Hurst and McVean 1997, 1998). These observations include postnatal imprinted effects (Itier et al. 1998), imprinted effects on sex chromosomes (Iwasa and Pomiankowski 1999, 2001), sex differences in imprinted expression (Hager et al. 2008), and ontogenic changes in imprinted status (Wolf et al. 2008). Such observations have inspired novel alternative hypotheses to explain the evolution of genomic imprinting (e.g., Iwasa and Pomiankowski 2001; Day and Bonduriansky 2004; Wolf and Hager 2006;Wolf 2009) and significant extensions of the conflict theory to generate predictions for the evolution of genomic imprinting under more complex social interactions (e.g., Haig 1997, 2000; Wilkins and Haig 2003a; Ùbeda 2008; Wild and West 2009).
I extend and refine predictions of the conflict theory to incorporate interactions within groups larger than those of matrisibs. I begin by examining the evolution of intragroup conflict without genomic imprinting. These results stand as a baseline expectation for conflict resolution within groups. Since imprinting can be costly (Hall 1990; Lalande 1996; Ùbeda and Wilkins 2008), imprinting is predicted only when results from the imprinting models differ substantially from the results of the biallelic models (Hurst 1999).
I then examine the evolution of imprinting as a result of competition among matrisibs as well as among paternal siblings (patrisibs). While previous models (Spencer et al. 1998, 2004; Mills and Moore 2004) have addressed the evolution of imprinting in populations where females have a specified, fixed number of mates, I generalize these results to populations with variation in the number of mates per female. Additionally, as the canonical version of the conflict theory concerns conflicts for maternal resources, the influence of competition among patrisibs on the evolution of imprinting is not well studied (but see Ùeda 2008) and the similarities and differences between competition among matrisibs and patrisibs is largely unknown. I find that competition among matrisibs and among patrisibs can favor the evolution of imprinting; however, the strength of intrafamilial conflict has differential sensitivity to variation in the number of mates per female and in male harem size. In a population of multiply mating females, the response to selection on allelic silencing that increases group fitness is dampened by the harmonic mean number of fathers per group. In contrast, when an interacting cohort is derived from multiple dams, the response to among-group selection is diminished by the arithmetic mean number of females per harem. This difference suggests that conflict among patrisibs may be stronger and conflict among matrisibs may be weaker than previously appreciated.
Finally, I extend my models to the X chromosome, as the functions of imprinted genes on the X chromosome have cast doubt on the generality of the conflict theory (Iwasa and Pomiankowski 1999, 2001). I show that the conflict theory predicts a relatively weak response to selection for silencing of paternally derived X chromosomes, while the silencing of the maternally derived X chromosome may respond rapidly to selection. These results suggest that if the evolution of genomic imprinting is driven by genetic conflicts, the X chromosome may harbor more maternally silenced alleles than paternally silenced ones.
The models developed in this article address “the key question for understanding the role of imprinting in sibling rivalry … the relative importance of competition with [patrisibs] and [matrisibs]” (Haig 2008, p. 271). I conclude by showing how the findings derived herein can explain the evolution of silencing of paternally derived genes that increase individual fitness while decreasing group fitness: a case predicted by the canonical version of the conflict theory to be maternally silenced.
The Model
I model the evolution of a modifier locus allele that silences a nearby gene in cis; that is, there is no allelic variation at the modified locus, but its alleles are differentially expressed on the basis of the action of the nearby regulator. I present three models, one examining the evolution of a regulator without imprinting, one in which a maternally derived regulator completely silences the coinherited maternal allele, and one in which the regulator, when paternally derived, silences the coinherited paternal allele. For each model, I allow the modifier alleles to be either autosomal or X linked. Because the single-locus X-linked case with a degenerate Y chromosome is mathematically equivalent to haplodiploidy, the sex chromosome model holds for haplodiploid insects, a group for which evidence of imprinted expression is compelling (Kronforst et al. 2008; Elango et al. 2009).
I describe a population that consists of many large groups that are founded by n♀ dams and the n♂ sires with which they mate. Competition among individuals born to a group is followed by random dispersal. I examine one biallelic locus with alleles A and a in frequencies p and q = 1 − p, respectively. I assume that paternal fitness increases linearly with the number of mates, and there is no reproductive skew within groups: that is, each sire fathers n♀ /n♂ offspring in a group, and each dam mothers 1/n♀ of the offspring in a group. I further assume random mating, nonoverlapping generations, and weak selection, such that genotypic frequencies after selection do not strongly differ from Hardy-Weinberg expectations and allele frequencies are approximately equal across the sexes. This method follows the approach of Wade (1978), which should be consulted for more details.
I begin with an allele with a direct effect on individual fitness (i) and a pleiotropic effect on group mean fitness (g); these values vary continuously and can be positive or negative. I do not impose any specific association between i and g or any bounds on their values aside from the constraint that individuals must have nonnegative fitnesses. An individual of genotype u has Iu influence on individual fitness and Gu influence on its group’s fitness. The fitness of the uth genotype in the tth group, wtu, is 1 + Iu + Gt., where Gt. is the mean genotypic effect on group fitness in the tth group (i.e., Σ ftuGu, summed across the u genotypes in the tth group, where ftu is frequency of the uth genotype in the tth group).
The recursive equation representing the selective change in allele frequency (Δp) equals the sum of the change in allele frequency by selection within (Δpwithin) and among (Δpamong) groups.
The within-group component is
| (1) |
in which wt. is the fitness of the tth group, ft. is its frequency, and Δpt. is the change in allele frequency within this group. For a given group t,
| (2) |
where pu|t is the frequency of the uth genotype in the tth group, fAu is the frequency of the A allele in the uth genotype, and wtu is the fitness of the uth genotype in the tth group.
The expression for the selective change in allele frequency among groups is
| (3) |
where w̄ represents the mean fitness of the population. I derived analytical solutions for all factorial combinations of n♂ and n♀ from 1 to 10 and to infinity and derived a general solution that holds true for each case.
