Abstract
The equilibrium sequence diversity of genes within a population and the rate of sequence divergence between populations or species depends on a variety of factors, including expression pattern, mutation rate, nature of selection, random drift, and mating system. Here, we extend population genetic theory developed for maternal-effect genes to predict the equilibrium polymorphism within species and sequence divergence among species for genes with social effects on fitness. We show how the fitness effects of genes, mating system, and genetic system affect predicted gene polymorphism. We find that, because genes with indirect social effects on fitness effectively experience weaker selection, they are expected to harbor higher levels of polymorphism relative to genes with direct fitness effects. The relative increase in polymorphism is proportional to the inverse of the genetic relatedness between individuals expressing the gene and their social partners that experience the fitness effects of the gene. We find a similar pattern of more rapid divergence between populations or species for genes with indirect social effects relative to genes with direct effects. We focus our discussion on the social insects, organisms with diverse indirect genetic effects, mating and genetic systems, and we suggest specific examples for testing our predictions with emerging sociogenomic tools.
Keywords: Indirect genetic effects, kin selection, mutation–selection balance, relaxed selective constraint, social evolution, sociogenomics
The equilibrium sequence diversity of a gene within a population is affected by many features of its population biology, including the rate of mutation, the nature of selection (e.g., balancing vs. directional), its pattern of expression (i.e., which individuals express the gene and its fitness effects), the effective population size, the mating system, and the genetic structure. For example, consider the different effects of recessive and dominant gene expression on the equilibrium polymorphism at mutation–selection balance. A deleterious recessive allele will achieve a higher equilibrium frequency than a deleterious dominant allele with the same selection coefficient, because, when rare, the majority of copies of the recessive are unexpressed and hidden away from natural selection in heterozygotes. Mating system interacts with expression because a deleterious recessive will reach a higher frequency in a randomly mating population than it will in an inbreeding population whereas a deleterious dominant is unaffected by inbreeding because its expression remains constant across mating systems. Similarly, a diploid gene expressed only by one sex, say females, will have twice the equilibrium heterozygosity as a gene expressed in both sexes, all else being equal, because of the number of unexpressed copies in males (Wright 1960; Whitlock and Wade 1995; Wade 1998, 2001; Barker et al. 2005; Demuth and Wade 2007; Wade et al. 2008). More generally, all factors that decrease the association between genotype and phenotype decrease the proportion of gene copies that are seen by selection and, thus, result in higher equilibrium polymorphism within populations at mutation–selection balance. When the phenotypic effects of a gene expressed in one individual are manifested in a different individual, as for example, when maternal genes affect offspring phenotype (i.e., maternal genetic effects), the association between genotype and phenotype is decreased and within-population polymorphism levels are increased (Wade 1998, 2001). In each of these cases, the higher level of polymorphism is the result of the relaxed selection, often called relaxed selective constraint (Demuth and Wade 2007; J. D. Van Dyken and M. J. Wade, unpubl. ms.) that attends the weaker association between genotype and phenotype.
Higher levels of polymorphism from relaxed selective constraint lead to higher levels of equilibrium additive genetic variance for fitness (Whitlock and Wade 1995; Wade 1998). When genetic variance within populations is converted to variance among populations by the combined action of random genetic drift and local selection, it results in higher rates of evolution measured as the number of fixed, nonsynonymous substitutions between two taxa (Demuth and Wade 2007; Cruickshank and Wade 2008; J. D. Van Dyken and M. J. Wade, unpubl. ms.). Divergence between species is often interpreted as evidence of adaptive evolution by diversifying natural selection (i.e., local selection), although relatively rapid divergence can also result from relaxed selective constraint (i.e., reduced effectiveness of purifying selection followed by divergence by random genetic drift). For example, maternally expressed genes can accumulate fixed differences between taxa several times more rapidly than a homologue with direct effects expressed in both sexes (Demuth and Wade 2007; Cruickshank and Wade 2008).
Here, we extend population genetic theory developed to predict polymorphism at maternal-effect genes to genes with other types of indirect effects arising from social interactions, such as kin selected effects. These indirect genetic effects, also known as associative effects, occur when the phenotype of an individual is affected by genes expressed in its social partners (Cheverud and Moore 1994; Moore et al. 1997; Wolf et al. 1998; Bijma et al. 2007). We are specifically interested in determining the “relative” levels of polymorphism at genes with direct effects, maternal effects, and sib effects, in haplo-diploid (HD) and diplo-diploid (DD) populations with both single and multiple mating. Thus, we are interested in how the patterns of gene expression, genetic system, and mating system affect expected sequence diversities. Higher levels of polymorphism within species for some genes allow these genes to respond more quickly to selection under changing conditions or to diversify more rapidly among populations when genetically subdivided.
Although our results apply to all social organisms in which phenotypes are influenced by the genotypes of interacting social partners, including model organisms usually considered to be solitary, for example, Drosophila (Wolf 2003) and Arabidopsis (Mutic and Wolf 2007), we focus our discussion on the social insects for three reasons: (1) Social interactions have special significance in social insect colonies; as described below, a variety of different indirect effects are theoretically important in the expression, evolution, and development of social insect phenotypes (Linksvayer and Wade 2005), and indeed recent empirical studies explicitly designed to estimate the relative contributions of direct and indirect genetic effects to variation for social insect phenotypes demonstrate the importance of indirect effects arising from social interactions (Linksvayer 2006, 2007; Wang et al. 2008; Linksvayer et al. 2009). (2) Social insects have diverse genetic and mating systems, so that our discussion focused on social insects applies broadly to organisms with divergent genetic and mating systems. (3) The emerging field of sociogenomics is rapidly growing (e.g., Robinson et al. 2005; Sinha et al. 2006; Barchuk et al. 2007; Toth et al. 2007), especially with the availability of the honey bee genome (Honey Bee Genome Sequencing Consortium 2006), and theory explicitly incorporating the complexities of patterns of gene expression, mating systems, and genetic systems in social organisms are required to make specific evolutionary genetic predictions to be tested with this new empirical resource.
In the social insects, phenotypes are influenced by a variety of different genetic and environmental pathways. The genetic pathways include genes with direct effects, expressed during the development of males and females, genes with maternal effects expressed only in queens or paternal effects expressed only in males, as well as genes with sib effects, expressed in adult workers or in sibling larvae (Linksvayer and Wade 2005; Linksvayer 2006, 2007). As we show here, much of the phenotypic variation among the social insects may be the result of relaxed selective constraint and its interaction with diversifying, adaptive selection. Furthermore, when worker phenotypes have indirect fitness effects on queen and male phenotypes, then genes affecting worker phenotypes and division of colony labor should harbor higher levels of polymorphism. This may be an important factor contributing to the well-known diversity of species and social phenotypes in these taxa.
