Abstract
Adaptive phenotypic plasticity allows organisms to cope with environmental variability, and yet, despite its adaptive significance, phenotypic plasticity is neither ubiquitous nor infinite. In this review, we merge developmental and population genetic perspectives to explore costs and limits on the evolution of plasticity. Specifically, we focus on the role of modularity in developmental genetic networks as a mechanism underlying phenotypic plasticity, and apply to it lessons learned from population genetic theory on the interplay between relaxed selection and mutation accumulation. We argue that the environmental specificity of gene expression and the associated reduction in pleiotropic constraints drive a fundamental tradeoff between the range of plasticity that can be accommodated and mutation accumulation in alternative developmental networks. This tradeoff has broad implications for understanding the origin and maintenance of plasticity and may contribute to a better understanding of the role of plasticity in the origin, diversification, and loss of phenotypic diversity.
Keywords: costs and limits, development, modularity, mutation accumulation, phenotypic plasticity, pleiotropy, relaxed selection
Introduction: costs and limits in the evolution of plasticity
Adaptive phenotypic plasticity allows organisms to cope with a range of environments varying over space and time.(1-3) Phenotypic plasticity is favored in fluctuating environments because it reduces variance in fitness from one generation to the next(4) and results in high geometric mean fitness.(5) Furthermore, phenotypic plasticity allows a population to invade multiple, disparate ecological niches, thus extending the geographic range and decreasing the probability of extinction caused by habitat loss or environmental stochasticity.(6)
Despite its benefits, plasticity is neither ubiquitous nor infinite, leading biologists to search for the costs and limits that constrain the evolution and development of plasticity.(3,7) DeWitt et al.,(8) in particular, outlined a broad range of costs and limits of phenotypic plasticity, and quantitative genetic methods to measure such costs. However, subsequent empirical work has failed to produce a consistent picture of the importance of these costs: while some studies have documented costs of plasticity,(9,10) more commonly, these costs are found to be reduced or absent, or limited to certain environments or components of plasticity.(11-13) Thus, although much progress has been made in the last decade, it is clear that our understanding of the costs and limits of plasticity remains incomplete.
We suggest that the costs and limits of plasticity may vary with the developmental mechanism of plasticity. Recent advances in genomics and developmental biology have opened this “black box” of plasticity, allowing unique insights into the costs and limits of plasticity, and providing a framework for predicting their relative importance. In particular, we are gaining insights into patterns of gene expression induced by a wide range of environmental factors(14) (Table 1), one of the principal mechanisms for a genotype to modify its phenotype and thus maintain high performance across a range of environments.(15-18)
Table 1.
Select evidence for differential gene expression between environments
| References | Environmenta | Species | # Genesb | |
|---|---|---|---|---|
| Stress | (96) | Salt | Bruguiera gymnorrhiza, mangrove | 287 (7,029) |
| (97) | Darkness | Euglena gracilis | 90 (610) | |
| (98) | Insecticide | Anopheles gambiae, mosquito | >77 (11,760) | |
| (99) | Lab rearing | Salmo salar, Atlantic salmon | 409 (4,575) | |
| (100) | Various | Saccharomyces cerevisiae, yeast | 900 (6,200) | |
| (101) | Various | Schizosaccharomyces pombe, fission yeast | >1,700 | |
| (102) | Sulfur | Arabidopsis thaliana, rockcress | 505 (15,442) | |
| (103) | Cold | Cyprinus carpio, European carp | 3,400 (13,440) | |
| (104) | Heat, cold | Arabidopsis thaliana, rockcress | 16,000 (22,746) | |
| (105) | Heat | Bacillus subtilis, hay bacilllus | 100 (4,100) | |
| (106) | Salt, cold | Arabidopsis thaliana, rockcress | 2,409 (8,100) | |
| Interactions | (107) | Pathogen | Plutella xylostella, diamondback moth | 44 (1,132) |
| (108) | Defenses | Arabidopsis thaliana, rockcress | 2,181 (23,750) | |
| (109) | Defenses | Arabidopsis thaliana, rockcress | 705 (2,375) | |
| (110) | Herbivory | Arabidopsis thaliana, rockcress | 90 (150) | |
| (111) | Host cuticle | Metarhizium anisopliae, a fungus | 560 (837) | |
| (112) | Herbivore | Solanum tuberosum, potato | 127 (11,421) | |
| Diet | (113) | Poor diet | Saccharomyces cerevisiae, yeast | 1,863 (5,258) |
| (114) | Limited nutrient | Saccharomyces cerevisiae, yeast | 1,881 (6,068) | |
| (115) | Limited nutrient | Escherichia coli | 400 (4,290) | |
| (116) | Larval diet | Drosophila melanogaster, fruit fly | 90 (6,000) | |
| (117) | Blood meal | Anopheles gambiae, mosquito | 4,924 (14,900) | |
| Polyphenism | (118) | Mating tactics | Salmo salar, Atlantic salmon | 432 (2,917) |
| (119) | Defense | Rana pirica, Hokkaidō frog | 300* | |
| (120) | Castes | Apis mellifera, honeybee | 240 (>6,000) | |
| (121) | Castes | Copidosoma floridanum, a parasitoid wasp | 195–230* | |
| (122) | Castes | Nasutitermes takasagoenis, a termite | 8* | |
| (123) | Castes | Melipona quadrifasciata, a stingless bee | 314* | |
| (124) | Castes | Reticulitermes flavipes, a termite | 153* | |
| (125) | Castes | Bombus terrestris, bumblebee | 12* | |
| (126) | Castes | Polistes canadensis, paper wasp | >10* | |
| (127) | Polyethism | Apis mellifera, honeybee | >4,000 |
“Environment” refers to factors such as temperature stress, different diets, interactions with predators or herbivores, or polyphenic morphs.
“# Genes” indicates the approximate number of differentially expressed genes identified. The number in parentheses indicates the total genes on a microarray; an asterisk indicates a non-array method such as subtractive hybridization or differential display.
In this review we argue that the degree of modularity in the developmental networks that underlie plasticity is of fundamental importance for understanding limits of developmental plasticity. We begin by reviewing the importance and ubiquity of modularity and environment-specific gene expression in the context of plasticity. We then introduce theoretical and empirical evidence suggesting that environment-specific gene expression, arising from modularity in developmental networks, drives a fundamental tradeoff between the reduction of pleiotropic constraints (i.e., the independent “refinement” of alternate phenotypes) and the degree of relaxed selection. Lastly, by expanding on existing theory and its application to empirical studies, we discuss implications for the evolution of plasticity. By integrating population genetic theory with the developmental genetic underpinnings of plasticity, we hope to contribute to a synthetic framework for the evolution of developmental plasticity. More specifically, we suggest that relaxed selection may impose lineage-level constraints – as opposed to individual-level fitness tradeoffs – that affect the evolutionary origin and maintenance of phenotypic plasticity. We focus our arguments on non-reversible phenotypic plasticity in coarse-grained environments, but we conclude by discussing how these ideas apply to reversible plasticity in fine-grained environments (Fig. 1).
Figure 1.