Model 1: Autosomal Genetics
On autosomes, a single biallelic locus generates four diploid genotypes (two homozygotes and reciprocal heterozygotes). With biallelic modification, reciprocal heterozygotes have equivalent phenotypic effects (see “Model 1.1: Autosomal Genetics: Biallelic Modifiers” and “Model 2.1: X Chromosomes: Biallelic Modifiers,” below). Since the phenotypic effect of an imprinting allele depends on its parent of origin, reciprocal heterozygotes are separated in “Model 1.2: Autosomal Genetics: Imprinting,” “Model 2.2: X Chromosomes: Maternal Imprinting,” and “Model 2.3: X Chromosomes: Paternal Imprinting.” Phenotypic effects of all autosomal genotypes are presented in table 1. Throughout this article, I list the maternally derived allele first, followed by the paternally derived allele.
Table 1.
Phenotypic effects of autosomal genotype
| Model, genotype (j) | ij | gj |
|---|---|---|
| Biallelic: | ||
| AA | i(1 + iAA) | g(1 + gAA) |
| Aa = aA | i(1 + hiiAA) | g(1 + hg gAA) |
| aa | i | g |
| Maternal silencing: | ||
| AA = Aa | i(1 + iA_) | g(1 + gA_) |
| aA = aa | i | g |
| Paternal silencing: | ||
| AA = aA | i(1 + i_A) | g(1 + g_A) |
| Aa = aa | i | g |
Note: Genotype, j, has an ij influence on individual fitness and a gj influence on group fitness. The maternally derived allele is before the paternally derived allele. The dominance coefficient h is a continuous variable. Complete additivity is a special case in which h = 0.5, and A is dominant when h = 1.0 and recessive when h = 0. Underdominance is represented by h < 0 and overdominance is represented by h > 1.
Model 1.1: Autosomal Genetics: Biallelic Modifiers
Without imprinting, aa individuals have i and g influence on individual fitness and group fitness, respectively. AA individuals multiplicatively modify i and g by 1 + iAA and 1 + gAA, respectively, while heterozygotes multiplicatively modify the effects i and g by 1 + hiiAA and 1 + hggAA respectively, where hi is the dominance coefficient (table 1). The evolution of this unimprinted modifier provides a reference point for the imprinting effects described below.
For any combination of n♂ and n♀, the change in allele frequency due to selection on individual and group fitness components is
| (4a) |
where
| (4b) |
| (4c) |
Thus,
| (4d) |
Note that although group structure changes the partitioning of individual fitness effects iiAA[p + hi(q − p)] to within- and among-group components (eqq. [4b], [4c]), the total response to selection on individual fitness effects is insensitive to group structure (eq. [4d]; see Okasha 2004 for discussion). In contrast, the response to selection on the group fitness component (ggAA/4)[p + hg(q − p)] is proportional to the sum of 1/n♀ and 1/n♂ in both the response to group selection (eq. [4c]) and the total response to selection (eq. [4d]). Thus, when social interactions occur among full sibs (i.e., n♀ = n♂ = 1, relatedness = 1/2), the response to group selection is half as strong as the response to individual selection. When competition is among genetically unrelated individuals (i.e., n♀ = n♂ = ∞), there is no response to group selection.
This system has one, nontrivial equilibrium, which is attained when
| (5) |
The stability of this equilibrium depends on the dominance relations. For example, when allele A does not indirectly influence fitness of other group members (g = 0), equation (5) becomes the classic result for a biallelic locus: that is, Δp = 0 when hi = p̂/(p̂ − q̂). This result is attainable only when hi > 1 (overdominance) or hi < 0 (underdominance), and only the former is stable (e.g., Crow and Kimura 1970, pp. 270–272).
When the modifier allele, A, is rare and fully dominant (i.e., hi = hg = 1), equation (4d) becomes
| (6a) |
By noting that pq/w̄ is nonnegative and then solving the inequality Δp > 0, I derive the condition in which a rare biallelic modifier mutant can increase in frequency:
| (6b) |
The correspondence of equation (6b) with Hamilton’s rule is straightforward: the left-hand side of the inequality is the relatedness coefficient (1/2 for full sibs, 1/4 for half sibs, etc.), the numerator of the right-hand side is the effect of the allele on individual fitness, and the denominator of the right-hand side is the effect on group fitness.
Variation in n♂ and n♀
When a population consists of more than one family type, n♂ and n♀ may not necessarily be replaceable by the mean number of mothers and fathers per group. Assuming that group productivity increases linearly with every additional mother but is insensitive to the number of fathers, the change in allele frequency in a population in which the frequency of a group with n♂ fathers and n♀ mothers is fn♂,n♀, and it equals
| (7) |
In the numerator, the summation is taken across all combinations of groups with n♂ fathers and n♀ mothers. In the denominator, the summation is taken with respect to all groups with n♀ mothers. Here, Δpn♂, n♀ is the total selective change in allele frequency expected in a population in which all groups have n♀ mothers and n♂ fathers (eqq. [4a]–[4d]).
Equation (7) is the frequency of each group type, multiplied by the expected change in allele frequency from this group, weighted by the number of offspring produced per group, summed across all groups. Applying equation (7) to an autosomal, biallelically expressed locus yields
| (8) |
where is the arithmetic mean number of mothers per group and Hn♂ is the harmonic mean number of fathers per group.
The response to individual selection is insensitive to n♀ and n♂, and it is therefore not surprising that it is unaffected by variation in these values. In contrast, the response to group selection is proportional to the sum of the reciprocal of the arithmetic and harmonic mean number of mothers and fathers per group, respectively (eq. [8]; see fig. 1 for a detailed example).