Maternal Genetic Effects Theory
Using a single locus population genetic model, Wade (1998) argued that, because selection on a maternal-effect gene is half as effective as selection on a direct-effect gene expressed within populations, nonsynonymous or replacement site diversity should be two times higher, at equilibrium, for a maternal-effect gene than for a direct-effect gene. This prediction assumes that the maternal- and direct-effect loci being compared experience similar strengths of selection when expressed, and furthermore that there have not been recent selective sweeps at the two loci or closely linked loci distorting the expected equilibrium. Among populations or lineages, for purifying selection, a greater rate of replacement site divergence is predicted for a maternal-effect gene than for a direct-effect gene because slightly deleterious mutations for maternal-effect genes are more likely to fix (Demuth and Wade 2007). Once again, this prediction assumes similar selection coefficients between the loci being compared as well as similar effective population sizes between the lineages being compared. When two genes with different expression patterns are compared for their relative levels of polymorphism within the same species or their relative levels of diversification across the same set of taxa, they necessarily have similar effective population sizes. Selection must be more than twice as strong on a maternal-effect gene in order for its diversification by positive selection to be more rapid than the rate of diversification by positive selection of a direct-effect gene.
In quantitative support of this theoretical prediction, Barker et al. (2005) compared sequence diversity of the maternal-effect gene, bicoid, with that of its zygotically expressed, tandem duplicate, zerknüllt (zen) within Drosophila melanogaster and D. simulans. They found that sequence diversity in the coding regions of bicoid is at least twice that of zen in both species as predicted. In contrast, the ratios of neutral, silent-site sequence diversity (third position base pairs and introns) were approximately one. This result is consistent with both loci experiencing similar strengths of purifying selection, but selection at the maternal effect bicoid being half as effective as selection at the direct effect zen, as predicted. For genes acting in early Drosophila development, Cruickshank and Wade (2008) found a twofold elevation in replacement site diversity for maternal-effect genes as a class relative to zygotic genes, once again consistent with these highly conserved genes experiencing on average similar strengths of purifying selection when expressed.
Following Kimura (1962), Demuth and Wade (2007, see their Fig. 1) quantify the rate of fixation of a maternal-effect gene relative to that of a direct-effect gene as a function of s, the average selection coefficient, assuming no dominance. For positively selected genes (where s > 0), the probability of fixation of a maternal-effect gene is always less than that of a direct-effect gene, with the relative rate asymptoting at a value of 0.50 as s exceeds +0.01. That is, positive selection acting on a maternal-effect gene is only half as effective as similar selection on a direct-effect gene. Conversely, for purifying selection acting against deleterious alleles, where the average s is negative (i.e., s < 0), the relative probability of fixation for a maternal-effect gene exceeds that of a direct-effect gene. For example, for s ~ −0.01, the probability of fixation is more than four times higher. Among taxa in the cyclorrhaphan flies (Stauber et al. 1999, 2002), the coding region of bicoid is evolving 1.67 times faster than zen (Demuth and Wade 2007), consistent with a predominance of purifying selection. However, among species within the genus Drosophila, where it is known that bicoid is under positive selection, it is evolving at a rate of only 0.68 that of zen, close to the limiting ratio of 0.50. Together these empirical tests demonstrate the utility of population genetic theory for predicting patterns of sequence variation within and between populations.
Figure 1.
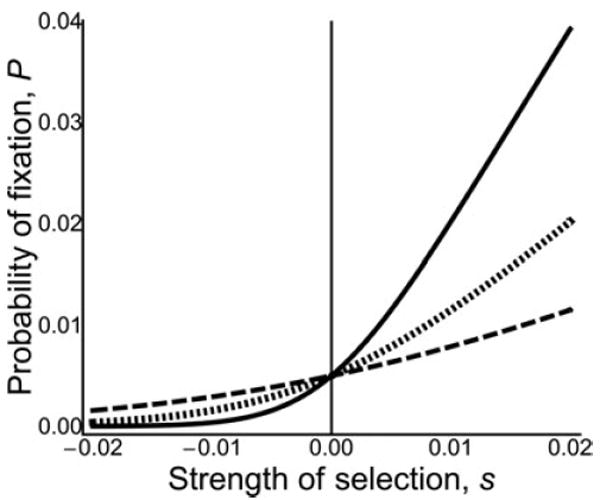
Probability of fixation of new mutants in a diplo-diploid population with different patterns of expression for weak negative to weak positive selection. Alleles with direct effects (solid line); alleles with maternal effects or sib effects in single-mating full-sibling family (dotted line); and alleles with sib effects in a multiple-mating half-sibling family (dashed line). Alleles with indirect effects are less likely to fix when beneficial and less likely to be lost when harmful. Ne = 100, p = 1/(2Ne).
Wade (2001) extended population genetic models for maternal-effect genes from diploid autosomal genes to X-linked and haplo-diploid genes. He showed that the effect of selection on maternal-effect genes is greater in haplo-diploids than in diplo-diploids and that it is not affected by inbreeding or mate numbers. However, these theoretical predictions have not yet been tested either with estimates of standing sequence diversity within populations or with estimates of the rates of phylogenetic divergence among taxa.
Social Genetics Theory
Sequence diversity of sib-effect genes in social insect populations is affected by the mating system and the number of colony queens (i.e., any factor that affects nestmate relatedness). In contrast, polymorphism of maternal-effect genes is insensitive to numbers of mates and the colony founding system does not affect those maternal genes whose expression impacts only their own progeny (Wade 1998, 2001). For genes with direct effects expressed in both sexes, equilibrium replacement site diversity should be lower given the same average intensity of selection, assuming predominantly purifying selection. For comparisons within species among genes with different expression patterns, we assume that the strength of selection, s and effective population size, Ne are the same (or essentially so) for all genes, regardless of expression pattern. That is, we assume that the genes experience approximately the same average distribution of s values for the period of comparison, and we ask what are the expected differences in sequence diversity? For the Hox genes and other highly conserved gene families experiencing strong purifying selection, this assumption is probably adequate. However, for positive selection on genes involved in the origin and refinement of adaptive novelty, this assumption is less likely to hold. Nevertheless, because the ratio of fixation probabilities for genes with different patterns of expression asymptotes for positive selection with values of s > 0.01 (Demuth and Wade 2007; see Fig. 3 below), some of our predictions are less sensitive to variations in s than others. Below, we will relax the assumption of equal distributions of s and examine its effect on the relative rates of fixation for the different kinds of gene expression patterns. As for the maternal effects theory described above, we also assume that the loci being compared are at equilibrium under mutation–selection balance and have not experienced recent selective sweeps disrupting this equilibrium.
Figure 3.