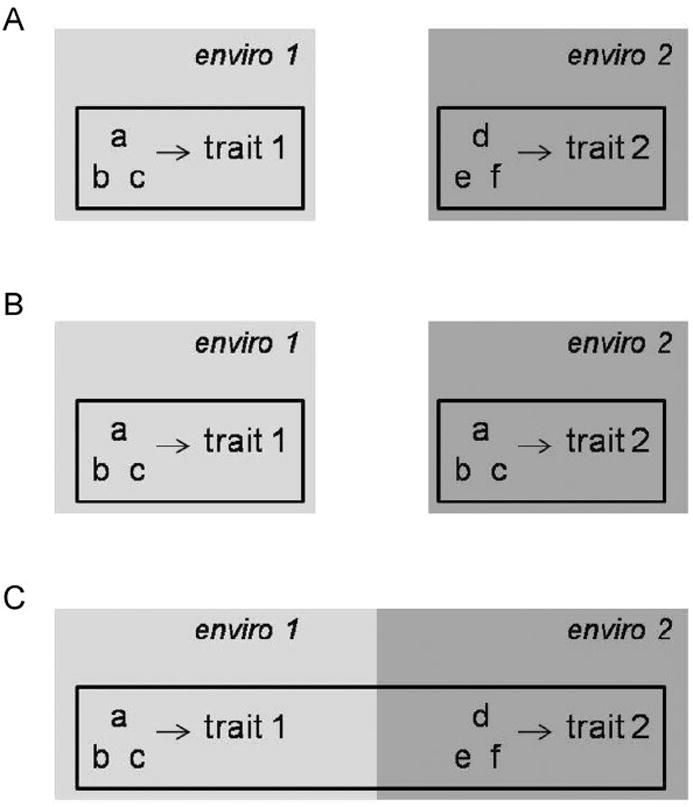
Conditions under which relaxed selection may limit the origin and maintenance of plasticity. In this figure, individuals are represented as outlined boxes, genes as lower-case letters, and environments as shaded boxes. (A): Relaxed selection may constrain the evolution of plasticity when environment-specific traits are associated with environment-specific expression of genes and individuals experience environmental variation in a coarse-grained manner. Relaxed selection will not constrain the evolution of plasticity when (B) environment-specific traits develop through mechanisms that do not require environment-specific gene expression (e.g., “hypervariable plasticity”), and (C) individuals experience environmental variation in a fine-grained manner and plastic traits are reversible.
Modularity and environment-specific gene expression in plasticity
Modularity is a pervasive concept in biology, from interacting genes and proteins to developmental interactions between traits. We argue that environment-specific gene expression that results from such modular organization comes with a tradeoff between increased evolvability – through the reduction of pleiotropic constraints – and relaxed selection – through a decreased effectiveness of both purifying and positive selection.
While there are diverse views on the precise definition of modularity, a common theme – and the definition that we adopt – is that modules are semi-independent, dissociable units (e.g., genes, proteins, and traits), where interactions are more tightly correlated within modules than between modules. Our definition thus encompasses the most common definitions of modularity used in different biological disciplines. For instance, the evo-devo definition of modularity, as used by Raff,(19) emphasizes how traits are underlain by different developmental networks, and postulates that such modular traits can be modified independently. Similarly, the developmental biology perspective of modularity emphasizes how transcription factor regulation can result in the independent expression of different sets of genes during development.(20) Finally, the population genetics view of modularity(21) is based on the genotype-to-phenotype map and the degree to which there are pleiotropic effects among components of a functional complex (“module”).
The common link between the various definitions of modularity is that pleiotropic constraints are lower between developmental modules than within modules, whether these modules consist of traits, regulatory interactions, or the genes themselves.(22) Moreover, regardless of the exact focus of each definition, they share the hypothesis that modularity increases evolvability by decreasing pleiotropic constraints: modules can be manipulated independently from one another, facilitating evolutionary change.(23)
Modular biological organization, whether on the level of genes, proteins, or traits, is predicted to evolve, at least in part, in response to variable environments.(24-27) Bacteria that can survive across a greater range of environments (e.g., soil bacteria or those with a broad host range) have more modular metabolic networks than those that are limited to less variable environments (e.g., obligate or specialized bacteria(28,29)). This theoretical and empirical work has thus raised the possibility that the ubiquity of variation in gene expression across environmental conditions, such as changes in temperature, social conditions, or nutrition (Table 1), may be underlain by the induction of alternate, modular developmental networks.
Environment-specific gene expression has long been appreciated to underlie plasticity in prokaryotes,(30) and has been inferred based on low genetic correlations in performance across a range of different environments.(31,32) Furthermore, hundreds of recent microarray studies – considering not only model organisms (Arabidopsis, Drosophila, and Escherichia coli) but also non-model organisms (mangroves, termites, and salmon) – indicate that environment-specific gene expression is an incredibly broad phenomenon (Table 1). For instance, 5–50% of the genome may vary in expression with environment, depending on the species and conditions considered (Table 1). The ubiquity of modular design of developmental networks and the hundreds of examples of environment-specific gene expression suggest that the induction of independent (modular) networks of genes is a common strategy to cope with environmental variation.
While modularity in developmental networks may or may not have evolved directly as a result of selection on plasticity in variable environments, once established, it favors the evolution of plasticity through reduction in pleiotropic constraints between alternative phenotypes. Modularity in development permits entire networks or sub-networks to be induced by specific environmental conditions, or cues, through switch-like processes.(33-35) Most importantly, however, modularity reduces pleiotropic constraints, permitting increased “fine-tuning” of different modules to different environments independent of each other.(36-40) It is well established that context-specific gene expression lessens pleiotropic constraints by limiting the effect of mutations to a specific context (e.g., tissues, sexes, and environments), potentially facilitating rapid sequence divergence.(41) Such adaptive facilitation may explain why protein evolutionary rates are higher in genes specific to sexes(42,43) and tissues,(44) although in some cases, variation in selection intensity might also explain these differences. Environment-specific gene expression by no means completely eliminates pleiotropic constraints, because many genes are expressed at a range of time periods and in a range of traits. However, environment-specific gene expression lessens pleiotropic constraints relative to shared or integrated patterns of gene expression across environments.
Environment-specific gene expression has a unique emergent property when compared to other instances of context-specific gene expression such as tissue-specific expression: in the latter, each gene is expressed somewhere in the soma of every individual in a population, whereas the expression of environment-specific genes is restricted to only those individuals within a population that experience the inducing environment. As detailed below, like in sex-limited expression, gene expression restricted to a subset of individuals effectively weakens positive and purifying selection, setting the stage for a fundamental tradeoff between reduced pleiotropy and enhanced plasticity on one side, and mutation accumulation and degradation of alternative pathways on the other. We begin with a population-genetic model perspective.
Relaxed selection and plasticity: theory
Here we review the theory that environment-specific genes, relative to genes expressed in all environments, potentially experience relaxed selection, with deleterious mutations having a higher probability of fixation and beneficial mutations having a lower chance of fixation.(45-51) While every individual in a population is subject to the forces of mutation and genetic drift, not all individuals experience selection in the same way, particularly in genetically or spatially sub-divided populations.(52-54) For coarse-grained environmental variability, an environment-specific gene, similar to a sex-specific gene, is expressed in only a fraction of the population over a given generation or time period, and so only those copies of the gene in this fraction of individuals experience selection. With sex-specific or environment-specific genes, we can partition the population into the fraction, ϕ that experiences selection on a gene, and the fraction (1 – ϕ) that does not. For autosomal, sex-specific genes, ϕ equals 1/2,(48,50,55) whereas for male-specific, X-linked genes, ϕ equals 1/3.(48,50,55) We would expect selection to be weakened by a factor, ϕ, compared to a gene under the same strength of selection but which is expressed in every individual in the population and every generation, as supported by theoretical work.(45-51,56,57)
The weakening of selection due to environment-specific expression allows deleterious mutations to accumulate in the population to a greater extent than for constitutively expressed traits or genes (Box 1).(45-51,56) The effect of such mutation accumulation on mean population fitness is measured by the “genetic load.”(58) A population will harbor 1/ϕ times as much polymorphic variation in a plastic gene as in a constitutively expressed gene under the same strength of selection, and this excess variation will primarily be maladaptive (Box 1).(56,57) For environment-specific traits, increased mutation accumulation due to conditional expression can lead to a potentially significant reduction in fitness of the trait in environments where it is required for survival and/or reproduction.(45-49) While the fitness of plastic genotypes may be reduced by this mutation accumulation, the structure of environmental variation will determine the overall performance of plastic, relative to specialist, genotypes. For instance, if a plastic genotype is competing with a genotype that specializes in this habitat, the mutation load will reduce the fitness of the plastic genotype species relative to non-plastic specialists that express the trait or gene in every individual and every generation. This has been shown to generate selection for non-plastic specialists relative to plastic generalists in constant environments.(46,49)
Box 1.