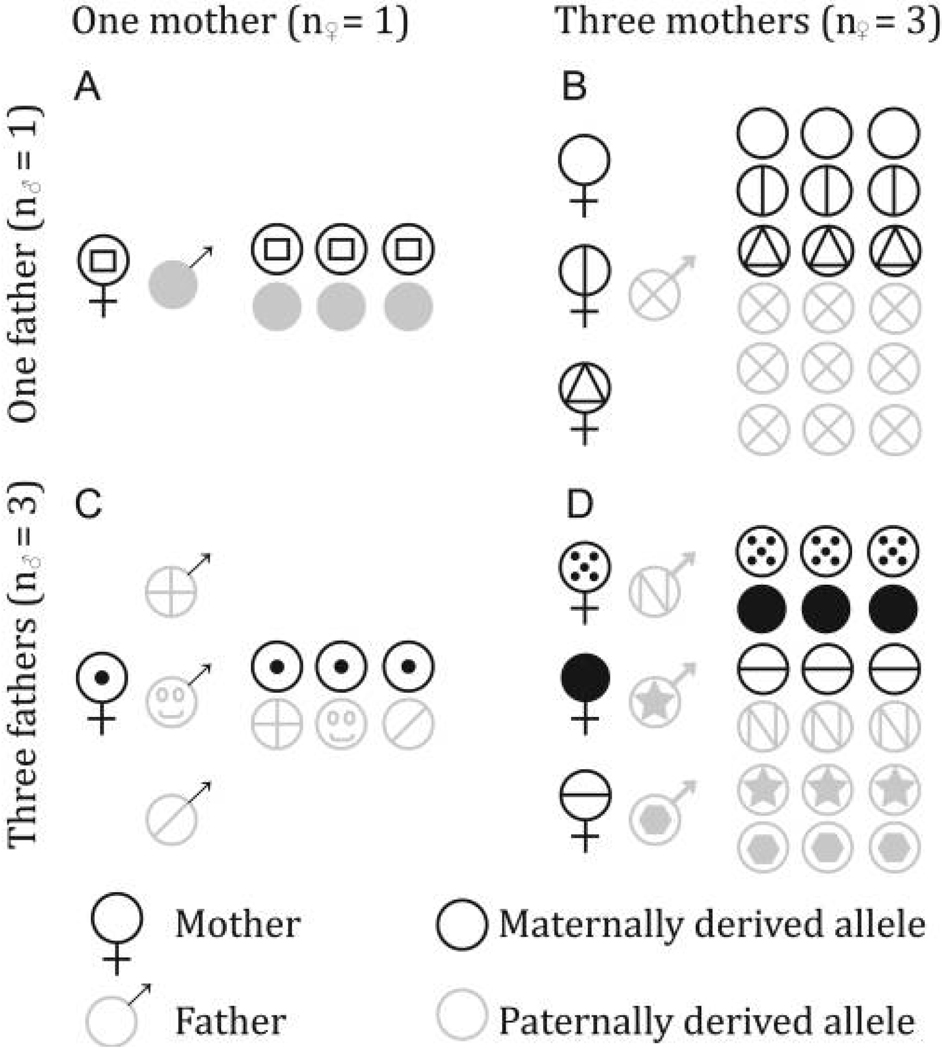
Figure 1.
A population consisting of four groups (A–D) with combinations of one or three mothers and one or three fathers. Each mother has equal reproductive output, while each father sires n♀/n♂ offspring. Half of paternally derived genes are in monopaternal groups (A and B). Of the remaining offspring, one-third of group members share fathers (matching gray symbols in C or D), while two-thirds do not (nonmatching gray symbols in C or D). Note that 1/2 × 1 + 1/2 × 1/3 = 2/3, the reciprocal of the harmonic mean number of fathers per group (1/Hn♂). In contrast, one-fourth of maternally derived genes are in monomaternal groups (A and C), while of the remaining maternally derived genes (3/4; B and D), only one-third of group members share a mother (matching black symbols in B or D). Note that 1/4 × 1 + 3/4 × 1/3 = 1/2, the reciprocal of the arithmetic mean number of mothers per group. This is the difference between the response to selection of a biallelic modifier or a maternal or paternal imprinting allele (eqq. [8], [9], and [10], respectively).
As a consequence of this asymmetric influence of variation in mate number, the efficacy of group selection is especially sensitive to groups with the least number of fathers. This result suggests that even when most females mate multiply, selection among groups may be quite potent so long is there is some variation in the number of mates per female. This result mirrors work on a kin-selection model with a variable number of matings per female (Wade 1982, 1985). In contrast, the response to among-group selection is divided by the arithmetic mean number of mothers per group, and it is therefore not particularly sensitive to a few males with exceptionally small harems.
Model 1.2: Autosomal Genetics: Imprinting
Here I investigate the evolution of maternal and paternal imprinting. For maternal silencing, both AA and Aa genotypes modify phenotypic influences on group (g) and individual (i) fitness by gA_ and iA_, respectively. Similarly, with paternal silencing, AA and aA individuals modify phenotypic influences on group (g) and individual (i) fitness by g_A and i_A, respectively (table 1).
The Δp recursions for maternal and paternal silencing alleles, incorporating variation in n♂ and n♀ following equation (7), are
| (9a) |
| (9b) |
These results are qualitatively similar to equations (6a) and (6b), representing the change in allele frequency of a rare, dominant modifier that is biallelically expressed, and they parallel previous work on the evolution of imprinting without conflict (Spencer 1997). Note that the evolution of maternal imprinting and that of paternal imprinting are very similar. Relative to the evolution of maternal silencing, in the paternal case, Hn♂ replaces , while i_A and g_A replace iA_ and gA_, respectively.