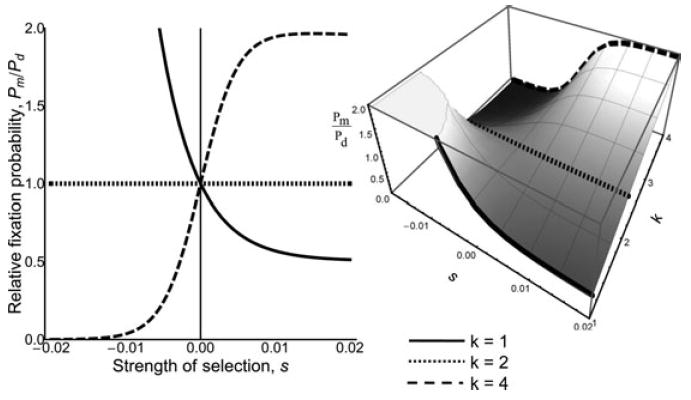
Relative fixation probability of maternal effect (or sib effect in a monandrous population) to direct effect loci in DD with weak negative to weak positive selection. The relative strength of selection experienced by the indirect effect locus, k = 1 (solid line), k = 2 (dotted line), and k = 4 (dashed line). The full three-dimensional graph of relative fixation probabilities is shown to the right. When k = 1, the direct and indirect effect loci experience the same strength of selection yet the direct effect locus is much more likely to be lost when it is deleterious (s < 0) and becomes twice as likely to be fixed when advantageous (s > 0) because the indirect effect locus experiences relaxed selective constraint. When k = 2, the increased selection experienced by the indirect effect locus is exactly balanced out by its relaxed selective constraint, as measured by the regression coefficient r, relative to the indirect effect locus, and the relative fixation probability of direct to indirect effects is equal (i.e., 2 × 1/2 = 1). When k = 4, the indirect effect locus effectively experiences selection twice as strong as the direct effect locus and the indirect effect locus becomes twice as likely to be fixed when advantageous and much more likely to be lost when deleterious.
For predictions of the relative levels of sequence diversity for genes within the same species, the assumption of equal Ne is straightforward as long as neither gene is closely linked to some other gene experiencing strong selection. However, among taxa, we can expect some variation in Ne and this may affect our predictions for the relative probabilities of fixation and rates of sequence evolution among the same set of taxa.
The Model
DIPLO-DIPLOIDS
Consider a diploid, autosomal locus, A, with two alternate alleles, A1 and A2 in frequencies p and q ( = 1 − p), respectively. If there are more than two alleles, we make the standard assumption that the frequency of A2 includes all alleles other than A1. Let A1 be expressed in individuals, their mothers, and their siblings, with the following additive effects on individual fitness; direct effect, sd, maternal effect, sm, and sib effect, ss. (Dominance deviations from additive expectation affect the Δp expressions below, but not their ratios which is our concern here.) Tables 1 and 2 summarize how the fitness of individuals depends on their own genotype and that of their family members, for families with a singly mated (monandrous) or multiply mated (polyandrous) mother. With these fitness effects, the change in allele frequency of A1 in the next generation after selection for a monandrous population is given by
| (1) |
where the mean fitness, W equals [1 + 2 p(sd + sm + ss)]. When sm and ss equal zero, equation (1) is the standard population genetic equation for the change in allele frequency for an allele with an additive direct effect, sd. For a polyandrous population, the coefficients of sd and sm remain constant, but that of ss is halved:
| (2) |
Table 1.
Fitness of offspring genotypes in families of a single-mating diplo-diploid population. Frequency (top of each cell) and fitness (bottom of each cell) of offspring depends on their own genotype and the genotypes of family members. Shaded boxes are offspring genotypes not produced. WDD monandry=1+2 p(sd+sm+ss). ΔpDD monandry=p q (sd+1/2 sm+1/2 ss)/WDD monandry.
| Mother | Father | A1A1 | A1A2 | A2A2 |
|---|---|---|---|---|
| A1A1 | A1A1 | 1 | ||
| 1+2 sd+2sm+2 ss | ||||
| A1A1 | A1A2 | ½ | ½ | |
| 1+2 sd+2sm+3/2 ss | 1+sd+2sm+3/2 ss | |||
| A1A1 | A2A2 | 1 | ||
| 1+sd+2sm | ||||
| A1A2 | A1A1 | ½ | ½ | |
| 1+2 sd+sm+3/2 ss | 1+sd+sm+3/2 ss | |||
| A1A2 | A1A2 | ¼ | ½ | ¼ |
| 1+2 sd+sm+ss | 1+sd+sm+ss | 1+sm+ss | ||
| A1A2 | A2A2 | ½ | ½ | |
| 1+sd+sm+1/2 ss | 1+sm+1/2 ss | |||
| A2A2 | A1A1 | 1 | ||
| 1+sd+ss | ||||
| A2A2 | A1A2 | ½ | ½ | |
| 1+sd+1/2 ss | 1+1/2 ss | |||
| A2A2 | A2A2 | 1 | ||
| 1 |
Table 2.
Fitness of offspring genotypes in families of a multiple-mating diplo-diploid population. WDD polyandry=1+2 p(sd+sm+ss). ΔpDD polyandry=p q (sd+1/2 sm+1/4 ss)/WDD polyandry.
| Mother | A1A1 | A1A2 | A2A2 |
|---|---|---|---|
| A1A1 | p | 1−p | |
| 1+2 sd+2 sm+(1+p) ss | 1+sd+2 sm+(1+p) ss | ||
| A1A2 | p/2 | ½ | (1−p)/2 |
| 1+2 sd+sm+(p+1/2) ss | 1+sd+sm+(p+1/2) ss | 1+sm+(p+1/2) ss | |
| A2A2 | P | 1−p | |
| 1+sd+p ss | 1+p ss |
The change in mating system from monandry to polyandry does not affect the contribution of either the direct effect or the maternal effect to the rate of evolution, but it does affect the coefficient of the sib effect, which decreases from ½ to ¼. The coefficients of sd, sm, and ss are the genetic regressions between the individual expressing the genes and the individual expressing its phenotypic (i.e., fitness) effects. For sm, the coefficient is ½, corresponding to the genetic relatedness between mother and offspring, and, for ss, the coefficient is ½ and ¼, the average relatedness between full-siblings and half-siblings, respectively, as determined by the mating system (see Fig. 1). For haplo-diploids, these genetic regressions change and, below, we describe how this affects the sequence diversity maintained at mutation–selection balance within populations, and how this contributes to sequence divergence between lineages.
HAPLO-DIPLOIDS
HD populations have an added complication because males and females must be modeled separately (Tables 3-6). One of the important considerations is how to assign fitnesses to males relative to females. Comparing Tables 3 and 4, note that we have assigned a fitness of (1 + 2 sd) to A1A1 females and a fitness of (1 + sd) to A1 males. This means that our derivation assumes no dosage compensation of the direct effects of alleles. An alternative, not explored here, is to assign haploid A1 males a fitness of (1 + 2sd). The equations for sex-specific changes in allele frequency per generation due to selection in a multiple-mating HD population are:
| (3) |
| (4) |
Table 3.