Increased genetic variation within species
Within a population, genetic variation is maintained by a balance between selection, mutation, and genetic drift. Beneficial mutations fix rapidly and so contribute very little to standing genetic variation.(128) In the absence of balancing selection, most polymorphism is due to the balance between recurrent mutation generating new variation, and purifying selection and drift removing this variation. For simplicity, consider a haploid population and assume that the rate of mutation from the deleterious to wild-type allele is negligible, the mean change in the frequency q of a deleterious allele in a single generation is written as, Δq = −q(1 − q)s/w̅ + u(1 − q), where s is the strength of selection against the allele, w̅ is the mean population fitness, and u is the deleterious mutation rate. Under standard assumptions,(129) the equilibrium allele frequency, q*, is u/s, and the expected polymorphism π at a locus is the sum of these frequencies over all sites
Note that, for small values of s, the equilibrium allele frequency, q*, is higher than it is for large values of s, hence the level of sequence polymorphism will be higher. Extending the argument to diploid systems, a gene with sex-specific or environment-specific expression should have twice the level of polymorphism as a gene experiencing the same selection but in both sexes or all environments.(50,55) In general, the average selection coefficient experienced by a gene, when only a fraction, ϕ, of its copies are expressed, is simply, sϕ + 0(1 − ϕ) or sϕ Now, consider the ratio of the expected π of a locus when conditionally expressed (ϕ) relative to another locus expressed constitutively (ϕ = 1). The relative increase in polymorphism due to conditional gene expression is
While the increased genetic load will increase heritability, the response to selection – the product of heritability and the selection differential – will be unaffected due to the decrease in the selection differential.(51)
These population genetic models are based on the assumption that some genes are “on” in some environments, and “off” in other environments. However, much empirical evidence (Table 1) suggests that many genes are expressed across environments, but at different levels. If selection is proportional to expression level, or if traits are induced at a particular threshold of gene expression, then these population genetic models – and their conclusions regarding relaxed selection – can be easily extended to many empirical examples of environment-specific gene expression. Regardless, more developmentally realistic models of selection are ripe for investigation, a point we will revisit near the end of our article.
Relaxed selection and plasticity: empirical evidence
Recent observations support the idea that relaxed selection may limit the evolution of phenotypic plasticity through relaxed selection on genes specific to different environments. First, there is growing support for the prediction that environment-specific genes should accumulate variation within species (Box 1). In bacteria, genes specific to particular population densities (quorum-induced genes), show increased levels of variation within species (Van Dyken and Wade, unpubl. data). In Drosophila, maternal effect genes (e.g., bicoid), for which only the maternal copy is expressed in offspring, also accumulate more variation within species relative to genes expressed at the same time of development, but by both sexes.(59)
Second, there is emerging support for the prediction that relaxed selection should result in increased divergence between species, because weakened purifying selection will overshadow weakened positive selection due to the higher input of deleterious mutations (Box 2). For instance, maternal effect genes are more divergent between Drosophila species compared to zygotic genes.(59-61) Furthermore, in horn-polyphenic beetles, genes that are more specific to horned and hornless morphs are more diverged relative to those with shared expression across morphs (Snell-Rood et al., unpubl. data). This accumulation of deleterious mutations may select against plastic generalists and may explain the abundance of specialists among taxa that experience alternate environments in a coarse-grained manner, such as insects on their host plants.(46,49)
Box 2.
Increased genetic divergence between species
Following from Box 1, the amount of divergence between two species is determined by the fixation process. The probability of fixation by random drift of a new deleterious allele with selection coefficient sϕ is(56,57)
For constitutively expressed genes, ϕ is 1, and deleterious alleles (s < 0) have a vanishingly small probability of fixing in a population and so do not contribute substantially to divergence. However, the range of effectively neutral mutations is extended by conditional gene expression from s < 1/2 Ne for constitutive mutations,(130) to s < 1/2 Neϕ, where ϕ is 1/2 for sex-specific autosomal genes, 1/3 for male-specific X-inked genes,(55) and 1/r for “kin selection” genes, where r is the genetic relatedness between performer and receiver of the social trait.(57) For environment-specific genes we let ϕ be the frequency with which the population experiences the inducing environment. As ϕ decreases, many more mutations with more severe deleterious effects become easier to fix between populations. On the other hand, beneficial mutations have a decreased probability of fixation(48)
Thus, limited gene expression (ϕ < 1) impedes adaptive evolution, but allows maladaptive mutations to accumulate both within and between species. Because deleterious mutations are significantly more common than beneficial mutations, the net effect of environment-specific expression is to increase genetic divergence between species.
Relaxed selection should result in not only the accumulation of deleterious mutations, but also a weakening of the strength of positive selection.(46-48) This prediction has been supported in studies of experimental evolution where adaptation occurs more rapidly in specialists evolving in constant environments than in generalists evolving in fluctuating environments.(62,63)
Additional evidence supporting a link between relaxed selection and mutation accumulation comes from contexts outside the phenotypic plasticity literature, where relaxed selection has long been hypothesized to limit adaptation to multiple environments. First, constraints on adaptation to multiple environments experienced over the lifetime of an individual have been hypothesized to be a major force behind the evolution of senescence: genes expressed later in life will not be expressed by all individuals in a population (because many will have since died) and are thus prone to accumulate mutations,(64,65) an idea that has been supported by some empirical data.(66,67) Second, in the ecology literature, relaxed selection has also been hypothesized to limit adaptation to multiple environments experienced throughout the range of a species. Relaxed selection may be a force that prohibits species from extending their ecological niche, leading to observed patterns of “niche conservatism”(52-54) and range limits.(68)
Relaxed selection should have long-term consequences for the evolution of plasticity. First, the evolution of adaptive environment-specific gene expression may be limited to environments that are experienced predictably and may be absent for rarely experienced environments. Some of the most environment-specific gene expression occurs between the sexes, “alternative phenotypes” expressed in each generation.(69,70) Regularly recurring selection on alternative networks through equal sex ratios may restrict mutation accumulation to low levels and allow highly modular alternative networks to persist. An interesting exception that may prove the rule occurs in aphids, where sexual and asexual generations alternate, such that males are expressed at most once every 10–20 female generations. Here, the accelerated divergence of male-specific genes is more consistent with relaxed selection than positive selection.(71)
Second, environment-specific gene expression may be restricted to a small number of alternate environments. Environmentally induced alternative morphs in insects rarely exceed two alternate forms (e.g., spring/summer morphs in insects, gregarious/solitary morphs in locusts, and sneaker/fighter morphs in beetles). Importantly, most exceptions to this rule are observed in eusocial insects (ants, bees, and termites) where all alternative morphs (castes: queens, workers, soldiers, etc.) are expressed for every round of (colony-level) selection.(72) Exceptions to this rule, such as facultative trimorphisms in non-social beetles,(73) may provide an ideal opportunity to test the importance of relaxed selection for limiting the evolution of plasticity.