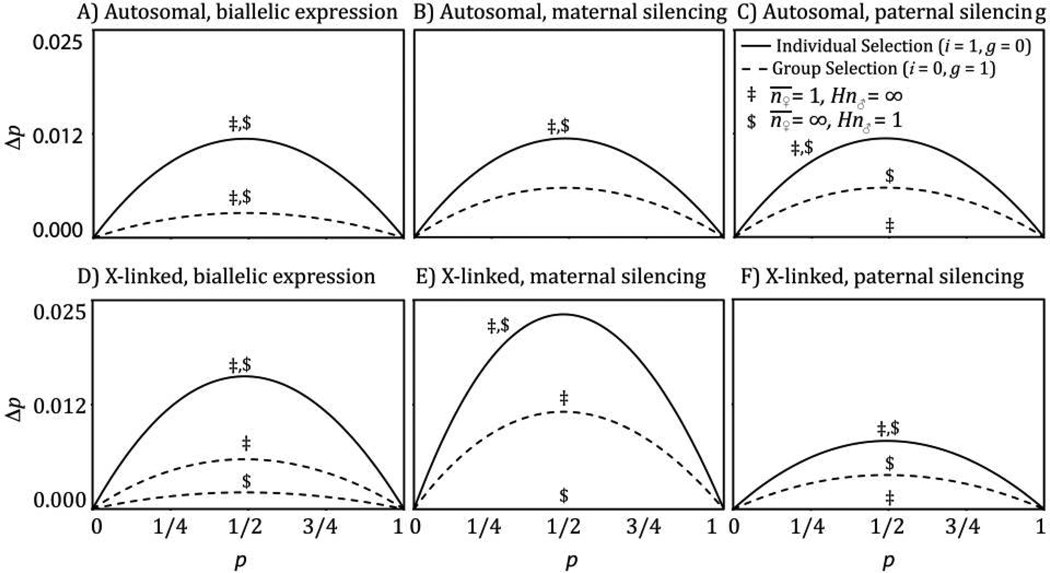
In figure 2A–2C, I present the relationship between p and Δp for an autosomal locus with a biallelically expressed, additive modifier (fig. 2A) or a maternal (fig. 2B) or a paternal (fig. 2C) silencer. Note that selection on maternal silencers is insensitive to n♂, and similarly, the evolution of paternal silencers is insensitive to n♀. Note further that the response to group selection in groups of maternal sibs born to infinitely mating females (, Hn♂ = ∞) is doubly as strong for maternal silencing as it is for biallelic modification, while this population structure favors only the silencing of paternally derived alleles when this silencing increases individual fitness (cf. fig. 2A, fig. 2B, and 2C).
Figure 2.
Δp as a function of p. Solid lines represent only effects on individuals (i = 1, g = 0), while dashed lines represent only effects on group fitness (i = 0, g = 1). Labels above dashed lines represent the mating structure. Allele increases individual or group fitness (iAA = gAA = iA_ = gA_ = 0.1), allelic effects are additive (hi = hg = 1/2), dosage compensation is complete (kAg = kag = kg = kAi = kai = ki = 1), and no expression doubles the effect of uniparental expression (i0 = 2iA_, g0 = 2gA_).
Here, the sign of Δp is independent of p. Thus, unlike the biallelic case and in contrast to the findings of Spencer et al. (1998), this model does not lead to a stable polymorphism. Therefore, the conditions in which a rare imprinting allele can increase in frequency are equivalent to the fixation conditions:
| (10a) |
| (10b) |
Although the model of Spencer et al. (1998) maintains a polymorphism, the absence of a stable polymorphism resembles previous work on the evolution of complete allelic silencing by imprinting, which precluded a stable polymorphism (Pearce and Spencer 1992).
Model 2: X Chromosomes
A biallelic, X-linked (or haplodiploid) locus is composed of six possible genotypes. As above, heterozygote mothers are treated equivalently, while heterozygote daughters are treated separately. Effects of the uth genotype on iu and gu are presented in table 2. Female genotypic effects are the same as those in the autosomal model. Male genotypic effects are scaled by the degree of dosage compensation kjl, where j is the haploid genotype and l is the trait. Thus, I allow influences on both group and individual fitness, as well as different genotypes, to have different degrees of dosage compensation.
Table 2.
Phenotypic effects of X-linked genotype
| Model, genotype (j) | ij | gj |
|---|---|---|
| No imprinting: | ||
| AA | i(1 + iAA) | g(1 + gAA) |
| Aa = aA | i(1 + hi iAA) | g(1 + hg gAA) |
| aa | i | g |
| Ay | i(1 + ki iAA) | g(1 + kg gAA) |
| ay | ikai | gkag |
| Maternal silencing: | ||
| AA = Aa | i(1 + iA_) | g(1 + gA_) |
| aA = aa | i | g |
| Ay | i(1 + i0) | g(1 + g0) |
| ay | iki | gkg |
| Paternal silencing: | ||
| AA = aA | i(1 + i_A) | g(1 + g_A) |
| aA = aa | i | g |
| Ay = ay | iki | gkg |
Note: Genotype, j, has an ij influence on individual fitness and a gj influence on group fitness. The maternally derived allele is listed before the paternally derived allele. The two male genotypes are written as Ay and ay, representing that males are heterogametic and inherit their Y chromosome paternally. Dosage compensation is represented by k, and it is allowed to vary by the genotype, which is denoted by a subscript. The scenarios where k = 1 represents complete dosage compensation (i.e., phenotypic effects are equal across the sexes); k = 1/2 represents no dosage compensation (male phenotypic effects are half as severe as female phenotypic effects).
Changes in allele frequencies are derived separately for males and females. Total Δp is calculated by scaling Δp♂ by 1/3 and Δp♀ by 2/3 (e.g., Owen 1986; Wade 2001), representing the fact that two of every three X-linked alleles occur in females. This approximation does not take into account any sex differences in departures from Hardy-Weinberg, and therefore it assumes weak selection.