Fitness of daughter genotypes in families of a single-mating haplo-diploid population. WHD♀monandry=1+2p(sd+sm+ss). ΔpHD♀monandry=p q (sd+1/2 sm+3/4 ss)/WHD♀monandry.
| Mother | Father | A1A1 | A1A2 | A2A2 |
|---|---|---|---|---|
| A1A1 | A1 | 1 | ||
| 1+2sd+2sm+2ss | ||||
| A1A1 | A2 | 1 | ||
| 1+sd+2sm+ss | ||||
| A1A2 | A1 | ½ | ½ | |
| 1+2sd+sm+3/2 ss | 1+sd+sm+3/2 ss | |||
| A1A2 | A2 | ½ | ½ | |
| 1+sd+sm+1/2 ss | 1+sm+1/2 ss | |||
| A2A2 | A1 | 1 | ||
| 1+sd+ss | ||||
| A2A2 | A2 | 1 | ||
| 1 |
Table 6.
Fitness of son genotypes in families of a multiple-mating haplo-diploid population. WHD♂polyandry=1+p (sd+2sm+2ss). Δ pHD♂polyandry=p q (sd+sm+1/2 ss)/WHD♂polyandry.
| Mother | A1 | A2 |
|---|---|---|
| A1A1 | 1 | |
| 1+sd+2sm+(1+p) ss | ||
| A1A2 | ½ | ½ |
| 1+sd+sm+(1+2 p)/2 ss | 1+sm+(1+2 p)/2 ss | |
| A2A2 | 1 | |
| 1+p ss |
Table 4.
Fitness of daughter genotypes in families of a multiple-mating haplo-diploid population. WHD♀polyandry=1+2p(sd+sm+ss). ΔpHD♀polyandry=p q (sd+1/2 sm+1/4 ss)/WHD♀polyandry.
| Mother | A1A1 | A1A2 | A2A2 |
|---|---|---|---|
| A1A1 | p | Q | |
| 1+2sd+2sm+(1+p) ss | 1+sd+2sm+(1+p) ss | ||
| A1A2 | p/2 | ½ | q/2 |
| 1+2sd+sm+(1+2 p)/2 ss | 1+sd+sm+(1+2 p)/2 ss | 1+sm+(1+2 p)/2 ss | |
| A2A2 | P | Q | |
| 1+sd+p ss | 1+p ss |
The mean fitness of haplo-diploid females, WHD♀polyandry, is [1 + 2p (sd + sm + ss)], identical to that of diplo-diploid females. With no dosage compensation, the mean fitness of haplo-diploid males, WHD♂monandry equals [1 + p (sd + 2sm + 2ss)]. The coefficient of sm, the maternal effect on offspring fitness, is 1 for males and ½ for females, representing the maternal-son and maternal-daughter genetic regressions, respectively, which do not change with maternal mate number. In contrast, the coefficient of ss, the sib-social effect on daughter fitness, does change from ½ (full-siblings) with monandry to ¼ (maternal half-siblings) with polyandry.
The total change in allele frequency in the population is the weighted average of that in each sex. The weightings equal the proportion of alleles found in males and females, that is, 1/3 and 2/3, respectively. Assuming weak selection, mean fitness is approximately equal in both sexes, and, we have
| (5) |
In a single-mating HD population, male and female change in allele frequency is:
| (6) |
| (7) |
Combining the weighted changes and assuming approximately equal fitness in males and females, we find that total change in allele frequency is
| (8) |
Comparing the coefficients of sm and ss in equations (5) and (8), we note that the coefficient of sm is 2/3 in both cases, equal to the coefficient of ss with monandry but twice as large as is the value of 1/3 with polyandry. Thus, a change in the mating system from monandry to polyandry will affect the relative rate of evolution of maternal to sib-effect genes.
EXPECTED POLYMORPHISM WITHIN POPULATIONS
Following Whitlock and Wade (1995), we compute the expected allele frequencies at mutation–selection balance (i.e., assuming purifying selection) given μ, the mutation rate per generation (Table 7). As shown by Wade (1998, 2001), maternal effect loci are predicted to be more polymorphic (twice as much for DD populations) than direct effect loci at mutation–selection balance. The expected polymorphism for maternal-effect genes does not depend on mating system. In contrast, for sib-effect loci, the predicted level of polymorphism maintained at mutation–selection balance within populations does depend on mating system. The most sequence variation is predicted to be maintained at sib-effect genes in multiple-mating populations. In DD, this is four times the amount of polymorphism relative to direct-effect genes and twice the polymorphism relative to maternal-effect genes. Generally, the predicted amount of polymorphism maintained within populations depends on the relatedness between the individual expressing the gene and the individual in which the phenotypic effects are manifested. For direct-effect genes, these individuals are one and the same and relatedness is one. The average relatedness between mothers and offspring is ½ in DD and 2/3 in HD, and the average relatedness between siblings depends on mating system (e.g., for DD, it is ½ with monandry and ¼ with polyandry).
Table 7.
Expected allele frequency at mutation–selection balance depending on pattern of expression (direct-, maternal-, or sib-effect loci), mating system (monandry or polyandry), and genetic system (haplo-diploid or diplo-diploid).
| Monandry DD | Polyandry DD | Monandry HD | Polyandry HD | |
|---|---|---|---|---|
| Direct effects | 1 μ/sd | 1 μ/sd | 1 μ/sd | 1 μ/sd |
| Maternal effects | 2 μ/sm | 2 μ/sm | 3/2 μ/sm | 3/2 μ/sm |
| Sib effects | 2 μ/ss | 4 μ/ss | 3/2 μ/ss | 3 μ/ss |
SEQUENCE VARIATION BETWEEN LINEAGES
Here we consider rates of sequence divergence between loci with different patterns of expression, estimated as probabilities of fixation. The rate of substitution of new allelic variants is governed by the mutation rate, μ and the probability of fixation, P. For alleles with additive effects, Kimura (1962) showed that the probability of fixation, Pd,DD of an allele at DD locus with a direct effect on fitness of s is
| (9) |
Substituting (s/2), the selection coefficient for a maternal effect relative to a direct-effect gene (i.e., half the strength of selection), Demuth and Wade (2007), following Kimura (1962), showed that the probability of fixation of a maternal effects allele in a DD population is
| (10) |
We can generalize the results of Demuth and Wade (2007) by substituting the appropriate regression coefficients for the different genetic systems and mating systems as derived above. We find that the probability of fixation of alleles with direct, maternal, or sib effects in a DD population is
| (11) |
where r, the weighted average of the genetic regression between the individual expressing the allele and the individuals receiving its fitness effects, takes a value of 1 for a direct effect allele, ½ for a maternal-effect allele in a DD population or a sib-effect allele in a DD monandrous population, and 1/4 for a sib-effect allele in a DD polyandrous population. These fixation probabilities for direct, maternal, and sib effects in DD populations are shown in Figure 1.