If relaxed selection acts as a constraint on adaptation to a range of environments, selection may favor mechanisms of plasticity other than modular developmental networks that result in environment-specific traits (Fig. 1). For instance, “hypervariable plasticity”, also known as somatic selection (39,74) and bet-hedging,(75) can produce wide ranges of alternate phenotypes without relying on environment-specific gene expression. For instance, genes involved in learning are general, but modification of particular synapses through common pathways is environment-specific. Determining selection on different mechanisms of plasticity is an important area open for future research.
Relaxed selection as a promoter and a constraint in the evolution of plasticity
Relaxed selection on environment-specific genes may serve as both a promoter and a constraint in the evolution of phenotypic plasticity, properties that may be seen most clearly during periods of environmental stasis. While positive selection is generally weakened on environment-specific genes, during periods of stasis, the potential strength of positive selection is restored. Prior periods of relaxed selection should facilitate evolutionary change under such conditions due to the accumulation of genetic variation and novel combinations of variants, both of which can fuel the rate of evolutionary response.(76-78) The release of cryptic genetic variation during shifts into novel or stressful environments has long been hypothesized to facilitate evolutionary change;(79,80) relaxed selection on environment-specific genes may provide a general mechanism by which this variability accumulates.
However, unexpressed genes or pathways may also degrade rapidly during periods of stasis due to further mutation accumulation.(81-83) For instance, sporulation, a complex response to stress in bacteria, involving the expression of over 200 genes, is predominantly lost through neutral processes of mutation accumulation instead of selection, when populations are not induced to sporulate for 6,000 generations.(84,85) Similarly, microbes in constant environments (e.g., within hosts) show massive reductions in genome size and the accumulation of pseudogenes through weakened selection; there is no evidence for active selection for smaller genomes due to higher replication rates.(86,87) Finally, a wide range of selection experiments in microbes supports the idea that mutation accumulation degrades adaptation to ancestral environments.(88-92) Thus, alternate developmental pathways that once supported phenotypic plasticity may be rapidly lost during periods of environmental stasis due to mutation accumulation on top of that already present in the “mutation load.”
Implications and future areas of research
When and where is relaxed selection promoting or constraining adaptive evolution?
Existing theoretical and empirical studies provide support for the notion that relaxed selection arising from environment-specific gene expression can bring about increased genetic variation, which may increase the probability of favorable combinations of mutations, and may fuel evolution (of expressed genes) under certain environmental conditions (e.g., periods of stasis). In contrast, both theoretical and empirical studies also support the notion that relaxed selection through phenotypic plasticity decreases the probability of fixation of adaptive alleles, and can bring about the deterioration of alternative pathways through the increased probability of fixation of deleterious alleles. Determining which of these two sets of consequences of relaxed selection predominates, and under what circumstances, will have to be a major focus to fully elucidate the costs, limits, and emerging properties of plasticity. For instance, does environment-specific gene expression facilitate adaptive evolution in the short term (e.g., during colonization of new habitats and radiation events) but impede it during cladogenesis? Does relaxed selection play an important role in the origin of novel traits but impede their subsequent diversification? Developing corresponding theoretical and empirical approaches to these questions will be critical to determine what, if any, role modular, environment-specific developmental networks play in the origin of new variants, traits, or species.
The fitness consequences of differential expression and the complexity of developmental regulation
Whether or not we will be able to address the above questions will depend in large part on the ability of population-genetic models to incorporate the continuous and complex nature of developmental regulatory mechanisms. Present models assume genes to be “on” or “off” in specific environments, yet this is rarely the case. More commonly, gene expression exhibits more or less significant differences across environments. In a subset of cases, such as the induction of traits at threshold levels of gene expression, “low” levels of expression may be functionally equivalent to “no” expression, causing genes to experience relaxed selection in “low-expression” environments. But how low is low enough for the strength of selection to vary significantly across gene classes and developmental contexts? To extend the framework presented here beyond on/off genes we will have to determine empirically how fitness relates to differential, environment-specific expression during development.
More important for further integration of the population and developmental genetic perspectives on the evolution of plasticity, is the incorporation of the complexity of developmental regulatory mechanisms into a population genetic framework. Differential gene expression, whether “on/off” or “high/low,” is brought about by mechanisms operating on a variety of levels of biological organization, including regulatory elements on the level of DNA, small-interfering RNAs on the RNA level, and transcription factors on the level of proteins, all of which may themselves be subject to relaxed selection under certain circumstances. Most importantly, many components of this machinery need to be functional for genes to be activated or silenced in an environment-specific manner. For instance, the lack of expression of a focal gene in one environment may only be possible through the induced expression of an inhibitory transcription factor recognizing and blocking a specific regulatory site. While the protein-coding portion of our focal gene experiences relaxed selection, the transcription factor and regulatory site do not. These roles may be reversed in the alternate environment.
The regulatory components associated with differential gene expression and thus relaxed selection may therefore vary substantially with network architecture. Our understanding of the population-genetic fate of these components will thus only be as good as our abilities to elucidate network architecture. A more thorough understanding of patterns of environment-specific gene expression may be particularly critical for understanding the few instances – in particular in RNA viruses – where organisms appear to adapt to a range of environments without suffering costs of mutation accumulation.(93,94)
Conditions limiting the evolution of environment-specific expression: the importance of environmental grain
The importance of relaxed selection in degrading alternate developmental pathways is entirely dependent on the grain of environmental variation (Fig. 1).(95) If each individual experiences all environments (fine-grained environment), environment-specific genes will be subject to selection in every individual in the population in each generation. Alternatively, each individual may experience only one of several environments during its lifetime (coarse-grained variation). Here, relaxed selection will constrain the evolution of environment-specific gene expression. For instance, individuals often experience a range of temperatures during their lifetime, facilitating the evolution of complex expression responses to temperature, but specific resources or dangers (e.g., insecticides) may be rare, selecting against a complex environment-specific response (see Table 1). Fine-grained environments go hand-in-hand with the evolution of tolerance or reversible plasticity, which likewise should be less limited by relaxed selection (provided alternate environments are experienced randomly with respect to age).
Our understanding of the evolution of plasticity is thus limited by our understanding of patterns of environmental variability: to what extent are different abiotic and biotic resources and stressors distributed in a fine-grained or coarse-grained manner? How is environmental variability structured for species with different dispersal abilities and life histories? Is the evolution of environment-specific expression determined by these patterns? Much of the theory and discussion on the topic of relaxed selection and plasticity focuses on coarse-grained environments due to their simplicity in modeling and their consequences for relaxed selection. However, in reality, environmental variation is often more complex, shifting between coarse and fine grained, and varying in a continuous instead of a discrete manner. We need not only better descriptions of patterns of environmental variation, but more biologically realistic modeling of this variation.
Conclusions
In conclusion, recent advances in developmental genetics and genomics open new doors for understanding relaxed selection as a limit to adaptation to multiple environments. In the end, relaxed selection may explain not only limits to evolutionary origin and maintenance of plasticity at the lineage level, but also the empirical paucity of costs of plasticity at the individual level. The more we understand the diverse developmental mechanisms that underlie plasticity, the more we can explain the varying costs and limits that affect the evolution of plasticity.
Outstanding questions box.
Evolution of development
-
1
What proportion of genes differ in absolute expression (“on”/“off”) between environments (or morphs) versus differential or relative expression? Is selection proportional to degree of differential expression such that population genetic models of relaxed selection (e.g., Boxes 1 and 2) can be easily extended to relative differences in expression (e.g., Table 1)?
-
2
What is the structure of networks that underlie plasticity? With respect to alternate phenotypes, are these networks modular or integrated? What evidence exists for non-modular networks underlying plasticity, where all genes are expressed in different environments, but different phenotypes result?