Model 2.1: X Chromosomes: Biallelic Modifiers
As in the autosomal case, the X-linked, unimprinted model describes a system in which a modifier influences the effect of an invariant allele. Incorporating variation in n♂ and n♀, the change in allele frequencies within each sex and across the entire population is
| (11a) |
| (11b) |
| (11c) |
| (11d) |
It may be somewhat counterintuitive that the response to selection in females is related to the degree of dosage compensation in males (gAAkAg − kag appears in eq. [11a]) and that the response to selection by males depends partly on the degree of dominance in females (gAA[p + hg(q − p)] appears in eq. [11b]). However, these results mirror recent discoveries regarding the efficacy of selection on male-specific mitochondrial effects (Unckless and Herren 2009; Wade and Brandvain 2009), in which sex-specific effects are indirectly selected in the other sex as a result of effects on group fitness. Note that the response to group selection in females is sensitive to both the number of mothers and the number of fathers per group, while the response to group selection in males is insensitive to the number of fathers per group. This result follows from the transmission genetics of sex chromosomes: males do not inherit paternally derived X alleles.
When rare, the A allele can invade if . As in the autosomal case, X chromosomes can maintain a polymorphism. The equilibrium allele frequency (which may be stable or unstable depending on dominance coefficients and dosage compensation) is
| (12) |
Model 2.2: X Chromosomes: Maternal Imprinting
Here I investigate the evolution of a maternal imprinting allele. AA and Aa females modify phenotypic effects on group (g) and individual (i) fitness by 1 + gA_ and 1 + iA_, respectively. In males, silencing of a maternally derived gene on the X chromosome results in no expression. I allow this lack of expression to alter both i and g by 1 + i0 and 1 + g0, respectively. The biological value of this null genotype is unclear, and it strongly influences my results. For example, zero expression of an essential gene could be nearly lethal, and in this case i0 and g0 would be nearly −1. In contrast, zero expression may double the effect of silencing one copy, such that i0 and g0 equal 2iA_ and 2gA_, respectively. The relationship between the unimprinted alleles (i.e., the a allele) and phenotype is much simpler, as ay males influence group and individual contributions to fitness by the standard dosage compensation, kg and ki, respectively (table 2).
The changes in allele frequencies due to selection within each sex and across the entire population are as follows:
| (13a) |
| (13b) |
| (13c) |
Since only half of the X chromosomes in females are maternally derived, while all X chromosomes in males are maternally derived, it is not surprising that the response to selection on maternal silencing of an allele on the X chromosome is approximately half as strong in females as it is in males.
As in the autosomal case, maternal imprinting on the X chromosome cannot sustain a stable polymorphism. Thus, the fixation criterion is equivalent to the invasion criterion, which is .
Model 2.3: X Chromosomes: Paternal Imprinting
When A silences a paternally derived allele in cis, AA and aA females modify phenotypic effects on group (g) and individual (i) fitness by 1 + gA_ and 1 + iA_, respectively (table 2). Silencing of a paternally derived gene on the X chromosome has no influence on males, and thus Δp♂ = 0. Thus, the total change in allele frequency of an X-linked paternal silencing allele is
| (14) |
This result is substantially less than the Δp recursions derived for biallelic modifiers, maternal imprinting, and paternal imprinting on autosomes. Figure 2D–2F shows Δp as a function of p for all forms of expression modification on the X chromosome. The modest response to selection for epigenetic modification of the X chromosome in fathers is apparent in figure 2F.
As only one-third of X chromosomes are paternally derived, the weak response to selection on epigenetic silencing of the paternal X mirrors recent empirical and theoretical developments in the evolutionary consequences of relaxed selection (Barker et al. 2005; Demuth and Wade 2007; Cruickshank and Wade 2008). As above, there is no stable equilibrium, and the invasion (fixation) criterion is 2Hn♂iiA_ > −ggA_.
Discussion
The differential expression of an allele on the basis of its parent of origin is known as genomic imprinting. I investigated the evolution of genomic imprinting when fitness is influenced by social interactions among cohorts. The findings presented herein have novel consequences for the evolution of genomic imprinting.
For example, by considering interactions within a cohort rather than focusing on direct transfer of nutrients from parent to offspring, my model suggests that genes influencing social behavior may be imprinted in a variety of species without direct resource transfer from mother to offspring. Limited support for this hypothesis comes from quantitative trait locus evidence suggesting that imprinted expression occurs in chickens (Campos et al. 2009; Rowe et al. 2009), a species with strong, heritable effects on individual and group fitness (Bijma et al. 2007). There will surely be many opportunities for experimental tests of this hypothesis, as advances in high-throughput expression arrays may uncover additional cases of genomic imprinting. Related extensions of the conflict theory to social insects (e.g., Queller 2003; Kronauer 2008; Wild and West 2009), in which there is growing evidence for genomic imprinting (Kronforst et al. 2008; Elango et al. 2009), also provide excellent tests for the explanatory power of the conflict theory to taxa without a placental-like habit.
Moreover, the arena of competitive social interactions can change over development, as early competition may occur among maternal siblings in a nest while later competition may occur among offspring sired by a limited number of males. Under this scenario, a gene that increases individual fitness at a cost to group fitness may be maternally silenced early in development but paternally silenced later. Some imprinted genes show this ontogenic pattern (e.g., Wolf et al. 2008) and will provide excellent tests for the theory presented in this study.
As in previous models (Spencer et al. 1998, 2004), the invasibility conditions derived herein show that even with single mating and one female per group, imprinting can evolve. However, in these cases, imprinting does not represent the sole mechanism of a response to selection, as unimprinted modification is also plausible (Hurst 1999). Unlike previous models of the conflict theory (Spencer et al. 1998, 2004), this derivation does not yield a stable polymorphism with imprinting loci stably coexisting with standard genic expression. Although the absence of a protected polymorphism is a difference from previous population-genetic models of the conflict theory, it is in agreement with population-genetic models of imprinting with complete inactivation (Pearce and Spencer 1992). Nonetheless, some imprinting loci are polymorphic with both imprinting expression and standard genic expression segregating within populations (Xu et al. 1993). It is plausible that my model is sufficient to explain these cases as transient polymorphisms; however, it is also possible that the observed polymorphisms suggest that this model may exclude some biologically meaningful regions of parameter space.