We can so determine the “relative” rate of divergence for diploid loci with different expression patterns using the ratio of fixation probabilities. For example, the relative rate of fixation for maternal effect alleles relative to direct effect alleles is
| (12) |
which simplifies to (Demuth and Wade 2007). Assuming that the loci have similar mutation rates, this equation describes the relative rates of divergence between maternal and direct-effect genes. We can relax the assumption of equal selection between differently expressed loci by assuming that selection on one locus is k times as strong as selection at the second locus, changing the ratio of fixation probabilities in equation (12) to
| (13) |
Here, P1,DD and P2,DD are the fixation probabilities for loci 1 and 2 with different patterns of expression, r1 and r2 are the relatedness coefficients for loci 1 and 2 that depend on expression pattern, and k is strength of selection on locus 1 (when expressed) relative to locus 2.
For haplo-diploid species, we need the change in the mean allele frequency by weak selection from equations (5) and (8) as well as the variance of the allele frequency change by random genetic drift to derive the probability of fixation using the diffusion methods of Kimura (1962). Assuming W ~ 1 for weak selection, the average change in frequency, Md, by selection for a gene with direct effect, sd, is pqsd and the variance owing to drift, V, is 2pq/3N (assuming equal numbers of males and females). The exponent in the equation for fixation probability is given by the ratio Md/(V/2) or 2Md /V. This gives us 3sdN for a gene with direct effects on male and female fitness. The resulting equation for fixation probability of a direct effect allele in a HD population is
| (14) |
For a gene with only a maternal effect, the ratio changes because the mean change by weak selection, Mm, is 2pqsm/ 3 whereas the variance caused by random drift remains the same, independent of how a gene is expressed. The ratio, 2Mm/V, equals 2smN
| (15) |
For a gene with only a sib effect, ss, the change in mean, Ms, varies with the mating system, from 2pqss/3 with monandry to pqss/3 with polyandry. Again, the variance owing to drift remains the same, so our exponent is 2ssN for monandry but only 1ssN for polyandry. As we did for a DD population (eq. 11), we can generalize these results to find the probability of fixation of alleles with direct, maternal, or sib effects in a HD population:
| (16) |
Where r equals 1 for a direct effect allele, 2/3 for a maternal effect allele or sib-effect allele with monandry, and 1/3 for a sib-effect allele with polyandry (see Figure 2).
Figure 2.
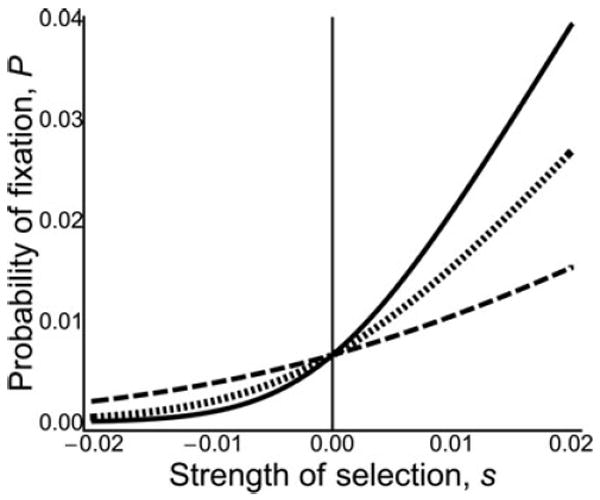
Probability of fixation of new mutants in a haplo-diploid population with different patterns of expression. Alleles with direct effects (solid line); alleles with maternal effects or sib effects in single-mating full-sibling family (dotted line); and alleles with sib effects in a multiple-mating half-sibling family (dashed line). Ne = 100, p = 2/(3Ne).
Similarly for the DD case above (eq. 14), the relative fixation rates of two loci, P1,HD and P2,HD with different patterns of expression is
| (17) |
Where, r1 and r2 are the relatedness coefficients for loci 1 and 2 that depend on expression pattern, and k is strength of selection on locus 1 (when expressed) relative to locus 2.
Assuming that new mutations at diploid loci initially occur at a frequency p = 1/2N, and N equals Ne, we show in Figure 3 how the relative fixation probabilities, and assuming equal mutation rates the relative rates of divergence, depend on pattern of expression and strength of selection. Notice that, changing a gene’s expression pattern via r is equivalent to changing the relative strength of selection experienced by a gene through k. As a result, two genes can experience the same effective strength of selection, and thus the same fixation probabilities, with different, but balancing patterns of expression and relative strength of selection. For example, in a diploid population, a maternal-effect gene experiencing twice the strength of selection as a direct-effect gene will have the same fixation probability as the direct-effect gene (i.e., the r of ½ will balance out the k of 2 for the maternal-effect gene relative to the r = k = 1 for the direct-effect gene; see Figs. 3 and 4).
Figure 4.
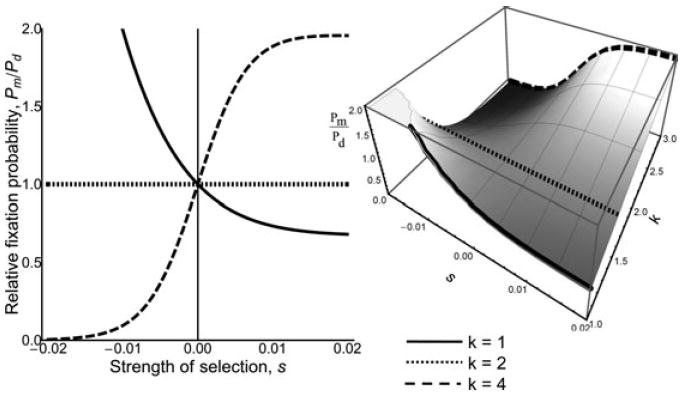
Relative fixation probability of maternal effect (or sib effect in a monandrous population) to direct effect loci in HD with weak negative to weak positive selection. The relative strength of selection experienced by the indirect effect locus, k = 1 (solid line), k = 1.5 (dotted line), and k = 3 (dashed line). The full three-dimensional graph of relative fixation probabilities is shown to the right. In contrast to the DD case, in HD, the relaxed selective constraint experienced by the indirect effect locus is exactly balanced by increased strength of selection when k = 3/2 (i.e., 3/2 * 2/3 = 1). And when k = 3, the indirect effect locus effectively experiences selection twice as strong as the direct effect locus (i.e., 3 * 2/3 = 2). The relatively flatter surface for HD relative to DD indicates that maternal versus direct expression has less of an effect on the probability of fixation in HD.