-
3
How do developmental networks underlying plasticity arise and diversify? Are particular genes such as duplicated genes,(113) or regulatory elements such as transacting factors, more likely to be co-opted in the development of alternate phenotypes or environment-specific expression?
Ecology and evolution of plasticity
-
4
To what extent does relaxed selection promote or constrain the evolution of plasticity through the accumulation of variation and the degradation of alternate pathways? To what extent does relaxed selection affect micro-evolutionary changes versus macro-evolutionary processes?
-
5
What is the role of environmental grain in the evolution of developmental mechanisms of plasticity? Are fine-grained environments more likely than coarse-grained environments to select for modular networks underlying plasticity?
-
6
Can lineage-level selection maintain complex alternate developmental pathways specific to different environments?
Acknowledgments
Many thanks to H. Maughan, R. L. Young, and M. Wund for comments on earlier versions of this manuscript. We appreciated comments from F. Nijhout, M.J. West-Eberhard, and G. Wagner during the development of this work. This manuscript was substantially improved by comments from several anonymous reviewers. E.S.R. was supported by an NIH-NRSA F32GM083830. Additional support was provided, in part, by National Science Foundation Grant IOS 0718522 to APM.
Glossary
- Adaptive phenotypic plasticity
The ability of a genotype to express phenotypes appropriate to local conditions and thus maintain high performance across a range of environments. We focus on irreversible, “developmental plasticity.”
- Coarse-grained environmental variation
The environment varies over space and time, but each individual spends its lifetime in one environment; fine-grained variation refers to spatial and/or temporal variability within an individual’s lifetime.
- Hypervariable plasticity
A developmental mechanism of plasticity whereby a range of variant phenotypes are produced and subsequently selected by experience with the environment, such as in learning or acquired immunity (also known as somatic, epigenetic, or developmental selection). In some contexts, “bet-hedging” may also be considered a form of hypervariable plasticity.
- Modularity
Independent evolutionary or developmental units; as related to plasticity – the induction of independent groups of genes or their regulators in different environments.
- Mutation load
The reduction in survival or reproduction – in particular in plastic genotypes – due to the accumulation of deleterious mutations in conditional or environment-specific traits or genes.
- Pleiotropic constraints
When a gene affects the development of at least two different traits, and thus evolutionary divergence between these traits is hampered.
- Relaxed selection
A weakening of both positive and purifying selection, resulting in an increased probability of fixing deleterious mutations and a decreased probability of fixing beneficial mutations.
References
- 1.Van Tienderen PH. Evolution of generalists and specialists in spatially heterogeneous environments. Evolution. 1991;45:1317–31. doi: 10.1111/j.1558-5646.1991.tb02638.x. [DOI] [PubMed] [Google Scholar]
- 2.Moran NA. The evolutionary maintenance of alternative phenotypes. Am Nat. 1992;139:971–89. [Google Scholar]
- 3.Schlichting CD, Pigliucci M. Phenotypic Evolution: A Reaction Norm Perspective. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 4.Gillespie JH, Langley CH. A general model to account for enzyme variation in natural populations. Genetics. 1974;76:837–84. doi: 10.1093/genetics/76.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewontin RC, Cohen D. On population growth in a randomly varying environment. Proc Natl Acad Sci USA. 1969;12:366–73. doi: 10.1073/pnas.62.4.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reddingius J. Gambling for existence: a discussion of some theoretical problems in animal population ecology. Acta Biotheor. 1971;22:1–208. [Google Scholar]
- 7.Callahan HS, Maughan H, Steiner UK. Phenotypic plasticity, costs of phenotypes, and costs of plasticity toward an integrative view. Ann NY Acad Sci. 2008;1133:44–66. doi: 10.1196/annals.1438.008. [DOI] [PubMed] [Google Scholar]
- 8.DeWitt TJ, Sih A, Wilson DS. Costs and limits of phenotypic plasticity. Trends Ecol Evol. 1998;13:77–81. doi: 10.1016/s0169-5347(97)01274-3. [DOI] [PubMed] [Google Scholar]
- 9.Van Kleunen M, Fischer M, Schmid B. Costs of plasticity in foraging characteristics of the clonal plant Ranunculus reptans. Evolution. 2000;54:1947–55. [PubMed] [Google Scholar]
- 10.Agrawal AA, Conner JK, Johnson MTJ, et al. Ecological genetics of an induced plant defense against herbivores: additive genetic variance and costs of phenotypic plasticity. Evolution. 2002;56:2206–13. doi: 10.1111/j.0014-3820.2002.tb00145.x. [DOI] [PubMed] [Google Scholar]
- 11.DeWitt TJ. Costs and limits of phenotypic plasticity: tests with predator-induced morphology and life history in a freshwater snail. J Evol Biol. 1998;11:465–80. [Google Scholar]
- 12.Scheiner SM, Berrigan D. The genetics of phenotypic plasticity. VII. The cost of plasticity in Daphnia pulex. Evolution. 1998;52:368–78. doi: 10.1111/j.1558-5646.1998.tb01638.x. [DOI] [PubMed] [Google Scholar]
- 13.Van Buskirk J, Steiner UK. The fitness cost of developmental canalization and plasticity. J Evol Biol. 2009;22:852–60. doi: 10.1111/j.1420-9101.2009.01685.x. [DOI] [PubMed] [Google Scholar]
- 14.Hodgins-Davis A, Townsend JP. Evolving gene expression: from G to E to Gx. Trends Ecol Evol. 2009;24:649–658. doi: 10.1016/j.tree.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aubin-Horth N, Renn SCP. Genomic reaction norms: using integrative biology to understand molecular mechanisms of phenotypic plasticity. Mol Ecol. 2009;18:3763–80. doi: 10.1111/j.1365-294X.2009.04313.x. [DOI] [PubMed] [Google Scholar]
- 16.Pigliucci M. How organisms respond to environmental changes: from phenotypes to molecules (and vice versa) Trends Ecol Evol. 1996;11:168–73. doi: 10.1016/0169-5347(96)10008-2. [DOI] [PubMed] [Google Scholar]
- 17.Schlichting CD, Smith H. Phenotypic plasticity: linking molecular mechanisms with evolutionary outcomes. Evol Ecol. 2002;16:189–211. [Google Scholar]
- 18.Promislow D. A regulatory network analysis of phenotypic plasticity in yeast. Am Nat. 2005;165:515–23. doi: 10.1086/429161. [DOI] [PubMed] [Google Scholar]
- 19.Raff RA. The Shape of Life: Genes, Development, and the Evolution of Animal Form. Chicago: University of Chicago Press; 1996. [Google Scholar]
- 20.Davidson EH. Genomic Regulatory Systems: Development and Evolution. San Diego: Academic Press; 2001. [Google Scholar]
- 21.Schlosser G, Wagner GP. Modularity in Development and Evolution. Chicago: University of Chicago Press; 2006. [Google Scholar]
- 22.Mezey JG. Modularity. In: Fox CW, Wolf JB, editors. Evolutionary Genetics: Concepts and Case Studies. Oxford: Oxford University Press; 2006. pp. 304–6. [Google Scholar]
- 23.Wagner GP, Altenberg L. Perspective: complex adaptations and the evolution of evolvability. Evolution. 1996;50:967–76. doi: 10.1111/j.1558-5646.1996.tb02339.x. [DOI] [PubMed] [Google Scholar]