Iwasa and Pomiankowski (1999, p. 488) state that “the conflict theory should apply equally to X-linked and autosome linked genes.” They survey imprinting data from X chromosomes, argue that the data are unsupportive of the conflict theory, and present a unique model to explain the evolution of X-linked imprinting. I show that although invasion conditions for imprinting on the X chromosome are similar to those on autosomes, selection on paternal imprinting on the X chromosome is a relatively meager evolutionary force. Depending on the phenotypic effect of a complete lack of expression in males, maternally derived X chromosomes may respond rapidly to selection for epigenetic modification, and therefore the X chromosome may host an overabundance of maternal imprinting. This result is missed by models that analyze evolutionary stability and ignore the dynamic attainment of equilibria, highlighting the importance of dynamically explicit genetic approaches in the analysis of evolutionary conflicts.
Standard renditions of the conflict theory assume that competition occurs strictly among maternal siblings and that offspring compete for maternal resources. In this case, n♀ = 1 and n♂ is the number of mates per female. A recent extension of the conflict theory considers competition for paternal resources among current and future paternal siblings when a male’s partner changes across litters (Ùbeda 2008). In some ways my research, particularly the case in which n♂ = 1 and n♀ > 1, is conceptually similar to that of Ùbeda (2008); however, there are some important differences in the underlying biology of the system, as well as different evolutionary consequences.
In Ùbeda’s (2008) model, the essential conflict occurs among paternal siblings over paternally derived resources. In the related special case of my model (when n♂ = 1 and n♀ > 1), social interactions among paternal siblings influence competitive fitness, but paternal resource allocation is not necessary for the evolution of imprinting. As such, my model predicts that paternal silencing of genes that decrease family fitness can occur in the absence of direct paternal care, such as in cases of interference competition for local resources, while Ùbeda’s (2008) model does not allow for this possibility.
Large harems influence the competitive environment of many offspring, while promiscuously sired broods are no different in size than monogamously sired broods. Consequentially, the strength of group selection on maternal imprinting is weakened by the reciprocal of the arithmetic mean number of females per harem, while the response to group selection for paternal imprinting is dampened by the harmonic mean number of fathers per group (or, in the case of competition among maternal sibs, mates per female). The harmonic mean is particularly sensitive to lower-bound values of the distribution. Thus, a few exceptionally promiscuous females do not strongly influence the evolution of paternal imprinting, while rare males with large harems greatly influence the evolution of maternal imprinting.
Important caveats to this result are the possibility that female fitness may increase with each additional mating and that the increase in harem productivity with each additional mother may be sublinear. As returns diminish with an increase in the number of mothers per group, the multiple-mothers case becomes more similar to the multiple-fathers case. An additional biological complication that is not included in this model is reproductive skew, that is, the variation in reproductive success among mothers or fathers in a group. Reproductive skew could be accommodated simply by replacing n♀ and n♂ with the “effective” n♀ and n♂ (e.g., Wade 1982).
The theory presented herein may shed some light on the evolutionary-genetic basis of Prader-Willi syndrome (PWS) and Angelman syndrome (AS). PWS is associated with a deletion of a portion of the paternally derived chromosome 15, while AS has different clinical phenotypes and is associated with a deletion of the same region in the maternally derived copy (Knoll et al. 1989).
PWS children have low birth weights and difficulty suckling, but after weaning they are characterized by food stealing, insatiable appetites, stubbornness, and oppositional behavior (Martin et al. 1998). Phenotypes of the later phase of PWS are opposite to standard theoretical predictions of the conflict theory (but see Haig and Wharton 2003;Ùbeda 2008), and therefore, this “biphasic” phenotype is a challenge to the conflict theory (Hurst and McVean 1997). If competition from early development through weaning is among current and future maternal sibs, but after weaning, competition is among a larger paternal sib-group, then the antisocial behavior associated with a lack of paternally derived regions of chromosome 15 would be broadly consistent with the extended version of the conflict theory presented herein.
AS is perhaps a more complex case, as it is mainly characterized by a few behavioral and mental abnormalities. Nonetheless, the behavioral phenotypes associated with AS display a striking correspondence to the novel predictions of this model. Children with AS display Duchenne laughter and smiles (Horsler and Oliver 2006), phenotypes that, according to Gervais and Wilson (2005), evolved as a product of group selection. This interpretation is broadly consistent with the notion that group selection on paternally derived genes is more efficacious than group selection on maternally derived genes, and it is in direct opposition to alternative constructions of the conflict theory, which have searched, perhaps in vain, for group fitness costs associated with AS and group fitness benefits associated with PWS (Haig 2000, 2008; Haig and Wharton 2003).
The model presented herein provides unique explanations of two human genetic diseases, and it calls for novel empirical investigations that can differentiate it from alternative explanations of the evolution of genomic imprinting as well as other instantiations of the conflict theory. I predict that with strong competition among paternal siblings of many dams, genes that increase group fitness at a cost to individual fitness will be silenced when maternally inherited. The best direct tests of the theory presented herein would investigate the genomic imprinting in species with intense local competition among paternal siblings, and particularly species in which males control large harems, as well as polygynandrous populations. Such investigation will allow for critical tests of the theory presented herein, as well as a strong test of the general importance of kin competition in the evolution of genomic imprinting.
Acknowledgments
This article benefited from comments by L. Moyle and M. J. Wade and members of their lab groups, as well as by L. Delph, N. Johnson, C. Lively, M. Patten, and T. Platt. This work was made possible by support from a National Institutes of Health (NIH) training grant, Common Themes in Reproductive Diversity; a National Science Foundation (NSF) predoctoral fellowship to Y.B.; an NSF Doctoral Dissertation Improvement grant to Y.B.; and NIH grant R01-GM084238 (to M. J. Wade).