Discussion
We show that all else being equal, selection acts less effectively on loci with indirect social effects on fitness than on loci with direct effects on fitness. As a result, at mutation–selection balance, indirect effect loci are expected to harbor higher levels of sequence polymorphism within populations. Furthermore, indirect effect loci are expected to diverge more rapidly between lineages than direct effect loci because slightly deleterious indirect effect alleles are more likely to drift to fixation and contribute to fixed differences than similarly deleterious direct effect alleles.
Generally, the relatedness between the individual expressing the gene and the individual manifesting its fitness effects determines the rate of change in allele frequency per generation, and as a result, the expected sequence polymorphism. Thus, decreasing relatedness between social partners is equivalent to weakening the average strength of selection experienced by genes with indirect fitness effects, and as a result increasing the standing genetic variation. This makes intuitive sense. As relatedness decreases, so does the association between genotype and phenotype. Direct-effect genes are expressed in the same individual in which the phenotypic effects are manifested and, barring factors that cause gene copies to be unexpressed (e.g., dominance, penetrance), there is a one-to-one correspondence between genotype and phenotype. Differently put, the genetic relatedness between an individual and itself is one. In contrast, maternal- and sib-effect genes are expressed in mothers and sibs, respectively, but have phenotypic effects on other individuals. The relatedness between mothers and offspring or between siblings is less than one. For example, the genetic relatedness between full-siblings and half-siblings in a randomly mating DD population is ½ and ¼, respectively. This results in selection half to one-fourth as weak and two to four times the amount of variation at mutation–selection balance for sib-effects genes relative to direct-effect genes (Table 7; Figs. 1 and 2). The fact that loci with indirect effects experience less effective selection than loci with direct effects is also captured by kin selection theory. In the well-known Hamilton’s Rule, rb − c > 0, the indirect fitness benefit to a social partner, b, is decremented by the relatedness between social partners, r, whereas the direct fitness cost to the actor, c, has a coefficient of one (Hamilton 1964). These two fitness effects are associated with a single gene in kin selection theory, whereas we have explored the relative rates of evolution and relative levels of polymorphism of pairs of genes with different types of direct or indirect fitness effects.
Mating system does not affect the relatedness between mothers and their offspring, so the expected sequence polymorphism for maternal-effect genes does not depend on mating system, all things being equal (Wade 1998). Mating system does affect the relatedness between siblings. In HD and DD populations with single mating, the expected sequence diversities for maternal and sib effects are the same, but for polyandrous DD and HD populations, sib-effect loci experience less selective constraint and hence accumulate more polymorphism at mutation–selection balance. Other factors that influence the relatedness between social partners, such as queen number and worker egg-laying, will also influence the predicted sequence diversity of caste- or worker-specific genes because such effects change the genetic regression between the genotype where the gene is expressed and the phenotype of the receiving individual.
As summarized above, what is important in terms of our theory is whether a gene has direct or indirect effects on fitness. In social insects, queens and males reproduce and workers typically do not (although in many HD social insects, workers can lay unfertilized male-destined eggs in certain situations). As a result, genes affecting most worker traits have mainly indirect fitness effects, dependent on the number and quality of new queens and males produced by colonies. Thus, genes expressed exclusively by workers are likely to have indirect sib effects on fitness; genes expressed by male and female sexuals are likely to have direct fitness effects; and queen genes that affect the quality of her sexual offspring have maternal fitness effects. (Genes that affect egg number and are sex-limited in their expression [i.e., expressed only by queens but also transmitted by males] also experience relaxed selection independent of mating system.)
The weaker purifying selection (i.e., relaxed constraint) on genes with indirect fitness effects potentially enables wider exploration of phenotypic and genotypic space both within and among populations. As a result, social insect populations may harbor more genetic and phenotypic variation within and between populations for worker traits relative to queen and male traits. However, worker and sexual traits are also likely to experience different selective regimes. For example, the traits of males and queens may often experience strong stabilizing selection whereas in some cases phenotypic variation for worker traits within a colony may be favorable. Generally, there is a tension between the reduced strength of selection that decreases the probability advantageous alleles spread, and relaxed selective constraint that enables the accumulation of genetic variation within population and facilitates rapid divergence between populations. The former applies to alleles that experience mainly positive selection whereas the latter applies to alleles that are mildly deleterious under most environmental conditions, experiencing mainly purifying selection.
All else equal, polyandrous populations should harbor more variation than monandrous populations for sib-effect genes expressed by workers. The discovery of genetic components to worker polymorphism in several highly polyandrous species (Hughes et al. 2003; Rheindt et al. 2005; Jaffe et al. 2007) lends tentative support to this prediction. Indeed alleles for caste determination may generally be mildly deleterious (see review by Anderson et al. 2008), but as explained above, genetically based variation in worker phenotypes may be adaptive in some cases, and not mainly governed by mutation–selection balance.
Given the diversity of genetic (DD or HD) and mating system found in social insects, it is relatively easy to select suitable study systems for testing our predictions. Mating frequency theoretically affects population sex ratio, and studies of sex-ratio evolution have identified several social insect species with populations differing primarily in mating frequency (Bourke and Franks 1995). Comparisons can also be made between lineages that differ in mating frequency, for example, between species in Pogonomyrmex harvester ants (Rheindt et al. 2004; Holbrook et al. 2007); between genera and species in fungus-growing ants (Villesen et al. 1999); or between species of stingless bees (Paxton et al. 1999). However, effective population size is likely a complicating factor in these comparisons. As such, it may often be easier to make comparisons of sequence variation within populations or sequence divergence between lineages using comparisons between loci as in equations (11-12), where both loci, although differing in expression pattern, are subject to the same variations in Ne.
Currently, an incisive test of our theory is limited by the absence of loci that can be defined as having indirect fitness effects. As explained above, genes expressed in adults of the worker caste are likely to have indirect fitness effects, whereas genes expressed in adult sexual females and males, or all larvae are likely to have direct fitness effects. Genes affecting traits involved in sib and maternal care, including provisioning and defense of brood, have sib and maternal effects, respectively (Linksvayer and Wade 2005). The simple predictions discussed here for relative polymorphism and sequence divergence between genes with different expression patterns assume each gene has exclusively direct, maternal, or sib fitness effects. However, in reality, genes may often have multiple pleiotropic effects and care must be taken to identify all the various fitness effects of genes of interest. Expected polymorphism and sequence divergence depends on the total strength of selection experienced by a gene (see eqs. 1-8). When genes have pleiotropic effects, coefficients for the various direct and indirect selection components must be added (assuming each effect experiences similar strengths of selection), and then expected polymorphism and divergence calculated. A good starting point for testing our predictions is focusing on the simplest case of genes with single types of direct or indirect fitness effects. The rapid growth of sociogenomics promises to provide a wealth of expression data in the near future and these data are the starting point for identifying differences such as sex- and caste-specific patterns of gene expression. Indeed a large number of genes with caste-specific patterns of expression have already been found in termites, ants, and especially honey bees (Miura et al. 1999; Evans and Wheeler 2001; Wheeler et al. 2006; Whitfield et al. 2006; Barchuk et al. 2007).