- 24.Schlichting CD. Phenotypic integration and environmental change – what are the consequences of differential phenotypic plasticity. BioScience. 1989;39:460–4. [Google Scholar]
- 25.Lipson H, Pollack JB, Suh NP. On the origin of modular variation. Evolution. 2002;56:1549–56. doi: 10.1111/j.0014-3820.2002.tb01466.x. [DOI] [PubMed] [Google Scholar]
- 26.Kashtan N, Alon U. Spontaneous evolution of modularity and network motifs. Proc Natl Acad Sci USA. 2005;102:13773–8. doi: 10.1073/pnas.0503610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kashtan N, Parter M, Dekel E, et al. Extinctions in heterogeneous environments and the evolution of modularity. Evolution. 2009;63:1964–75. doi: 10.1111/j.1558-5646.2009.00684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parter M, Kashtan N, Alon U. Environmental variability and modularity of bacterial metabolic networks. BMC Evol Biol. 2007;7:169. doi: 10.1186/1471-2148-7-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kreimer A, Borenstein E, Gophna U, et al. The evolution of modularity in bacterial metabolic networks. Proc Natl Acad Sci USA. 2008;105:6976–81. doi: 10.1073/pnas.0712149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–56. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 31.Bell G. The ecology and genetics of fitness in Chlamydomonas. 5. The relationship between genetic correlation and environmental variance. Evolution. 1992;46:561–6. doi: 10.1111/j.1558-5646.1992.tb02060.x. [DOI] [PubMed] [Google Scholar]
- 32.Fry JD. On the maintenance of genetic variation by disruptive selection among hosts in a phytophagous mite. Evolution. 1992;46:279–83. doi: 10.1111/j.1558-5646.1992.tb02003.x. [DOI] [PubMed] [Google Scholar]
- 33.Evans JD, Wheeler DE. Gene expression and the evolution of insect polyphenisms. BioEssays. 2001;23:62–8. doi: 10.1002/1521-1878(200101)23:1<62::AID-BIES1008>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 34.Segal E, Shapira M, Regev A, et al. Module networks: identifying regulatory modules and then condition-specific regulators from gene expression data. Nat Genet. 2003;34:166–76. doi: 10.1038/ng1165. [DOI] [PubMed] [Google Scholar]
- 35.Balázsi G, Barabasi AL, Oltvai ZN. Topological units of environmental signal processing in the transcriptional regulatory network of Escherichia coli. Proc Natl Acad Sci USA. 2005;102:7841–6. doi: 10.1073/pnas.0500365102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartwell LH, Hopfield JJ, Leibler S, et al. From molecular to modular biology. Nature. 1999;402:C47–52. doi: 10.1038/35011540. [DOI] [PubMed] [Google Scholar]
- 37.Schlosser G. Modularity and the units of evolution. Theor Biosci. 2002;121:1–80. [Google Scholar]
- 38.Waxman D, Peck JR. Pleiotropy and the preservation of perfection. Science. 1998;279:1210–3. [PubMed] [Google Scholar]
- 39.West-Eberhard MJ. Developmental Plasticity and Evolution. Oxford: Oxford University Press; 2003. [Google Scholar]
- 40.Otto SP. Two steps forward, one step back: the pleiotropic effects of favoured alleles. Proc R Soc Lond B Biol Sci. 2004;271:705–14. doi: 10.1098/rspb.2003.2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pál C, Papp B, Lercher MJ. An integrated view of protein evolution. Nat Rev Genet. 2006;7:337–48. doi: 10.1038/nrg1838. [DOI] [PubMed] [Google Scholar]
- 42.Larracuente AM, Sackton TB, Greenberg AJ, et al. Evolution of protein-coding genes in Drosophila. Trends Genet. 2008;23:114–23. doi: 10.1016/j.tig.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 43.Ellegren H, Parsch J. The evolution of sex-biased genes and sex-biased gene expression. Nat Rev Genet. 2007;8:689–98. doi: 10.1038/nrg2167. [DOI] [PubMed] [Google Scholar]
- 44.Duret L, Mouchiroud D. Determinants of substitution rates in mammalian genes: expression pattern affects selection intensity but not mutation rate. Mol Biol Evol. 2000;17:68–74. doi: 10.1093/oxfordjournals.molbev.a026239. [DOI] [PubMed] [Google Scholar]
- 45.Van Dyken JD, Wade MJ. Quantifying the evolutionary consequences of conditional gene expression in time and space. In Review. [Google Scholar]
- 46.Kawecki TJ. Accumulation of deleterious mutations and the evolutionary cost of being a generalist. Am Nat. 1994;144:833–8. [Google Scholar]
- 47.Fry JD. The evolution of host specialization: Are trade-offs overrated? Am Nat. 1996;148:S84–107. [Google Scholar]
- 48.Whitlock MC. The red queen beats the jack-of-all-trades: the limitations on the evolution of phenotypic plasticity and niche breadth. Am Nat. 1996;148:S65–77. [Google Scholar]
- 49.Kawecki TJ, Barton NH, Fry JD. Mutational collapse of fitness in marginal habitats and the evolution of ecological specialization. J Evol Biol. 1997;10:407–29. [Google Scholar]
- 50.Wade MJ. The evolutionary genetics of maternal effects. In: Mousseau TA, Fox CW, editors. Maternal Effects as Adaptations. Oxford: Oxford University Press; 1998. pp. 5–21. [Google Scholar]
- 51.Whitlock MC, Wade MJ. Speciation: founder events and their effects on X-linked and autosomal genes. Am Nat. 1995;145:676–85. [Google Scholar]
- 52.Rosenzweig ML. Habitat selection as a source of biological diversity. Evol Ecol. 1989;1:315–30. [Google Scholar]
- 53.Holt R, Gaines M. Analysis of adaptation in heterogeneous landscapes: implications for the evolution of fundamental niches. Evol Ecol. 1992;6:433–47. [Google Scholar]
- 54.Holt RD. Demographic constraints in evolution: towards unifying the evolutionary theories of senescence and niche conservatism. Evol Ecol. 1996;6:433–47. [Google Scholar]
- 55.Wade MJ, Priest NK, Cruickshank TE. A theoretical overview of genetic maternal effects: evolutionary predictions and empirical tests with mammalian data. In: Maestripieri D, Mateo JM, editors. Maternal Effects in Mammals. Chicago: University of Chicago Press; 2009. pp. 38–63. [Google Scholar]
- 56.Kimura M. On the probability of fixation of mutant genes in populations. Genetics. 1962;47:713–9. doi: 10.1093/genetics/47.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Linksvayer TA, Wade MJ. Genes with social effects are expected to harbor more sequence variation within and between species. Evolution. 2009;63:1685–96. doi: 10.1111/j.1558-5646.2009.00670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller HJ. Our load of mutations. Am J Hum Genet. 1950;2:111–76. [PMC free article] [PubMed] [Google Scholar]
- 59.Cruickshank T, Wade MJ. Microevolutionary support for a developmental hourglass: gene expression patterns shape sequence variation and divergence in Drosophila. Evol Dev. 2008;10:583–90. doi: 10.1111/j.1525-142X.2008.00273.x. [DOI] [PubMed] [Google Scholar]
- 60.Barker MS, Demuth JP, Wade MJ. Maternal expression relaxes constraint on innovation of the anterior determinant, bicoid. PLoS Genet. 2005;1:527–30. doi: 10.1371/journal.pgen.0010057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Demuth JP, Wade MJ. Maternal expression increases the rate of bicoid evolution by relaxing selective constraint. Genetica. 2007;129:37–43. doi: 10.1007/s10709-006-0031-4. [DOI] [PubMed] [Google Scholar]