Literature Cited
- Barker MS, Demuth JP, Wade MJ. Maternal expression relaxes constraint on innovation of the anterior determinant, bicoid. PLoS Genetics. 2005;1:527–530. doi: 10.1371/journal.pgen.0010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS. Genomic imprinting: employing and avoiding epigenetic processes. Genes and Development. 2009;23:2124–2133. doi: 10.1101/gad.1841409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger F, Chaudhury A. Parental memories shape seeds. Trends in Plant Science. 2009;14:550–556. doi: 10.1016/j.tplants.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Bijma P, Muir WM, Ellen ED, Wolf JB, van Arendonk JAM. Multilevel selection 2: estimating the genetic parameters determining inheritance and response to selection. Genetics. 2007;175:289–299. doi: 10.1534/genetics.106.062729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos RLR, Nones K, Ledur MC, Moura A, Pinto LFB, Ambo M, Boschiero C, et al. Quantitative trait loci associated associated with fatness in a broiler-layer cross. Animal Genetics. 2009;40:729–736. doi: 10.1111/j.1365-2052.2009.01910.x. [DOI] [PubMed] [Google Scholar]
- Crow JF, Kimura M. An introduction to population genetics theory. New York: Harper & Row; 1970. [Google Scholar]
- Cruickshank T, Wade MJ. Microevolutionary support for a developmental hourglass: gene expression patterns shape sequence variation and divergence in Drosophila. Evolution and Development. 2008;10:583–590. doi: 10.1111/j.1525-142X.2008.00273.x. [DOI] [PubMed] [Google Scholar]
- Day T, Bonduriansky R. Intralocus sexual conflict can drive the evolution of genomic imprinting. Genetics. 2004;167:1537–1546. doi: 10.1534/genetics.103.026211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuth JP, Wade MJ. Maternal expression increases the rate of bicoid evolution by relaxing selective constraint. Genetica. 2007;129:37–43. doi: 10.1007/s10709-006-0031-4. [DOI] [PubMed] [Google Scholar]
- Elango N, Hunt BG, Goodisman MAD, Yi SV. DNA methylation is widespread and associated with differential gene expression in castes of the honeybee, Apis mellifera. Proceedings of the National Academy of Sciences of the USA. 2009;106:11206–11211. doi: 10.1073/pnas.0900301106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervais M, Wilson DS. The evolution and functions of laughter and humor: a synthetic approach. Quarterly Review of Biology. 2005;80:395–430. doi: 10.1086/498281. [DOI] [PubMed] [Google Scholar]
- Hager R, Cheverud JM, Leamy LJ, Wolf JB. Sex dependent imprinting effects on complex traits in mice. BMC Evolutionary Biology. 2008;8:9. doi: 10.1186/1471-2148-8-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. Parental antagonism, relatedness asymmetries, and genomic imprinting. Proceedings of the Royal Society B: Biological Sciences. 1997;264:1657–1662. doi: 10.1098/rspb.1997.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D. Genomic imprinting, sex-biased dispersal, and social behavior. Annals of the New York Academy of Sciences. 2000;907:149–163. doi: 10.1111/j.1749-6632.2000.tb06621.x. [DOI] [PubMed] [Google Scholar]
- Haig D. Kinship asymmetries and the divided self. Behavioral and Brain Sciences. 2008;31:271–272. [Google Scholar]
- Haig D, Wharton R. Prader-Willi syndrome and the evolution of human childhood. American Journal of Human Biology. 2003;15:320–329. doi: 10.1002/ajhb.10150. [DOI] [PubMed] [Google Scholar]
- Hall JG. Genomic imprinting: review and relevance to human diseases. American Journal of Human Genetics. 1990;46:857–873. [PMC free article] [PubMed] [Google Scholar]
- Horsler K, Oliver C. The behavioural phenotype of Angelman syndrome. Journal of Intellectual Disability Research. 2006;50:33–53. doi: 10.1111/j.1365-2788.2005.00730.x. [DOI] [PubMed] [Google Scholar]
- Hurst LD. Is multiple paternity necessary for the evolution of genomic imprinting? Genetics. 1999;153:509–512. doi: 10.1093/genetics/153.1.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, McVean GT. Growth effects of uniparental disomies and the conflict theory of genomic imprinting. Trends in Genetics. 1997;13:436–443. doi: 10.1016/s0168-9525(97)01273-0. [DOI] [PubMed] [Google Scholar]
- Hurst LD. Do we understand the evolution of genomic imprinting? Current Opinion in Genetics and Development. 1998;8:701–708. doi: 10.1016/s0959-437x(98)80040-3. [DOI] [PubMed] [Google Scholar]
- Itier JM, Tremp GL, Leonard JF, Multon MC, Ret G, Schweighoffer F, Tocque B, Bluet-Pajot MT, Cormier V, Dautry F. Imprinted gene in postnatal growth role. Nature. 1998;393:125–126. doi: 10.1038/30120. [DOI] [PubMed] [Google Scholar]
- Iwasa Y, Pomiankowski A. Sex specific X chromosome expression caused by genomic imprinting. Journal of Theoretical Biology. 1999;197:487–495. doi: 10.1006/jtbi.1998.0888. [DOI] [PubMed] [Google Scholar]
- Iwasa Y. The evolution of X-linked genomic imprinting. Genetics. 2001;158:1801–1809. doi: 10.1093/genetics/158.4.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoll JHM, Nicholls RD, Magenis RE, Graham JM, Lalande M, Latt SA. Angelman and Prader-Willi syndromes share a common chromosome-15 deletion but differ in parental origin of the deletion. American Journal of Medical Genetics. 1989;32:285–290. doi: 10.1002/ajmg.1320320235. [DOI] [PubMed] [Google Scholar]
- Kronauer DJC. Genomic imprinting and kinship in the social Hymenoptera: what are the predictions? Journal of Theoretical Biology. 2008;254:737–740. doi: 10.1016/j.jtbi.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Kronforst MR, Gilley DC, Strassmann JE, Queller DC. DNA methylation is widespread across social Hymenoptera. Current Biology. 2008;18:R287–R288. doi: 10.1016/j.cub.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Lalande M. Parental imprinting and human disease. Annual Review of Genetics. 1996;30:173–195. doi: 10.1146/annurev.genet.30.1.173. [DOI] [PubMed] [Google Scholar]