A notable set of candidate genes for testing our predictions are the major royal jelly protein genes (mrjp1–9) found in honey bees. These genes are thought to have arisen via tandem duplication events from the yellow genes found in Drosophila and other insects (Drapeau et al. 2006). Some are expressed primarily in the hypopharyngeal glands of nurse workers, the site of synthesis of royal jelly proteins that are fed by nurses to larvae and influence whether the larvae develop into queens or workers. These genes thus have indirect sib effects on fitness. Other major royal jelly protein genes are expressed in different body parts of developing queen, worker, and male larvae (summarized in Drapeau et al. 2006, Table 3). These can be considered to be direct-effect genes because they are expressed in both sexes as well as in workers. Comparisons of sequence diversity within populations and divergence between Apis species for the different major royal jelly protein loci that differ in patterns of expression may provide a suitable test of our predictions.
Socially parasitic species in which the worker caste is rarely or never expressed also provide a very promising test case. Because genes affecting worker traits are rarely/never expressed, these genes should experience drastically reduced selective constraint and should harbor even more sequence variation than sib-effect genes in normal social insect populations. Furthermore, the closest relative of socially parasitic species is often the host (Emery’s Rule, Buschinger 1990; Sumner et al. 2004), offering a unique opportunity for paired comparison of sequence variation for direct and indirect-effect genes in parasite and host.
Table 5.
Fitness of son genotypes in families of a single-mating haplo-diploid population. WHD♂monandry=1+p(sd+2sm+2ss). ΔpHD♂monandry=p q (sd+sm+1/2 ss)/WHD♂monandry.
| Mother | Father | A1 | A2 |
|---|---|---|---|
| A1A1 | A1 | 1 | |
| 1+sd+2sm+2 ss | |||
| A1A1 | A2 | 1 | |
| 1+sd+2sm+ss | |||
| A1A2 | A1 | ½ | ½ |
| 1+sd+sm+3/2 ss | 1+sm+3/2 ss | ||
| A1A2 | A2 | ½ | ½ |
| 1+sd+sm+1/2 ss | 1+sm+1/2 ss | ||
| A2A2 | A1 | 1 | |
| 1+ss | |||
| A2A2 | A2 | 1 | |
| 1 |
Acknowledgments
We thank two anonymous reviewers and the editor for helpful comments on the manuscript as well as D. van Dyken, Y. Brandvain, and T. Cruickshank. TAL was funded by a National Science Foundation postdoctoral fellowship in biological informatics; MJW was supported by National Institutes of Health 1R01GM084238-01.
LITERATURE CITED
- Anderson KE, Linksvayer TA, Smith CR. The causes and consequences of genetic caste determination in ants (Hymenoptera: Formicidae) Myrmecol News. 2008;11:119–132. [Google Scholar]
- Barchuk AR, Cristino AS, Kucharski R, Costa LF, Simoes ZLP, Maleszka R. Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev Biol. 2007;7:70. doi: 10.1186/1471-213X-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker MS, Demuth JP, Wade MJ. Maternal expression relaxes constraint on innovation of the anterior determinant, bicoid. PLoS Genet. 2005;1:527–530. doi: 10.1371/journal.pgen.0010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bijma P, Muir WA, Van Arendonk JAM. Multilevel selection 1: quantitative genetics of inheritance and response to selection. Genetics. 2007;175:277–288. doi: 10.1534/genetics.106.062711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke AFG, Franks NR. Social evolution in ants. Princeton Univ Press; Princeton, NJ: 1995. [Google Scholar]
- Buschinger A. Sympatric speciation and radiative evolution of socially parasitic ants: heretic hypotheses and their factual background. J Zoolog Syst Evol Res. 1990;28:241–260. [Google Scholar]
- Cheverud JM, Moore AJ. Quantitative genetics and the role of the environment provided by relatives in behavioral evolution. In: Boake CRB, editor. Quantitative genetic studies of behavioral evolution. Univ of Chicago Press; Chicago, IL: 1994. pp. 67–100. [Google Scholar]
- Cruickshank T, Wade MJ. Microevolutionary support for a developmental hourglass: gene expression patterns shape sequence variation and divergence in Drosophila. Evol Dev. 2008;10:583–590. doi: 10.1111/j.1525-142X.2008.00273.x. [DOI] [PubMed] [Google Scholar]
- Demuth JP, Wade MJ. Maternal expression increases the rate of bicoid evolution by relaxing selective constraint. Genetica. 2007;129:37–43. doi: 10.1007/s10709-006-0031-4. [DOI] [PubMed] [Google Scholar]
- Drapeau MD, Albert S, Kucharski R, Prusko C, Maleszka R. Evolution of the Yellow/Major Royal Jelly Protein family and the emergence of social behavior in honey bees. Genome Res. 2006;16:1385–1394. doi: 10.1101/gr.5012006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans JD, Wheeler DE. Gene expression and the evolution of insect polyphenisms. Bioessays. 2001;23:62–68. doi: 10.1002/1521-1878(200101)23:1<62::AID-BIES1008>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Hamilton WD. The genetical evolution of social behaviour. I, II. Journal of Theoretical Biology. 1964;7:1–16. 17–52. doi: 10.1016/0022-5193(64)90038-4. [DOI] [PubMed] [Google Scholar]
- Holbrook CT, Strehl CP, Johnson RA, Gadau J. Low queen mating frequency in the seed-harvester ant Pogonomyrmex (Ephebomyrmex) pima: implications for the evolution of polyandry. Behav Ecol Sociobiol. 2007;62:229–236. [Google Scholar]
- Honey Bee Genome Sequencing Consortium. Insights into social insects from the genome of the honeybee Apis mellifera. Nature. 2006;443:931–949. doi: 10.1038/nature05260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes WOH, Sumner S, Van Borm S, Boomsma JJ. Worker caste polymorphism has a genetic basis in Acromyrmex leaf-cutting ants. Proc Natl Acad Sci USA. 2003;100:9394–9397. doi: 10.1073/pnas.1633701100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe R, Kronauer DJC, Kraus FB, Boomsma JJ, Moritz RFA. Worker caste determination in the army ant Eciton burchellii. Biol Lett. 2007;3:513–516. doi: 10.1098/rsbl.2007.0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. On the probability of fixation of mutant genes in a population. Genetics. 1962;47:713–719. doi: 10.1093/genetics/47.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linksvayer TA. Direct, maternal, and sibsocial genetic effects on individual and colony traits in an ant. Evolution. 2006;60:2552–2561. doi: 10.1554/06-011.1. [DOI] [PubMed] [Google Scholar]