- 62.Bennett AF, Lenski RE, Mittler JE. Evolutionary adaptation to temperature 1. Fitness responses of Escherichia coli to changes in its thermal environment. Evolution. 1992;46:16–30. doi: 10.1111/j.1558-5646.1992.tb01981.x. [DOI] [PubMed] [Google Scholar]
- 63.Kassen B, Bell G. Experimental evolution in Chlamydomonas. IV. Selection in environments that vary through time at different scales. Heredity. 1998;80:732–41. [Google Scholar]
- 64.Medawar PB. An Unsolved Problem of Biology. London: H.K Lewis; 1952. [Google Scholar]
- 65.Charlesworth B. Evolution in age-structured populations. Cambridge Stud Math Biol. 1994;13:1–306. [Google Scholar]
- 66.Hughes KA, Charlesworth B. A genetic analysis of senescence in Drosophila. Nature. 1994;367:64–6. doi: 10.1038/367064a0. [DOI] [PubMed] [Google Scholar]
- 67.Cutter AD, Ward S. Sexual and temporal dynamics of molecular evolution in C. elegans development. Mol Biol Evol. 2005;22:178–88. doi: 10.1093/molbev/msh267. [DOI] [PubMed] [Google Scholar]
- 68.Angert AL, Schemske DW. The evolution of species’ distributions: reciprocal transplants across the elevation ranges of Mimulus cardinalis and M. lewisii. Evolution. 2005;59:1671–84. [PubMed] [Google Scholar]
- 69.Jagadeeshan S, Singh RS. Rapidly evolving genes of Drosophila: differing levels of selective pressure in testis, ovary, and head tissues between sibling species. Mol Biol Evol. 2005;22:1793–801. doi: 10.1093/molbev/msi175. [DOI] [PubMed] [Google Scholar]
- 70.Haerty W, Jagadeeshan S, Kulathinal RJ, et al. Evolution in the fast lane: rapidly evolving sex-related genes in Drosophila. Genetics. 2007;177:1321–35. doi: 10.1534/genetics.107.078865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brisson JA, Nuzhdin SV. Rarity of males in pea aphids results in mutational decay. Science. 2008;319:58. doi: 10.1126/science.1147919. [DOI] [PubMed] [Google Scholar]
- 72.Evans JD, Wheeler DE. Differential gene expression between developing queens and workers in the honey bee, Apis mellifera. Proc Natl Acad Sci USA. 1999;96:5575–80. doi: 10.1073/pnas.96.10.5575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rowland JM, Emlen DJ. Two thresholds, three male forms result in facultative male trimorphism in beetles. Science. 2009;323:773–6. doi: 10.1126/science.1167345. [DOI] [PubMed] [Google Scholar]
- 74.Wade MJ. The evolutionary genetics of maternal effects. In: Mousseau TA, Fox CW, editors. Maternal Effects as Adaptations. Oxford: Oxford University Press; 1998. pp. 5–21. [Google Scholar]
- 75.Cooper WS, Kaplan RH. Adaptive coin-flipping: a decision-theoretic examination of natural selection for random individual variation. J Theor Biol. 1982;94:135–51. doi: 10.1016/0022-5193(82)90336-8. [DOI] [PubMed] [Google Scholar]
- 76.Mao EF, Lane L, Lee J, et al. Proliferation of mutators in a cell population. J Bacteriol. 1997;179:417–22. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–5. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- 78.Tenaillon O, Toupance B, Le Nagard H, et al. Mutators, population size, adaptive landscape and the adaptation of asexual populations of bacteria. Genetics. 1999;152:485–93. doi: 10.1093/genetics/152.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gibson G, Dworkin I. Uncovering cryptic genetic variation. Nat Rev Genet. 2004;5:681–90. doi: 10.1038/nrg1426. [DOI] [PubMed] [Google Scholar]
- 80.Schlichting CD. Hidden reaction norms, cryptic genetic variation and evolvability. Ann NY Acad Sci. 2009;1133:187–203. doi: 10.1196/annals.1438.010. [DOI] [PubMed] [Google Scholar]
- 81.Wagner A. Risk management in biological evolution. J Theor Biol. 2003;225:45–57. doi: 10.1016/s0022-5193(03)00219-4. [DOI] [PubMed] [Google Scholar]
- 82.Masel J, King OD, Maughan H. The loss of adaptive plasticity during long periods of environmental stasis. Am Nat. 2007;169:38–46. doi: 10.1086/510212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lahti DC, Johnson NA, Ajie BC, et al. Relaxed selection in the wild. Trends Ecol Evol. 2009;9:487–96. doi: 10.1016/j.tree.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 84.Maughan H, Masel J, Birky CW, et al. The roles of mutation accumulation and selection in loss of sporulation in experimental populations of Bacillus subtilis. Genetics. 2007;177:937–48. doi: 10.1534/genetics.107.075663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maughan H, Birky CW, Nicholson WL. Transcriptome divergence and the loss of plasticity in Bacillus subtilis after 6,000 generations of evolution under relaxed selection for sporulation. J Bacteriol. 2009;191:428–33. doi: 10.1128/JB.01234-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–96. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 87.Bentley SD, Parkhill J. Comparative genomic structure of prokaryotes. Annu Rev Genet. 2004;38:771–91. doi: 10.1146/annurev.genet.38.072902.094318. [DOI] [PubMed] [Google Scholar]
- 88.Reboud X, Bell G. Experimental evolution in Chlamydomonas. 3. Evolution of specialist and generalist types in environments that vary in space and time. Heredity. 1997;78:507–14. [Google Scholar]
- 89.Giraud A, Matic I, Tenaillon O, et al. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science. 2001;291:2606–8. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- 90.MacLean RC, Bell G. Experimental adaptive radiation in Pseudomonas. Am Nat. 2002;160:569–81. doi: 10.1086/342816. [DOI] [PubMed] [Google Scholar]
- 91.Collins S, Bell G. Phenotypic consequences of 1,000 generations of selection at elevated CO2 in a green alga. Nature. 2004;431:566–9. doi: 10.1038/nature02945. [DOI] [PubMed] [Google Scholar]
- 92.MacLean RC. Pleiotropy and GAL pathway degeneration in yeast. J Evol Biol. 2007;20:1333–8. doi: 10.1111/j.1420-9101.2007.01351.x. [DOI] [PubMed] [Google Scholar]
- 93.Turner PE, Elena SF. Cost of host radiation in an RNA virus. Genetics. 2000;156:1465–70. doi: 10.1093/genetics/156.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Elena SF. Restrictions to RNA virus adaptation: an experimental approach. J Gen Mol Microbiol. 2002;81:135–42. doi: 10.1023/a:1020589929125. [DOI] [PubMed] [Google Scholar]
- 95.Levins R. Theory of fitness in a heterogeneous environment. II. Developmental flexibility and niche selection. Am Nat. 1963;839:75–90. [Google Scholar]
- 96.Miyama M, Hanagata N. Microarray analysis of 7029 gene expression patterns in Burma mangrove under high-salinity stress. Plant Sci. 2007;172:948–57. [Google Scholar]
- 97.Ferreira VD, Rocchetta I, Conforti V, et al. Gene expression patterns in Euglena gracilis: insights into the cellular response to environmental stress. Gene. 2007;389:136–45. doi: 10.1016/j.gene.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 98.Vontas J, Blass C, Koutsos AC, et al. Gene expression in insecticide resistant and susceptible Anopheles gambiae strains constitutively or after insecticide exposure. Insect Mol Biol. 2005;14:509–21. doi: 10.1111/j.1365-2583.2005.00582.x. [DOI] [PubMed] [Google Scholar]
- 99.Aubin-Horth N. Interaction of rearing environment and reproductive tactic on gene expression profiles in Atlantic salmon. J Hered. 2005;96:261–78. doi: 10.1093/jhered/esi030. [DOI] [PubMed] [Google Scholar]