- Martin A, State M, Koenig K, Schultz R, Dykens EM, Cassidy SB, Leckman JF. Prader-Willi syndrome. American Journal of Psychiatry. 1998;155:1265–1273. doi: 10.1176/ajp.155.9.1265. [DOI] [PubMed] [Google Scholar]
- Mills W, Moore T. Polyandry, life-history trade-offs and the evolution of imprinting at Mendelian loci. Genetics. 2004;168:2317–2327. doi: 10.1534/genetics.104.030098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, Haig D. Genomic imprinting in mammalian development: a parental tug-of-war. Trends in Genetics. 1991;7:45–49. doi: 10.1016/0168-9525(91)90230-N. [DOI] [PubMed] [Google Scholar]
- Morison IM, Ramsay JP, Spencer HG. A census of mammalian imprinting. Trends in Genetics. 2005;21:457–465. doi: 10.1016/j.tig.2005.06.008. [DOI] [PubMed] [Google Scholar]
- Okasha S. Multi-level selection and the partitioning of covariance: a comparison of three approaches. Evolution. 2004;58:486–494. [PubMed] [Google Scholar]
- Pearce GP, Spencer HG. Population genetic models of genomic imprinting. Genetics. 1992;130:899–907. doi: 10.1093/genetics/130.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen RE. Gene frequency clines at X-linked or haplodiploid loci. Heredity. 1986;57:209–219. doi: 10.1038/hdy.1986.111. [DOI] [PubMed] [Google Scholar]
- Queller DC. Theory of genomic imprinting conflict in the social insects. BMC Evolutionary Biology. 2003;3:15. doi: 10.1186/1471-2148-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe SJ, Pong-Wong R, Haley CS, Knott SA, De Koning DJ. Detecting parent of origin and dominant QTL in a two-generation commercial poultry pedigree using variance component methodology. Genetics Selection Evolution. 2009;41:6. doi: 10.1186/1297-9686-41-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer HG. Mutation-selection balance under genomic imprinting at an autosomal locus. Genetics. 1997;147:281–287. doi: 10.1093/genetics/147.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer HG, Feldman MW, Clark AG. Genetic conflicts, multiple paternity and the evolution of genomic imprinting. Genetics. 1998;148:893–904. doi: 10.1093/genetics/148.2.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer HG, Feldman MW, Clark AG, Weisstein AE. The effect of genetic conflict on genomic imprinting and modification of expression at a sex-linked locus. Genetics. 2004;166:565–579. doi: 10.1534/genetics.166.1.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ùbeda F. Evolution of genomic imprinting with biparental care: implications for Prader-Willi and Angelman syndromes. PLoS Biology. 2008;6:1678–1692. doi: 10.1371/journal.pbio.0060208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ùbeda F, Wilkins JF. Imprinted genes and human disease: an evolutionary perspective. Advances in Experimental Medicine and Biology. 2008;626:101–115. [PubMed] [Google Scholar]
- Unckless RL, Herren JK. Population genetics of sexually antagonistic mitochondrial mutants under inbreeding. Journal of Theoretical Biology. 2009;260:132–136. doi: 10.1016/j.jtbi.2009.06.004. [DOI] [PubMed] [Google Scholar]
- Wade MJ. Kin selection: classical approach and a general solution. Proceedings of the National Academy of Sciences of the USA. 1978;75:6154–6158. doi: 10.1073/pnas.75.12.6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade MJ. The effect of multiple inseminations on the evolution of social behaviors in diploid and haplodiploid organisms. Journal of Theoretical Biology. 1982;95:351–368. doi: 10.1016/0022-5193(82)90250-8. [DOI] [PubMed] [Google Scholar]
- Wade MJ. The influence of multiple inseminations and multiple foundresses on social evolution. Journal of Theoretical Biology. 1985;112:109–121. doi: 10.1016/s0022-5193(85)80119-3. [DOI] [PubMed] [Google Scholar]
- Wade MJ. Maternal effect genes and the evolution of sociality in haplo-diploid organisms. Evolution. 2001;55:453–458. doi: 10.1554/0014-3820(2001)055[0453:megate]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Wade MJ, Brandvain Y. Reversing mother’s curse: selection on male mitochondrial fitness effects. Evolution. 2009;63:1084–1089. doi: 10.1111/j.1558-5646.2009.00614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild G, West SA. Genomic imprinting and sex allocation. American Naturalist. 2009;173:E1–E14. doi: 10.1086/593305. [DOI] [PubMed] [Google Scholar]
- Wilkins JF, Haig D. Inbreeding, maternal care and genomic imprinting. Journal of Theoretical Biology. 2003a;221:559–564. doi: 10.1006/jtbi.2003.3206. [DOI] [PubMed] [Google Scholar]
- Wilkins JF. What good is genomic imprinting: the function of parent-specific gene expression. Nature Reviews Genetics. 2003b;4:359–368. doi: 10.1038/nrg1062. [DOI] [PubMed] [Google Scholar]
- Wolf JB. Cytonuclear interactions can favor the evolution of genomic imprinting. Evolution. 2009;63:1364–1371. doi: 10.1111/j.1558-5646.2009.00632.x. [DOI] [PubMed] [Google Scholar]
- Wolf JB, Hager R. A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biology. 2006;4:2238–2243. doi: 10.1371/journal.pbio.0040380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Cheverud JM, Roseman C, Hager R. Genome-wide analysis reveals a complex pattern of genomic imprinting in mice. PLoS Genetics. 2008;4:e1000091. doi: 10.1371/journal.pgen.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YQ, Goodyer CG, Deal C, Polychronakos C. Functional polymorphism in the parental imprinting of the human IGF2R gene. Biochemical and Biophysical Research Communications. 1993;197:747–754. doi: 10.1006/bbrc.1993.2542. [DOI] [PubMed] [Google Scholar]