- Linksvayer TA. Ant species differences determined by epistasis between brood and worker genomes. PLoS ONE. 2007;2:e994. doi: 10.1371/journal.pone.0000994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linksvayer TA, Wade MJ. The evolutionary origin and elaboration of sociality in the aculeate Hymenoptera: maternal effects, sib-social effects, and heterochrony. Q Rev Biol. 2005;80:317–336. doi: 10.1086/432266. [DOI] [PubMed] [Google Scholar]
- Linksvayer TA, Fondrk MK, Page RE., Jr Honey bee social regulatory networks are shaped by colony-level selection. Am Nat. 2009;173:E99–E107. doi: 10.1086/596527. [DOI] [PubMed] [Google Scholar]
- Miura T, Kamikouchi A, Sawata M, Takeuchi H, Natori S, Kubo T, Matsumoto T. Soldier caste-specific gene expression in the mandibular glands of Hodotermopsis japonica (Isoptera : Termopsidae) Proc Natl Acad Sci USA. 1999;96:13874–13879. doi: 10.1073/pnas.96.24.13874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore AJ, Brodie ED, III, Wolf JB. Interacting phenotypes and the evolutionary process .1. Direct and indirect genetic effects of social interactions. Evolution. 1997;51:1352–1362. doi: 10.1111/j.1558-5646.1997.tb01458.x. [DOI] [PubMed] [Google Scholar]
- Mutic JJ, Wolf JB. Indirect genetic effects from ecological interactions in Arabidopsis thaliana. Mol Ecol. 2007;16:2371–2381. doi: 10.1111/j.1365-294X.2007.03259.x. [DOI] [PubMed] [Google Scholar]
- Paxton RJ, Weissschuh N, Engels W, Hartfelder K, Javier J, Quezada-Euan G. Not only single mating in stingless bees. Naturwissenschaften. 1999;86:143–146. [Google Scholar]
- Rheindt FE, Gadau J, Strehl CP, Holldobler B. Extremely high mating frequency in the Florida harvester ant (Pogonomyrmex badius) Behav Ecol Sociobiol. 2004;56:472–481. [Google Scholar]
- Rheindt FE, Strehl CP, Gadau J. A genetic component in the determination of worker polymorphism in the Florida harvester ant Pogonomyrmex badius. Insectes Soc. 2005;52:163–168. [Google Scholar]
- Robinson GE, Grozinger CM, Whitfield CW. Sociogenomics: social life in molecular terms. Nat Rev Genet. 2005;6:257–270. doi: 10.1038/nrg1575. [DOI] [PubMed] [Google Scholar]
- Sinha S, Ling X, Whitfield CW, Zhai CX, Robinson GE. Genome scan for cis-regulatory DNA motifs associated with social behavior in honey bees. Proc Natl Acad Sci USA. 2006;103:16352–16357. doi: 10.1073/pnas.0607448103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber M, Jackle H, Schmidt-Ott U. The anterior determinant bicoid of Drosophila is a derived Hox class 3 gene. Proc Natl Acad Sci USA. 1999;96:3786–3789. doi: 10.1073/pnas.96.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber M, Prell A, Schmidt-Ott U. A single Hox3 gene with composite bicoid and zerknullt expression characteristics in non-cyclorrhaphan flies. Proc Natl Acad Sci USA. 2002;99:274–279. doi: 10.1073/pnas.012292899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumner S, Aanen DK, Delabie J, Boomsma JJ. The evolution of social parasitism in Acromyrmex leaf-cutting ants: a test of Emery’s rule. Insectes Soc. 2004;51:37–42. [Google Scholar]
- Toth AL, Varala K, Newman TC, Miguez FE, Hutchison SK, Willoughby DA, Simons JF, Egholm M, Hunt JH, Hudson ME, Robinson GE. Wasp gene expression supports an evolutionary link between maternal behavior and eusociality. Science. 2007;318:441–444. doi: 10.1126/science.1146647. [DOI] [PubMed] [Google Scholar]
- Villesen P, Gertsch PJ, Frydenberg J, Mueller UG, Boomsma JJ. Evolutionary transition from single to multiple mating in fungus-growing ants. Mol Ecol. 1999;8:1819–1825. doi: 10.1046/j.1365-294x.1999.00767.x. [DOI] [PubMed] [Google Scholar]
- Wade MJ. The evolutionary genetics of maternal effects. In: Mousseau TA, Fox CW, editors. Maternal effects as adaptations. Oxford Univ Press; New York: 1998. pp. 5–21. [Google Scholar]
- Wade MJ. Maternal effect genes and the evolution of sociality in haplo-diploid organisms. Evolution. 2001;55:453–458. doi: 10.1554/0014-3820(2001)055[0453:megate]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Wade MJ, Priest NK, Cruickshank T. Maternal effects in mammals. Univ of Chicago Press; Chicago, IL: 2008. A theoretical overview of maternal genetic effects: evolutionary predictions and empirical tests using sequence data within and across mammalian taxa. [Google Scholar]
- Wang J, Ross KG, Keller L. Genome-wide expression patterns and the genetic architecture of a fundamental social trait. PLoS Genetics. 2008:47–e1000127. doi: 10.1371/journal.pgen.1000127. Available at http://www.plosgenetics.org/article/info:doi%2F10.1371%2Fjournal.pgen.1000127. [DOI] [PMC free article] [PubMed]
- Wheeler DE, Buck N, Evans JD. Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Insect Mol Biol. 2006;15:597–602. doi: 10.1111/j.1365-2583.2006.00681.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield CW, Ben-Shahar Y, Brillet C, Leoncini I, Crauser D, LeConte Y, Rodriguez-Zas S, Robinson GE. Genomic dissection of behavioral maturation in the honey bee. Proc Natl Acad Sci USA. 2006;103:16068–16075. doi: 10.1073/pnas.0606909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC, Wade MJ. Speciation: founder events and their effects on X-linked and autosomal genes. Am Nat. 1995;145:676–685. [Google Scholar]
- Wolf JB. Genetic architecture and evolutionary constraint when the environment contains genes. Proc Natl Acad Sci USA. 2003;100:4655–4660. doi: 10.1073/pnas.0635741100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JB, Brodie ED, III, Cheverud JM, Moore AJ, Wade MJ. Evolutionary consequences of indirect genetic effects. Trends Ecol Evol. 1998;13:64–69. doi: 10.1016/s0169-5347(97)01233-0. [DOI] [PubMed] [Google Scholar]
- Wright S. On the number of self-incompatibility alleles maintained in equilibrium by a given mutation rate in a population of given size: a reexamination. Biometrics. 1960;16:61–85. [Google Scholar]