- 100.Gasch AP. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–57. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chen DR. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell. 2003;14:214–29. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nikiforova VJ, Gakiere B, Kempa S, et al. Towards dissecting a nutrient metabolism in plants: a systems biology case study on sulphur metabolism. J Exp Bot. 2004;55:1861–70. doi: 10.1093/jxb/erh177. [DOI] [PubMed] [Google Scholar]
- 103.Gracey AY, Fraser EJ, Li WZ, et al. Coping with cold: an integrative, multitissue analysis of the transcriptome of a poikilothermic vertebrate. Proc Natl Acad Sci USA. 2004;101:16970–5. doi: 10.1073/pnas.0403627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Swindell WR, Huebner M, Weber AP. Plastic and adaptive gene expression patterns associated with temperature stress in Arabidopsis thaliana. Heredity. 2007;99:143–50. doi: 10.1038/sj.hdy.6800975. [DOI] [PubMed] [Google Scholar]
- 105.Helmann JD, Wu MFW, Kobel PA, et al. Global transcriptional response of Bacillus subtilis to heat shock. J Bacteriol. 2001;183:7318–28. doi: 10.1128/JB.183.24.7318-7328.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kreps JA, Wu YJ, Chang HS, et al. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–41. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eum JH, Seo YR, Yoe SM, et al. Analysis of the immune-inducible genes of Plutella xylostella using expressed sequence tags and cDNA microarray. Dev Comp Immunol. 2007;31:1107–20. doi: 10.1016/j.dci.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 108.De Vos M, Van Oosten VR, Van Poecke RMP, et al. Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact. 2005;18:923–37. doi: 10.1094/MPMI-18-0923. [DOI] [PubMed] [Google Scholar]
- 109.Schenk PM, Kazan K, Wilson I, et al. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA. 2000;97:11655–60. doi: 10.1073/pnas.97.21.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Reymond P, Weber H, Damond M, et al. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell. 2000;12:707–19. doi: 10.1105/tpc.12.5.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Freimoser FM, Hu G, St Leger RJ. Variation in gene expression patterns as the insect pathogen Metarhizium anisopliae adapts to different host cuticles or nutrient deprivation in vitro. Microbiol-SGM. 2005;151:361–71. doi: 10.1099/mic.0.27560-0. [DOI] [PubMed] [Google Scholar]
- 112.Lawrence SD, Novak NG, Ju CJT, et al. Potato, Solanum tuberosum, defense against Colorado potato beetle, Leptinotarsa decemlineata (Say): microarray gene expression profiling of potato by Colorado potato beetle regurgitant treatment of wounded leaves. J Chem Ecol. 2008;34:1013–25. doi: 10.1007/s10886-008-9507-2. [DOI] [PubMed] [Google Scholar]
- 113.Landry CR, Oh J, Hartl DL, et al. Genome-wide scan reveals that genetic variation for transcriptional plasticity in yeast is biased towards multi-copy and dispensable genes. Gene. 2006;366:343–51. doi: 10.1016/j.gene.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 114.Boer VM, de Winde JH, Pronk JT, et al. The genome-wide transcriptional responses of Saccharomyces cerevisiae grown on glucose in aerobic chemostat cultures limited for carbon, nitrogen, phosphorus or sulfur. J Biol Chem. 2003;278:3265–74. doi: 10.1074/jbc.M209759200. [DOI] [PubMed] [Google Scholar]
- 115.Tani TH, Khodursky A, Blumenthal RM, et al. Adaptation to famine: a family of stationary-phase genes revealed by microarray analysis. Proc Natl Acad Sci USA. 2002;99:13471–6. doi: 10.1073/pnas.212510999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Carsten LD, Watts T, Markow TA. Gene expression patterns accompanying a dietary shift in Drosophila melanogaster. Mol Ecol. 2005;14:3203–8. doi: 10.1111/j.1365-294X.2005.02654.x. [DOI] [PubMed] [Google Scholar]
- 117.Marinotti O, Nguyen QK, Calvo E, et al. Microarray analysis of genes showing variable expression following a blood meal in Anopheles gambiae. Insect Mol Biol. 2005;14:365–73. doi: 10.1111/j.1365-2583.2005.00567.x. [DOI] [PubMed] [Google Scholar]
- 118.Aubin-Horth N. Alternative life histories shape brain gene expression profiles in males of the same population. Proc R Soc Lond B Biol Sci. 2005;272:1655–62. doi: 10.1098/rspb.2005.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Mori T, Hiraka I, Kurata Y, et al. Genetic basis of phenotypic plasticity for predator-induced morphological defenses in anuran tadpole, Rana pirica, using cDNA subtraction and microarray analysis. Biochem Biophys Res Commun. 2005;330:1138–45. doi: 10.1016/j.bbrc.2005.03.091. [DOI] [PubMed] [Google Scholar]
- 120.Barchuk AR, Cristino AS, Kucharski R, et al. Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Dev Biol. 2007;7:70. doi: 10.1186/1471-213X-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Donnell DM, Strand MR. Caste-based differences in gene expression in the polyembryonic wasp Copidosoma floridanum. Insect Biochem Mol Biol. 2006;36:141–53. doi: 10.1016/j.ibmb.2005.11.009. [DOI] [PubMed] [Google Scholar]
- 122.Hojo M, Koshikawa S, Cornette R, et al. Identification of soldier-specific genes in the nasute termite Nasutitermes takasagoensis (Isoptera: Termitidae) Entomol Sci. 2005;8:379–87. [Google Scholar]
- 123.Judice CC, Carazzole MF, Festa F, et al. Gene expression profiles underlying alternative caste phenotypes in a highly eusocial bee, Melipona quadrifasciata. Insect Mol Biol. 2006;15:33–44. doi: 10.1111/j.1365-2583.2005.00605.x. [DOI] [PubMed] [Google Scholar]
- 124.Scharf ME, Wu-Scharf D, Pittendrigh BR, et al. Caste- and development-associated gene expression in a lower termite. Genome Biol. 2003;4:R62. doi: 10.1186/gb-2003-4-10-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Pereboom JJM, Jordan WC, Sumner S, et al. Differential gene expression in queen-worker caste determination in bumble-bees. Proc R Soc Lond B Biol Sci. 2005;272:1145–52. doi: 10.1098/rspb.2005.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sumner S, Pereboom JJM, Jordan WC. Differential gene expression and phenotypic plasticity in behavioural castes of the primitively eusocial wasp, Polistes canadensis. Proc R Soc Lond B Biol Sci. 2006;273:19–26. doi: 10.1098/rspb.2005.3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Whitfield CW, Ben-Shahar Y, Brillet C, et al. Genomic dissection of behavioral maturation in the honey bee. Proc Natl Acad Sci USA. 2006;103:16068–75. doi: 10.1073/pnas.0606909103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.McDonald JH, Kreitman M. Adaptive protein evolution at the ADH locus in Drosophila. Nature. 1991;351:652–4. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 129.Crow JF, Kimura M. An Introduction to Population Genetics Theory. New York: Harper and Row; 1970. [Google Scholar]
- 130.Kimura M, Ohta T. Protein polymorphism as a phase of molecular evolution. Nature. 1971;229:467–9. doi: 10.1038/229467a0. [DOI] [PubMed] [Google Scholar]
- 131.Tirosh I, Reikhav S, Levy AA, et al. A yeast hybrid provides insight into the evolution of gene expression regulation. Science. 2009;324:659–62. doi: 10.1126/science.1169766. [DOI] [PubMed] [Google Scholar]
- 132.Scheiner SM. The genetics of phenotypic plasticity. VII. Evolution in a spatially-structured environment. J Evol Biol. 1998;11:303–20. [Google Scholar]


