Abstract
The title compound, [Mn2O2(C10H24N4)2](C24H20B)2Cl·2CH3CN, is a mixed-valent MnIII/MnIV oxide-bridged manganese dimer with one chloride and two tetraphenylborate counter-anions. There are two non-coordinated molecules of acetonitrile in the formula unit. A center of inversion is present between the two metal atoms, and, consequently, there is no distinction between MnIII and MnIV metal centers. In the Mn2O2 core, the Mn—O distances are 1.817 (3) and 1.821 (3) Å. The cyclam ligand is in the cis configuration. The chloride counter-anion resides on a center of symmetry, whereas the tetraphenylborate counter-anion is in a general position. The cyclam ligand is hydrogen bonded to the acetonitrile as well as to the chloride anion. One of the phenyl rings of the anion and the acetonitrile solvent molecule are each disordered over two sets of sites.
Related literature
For structures of different salts containing the disordered mixed-valent {[(cyclam)MnO]2}3+ cation, see: Goodson et al. (1990 ▶); Lu et al. (2001 ▶). For structures of the non-disordered MnIII—MnIVO2 core, see: Brewer et al. (1989 ▶); Levaton & Olmstead (2010 ▶). For cyclam configurations, see: Bosnich et al. (1965 ▶).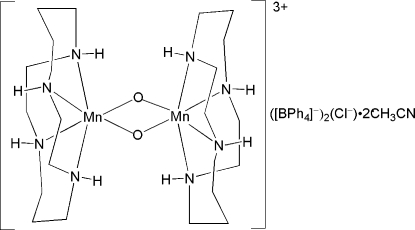
Experimental
Crystal data
[Mn2O2(C10H24N4)2](C24H20B)2Cl·2C2H3N
M r = 1298.52
Triclinic,
a = 11.437 (2) Å
b = 11.713 (2) Å
c = 13.967 (3) Å
α = 104.136 (3)°
β = 97.697 (3)°
γ = 107.627 (3)°
V = 1684.9 (6) Å3
Z = 1
Mo Kα radiation
μ = 0.47 mm−1
T = 90 K
0.15 × 0.11 × 0.08 mm
Data collection
Bruker SMART APEXII diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.952, T max = 0.972
17931 measured reflections
6092 independent reflections
4083 reflections with I > 2σ(I)
R int = 0.064
Refinement
R[F 2 > 2σ(F 2)] = 0.061
wR(F 2) = 0.173
S = 1.03
6092 reflections
433 parameters
258 restraints
H-atom parameters constrained
Δρmax = 0.54 e Å−3
Δρmin = −0.81 e Å−3
Data collection: APEX2 (Bruker, 2007 ▶); cell refinement: SAINT (Bruker, 2007 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: XP in SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXL97.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811019829/hg5039sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811019829/hg5039Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Selected bond lengths (Å).
| Mn1—O1 | 1.817 (3) |
| Mn1—O1i | 1.821 (3) |
| Mn1—N1 | 2.187 (5) |
| Mn1—N2 | 2.092 (3) |
| Mn1—N3 | 2.178 (3) |
| Mn1—N4 | 2.116 (3) |
Symmetry code: (i) 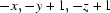 .
.
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1⋯N5 | 0.93 | 2.42 | 3.313 (11) | 160 |
| N1—H1⋯N5B | 0.93 | 1.98 | 2.832 (10) | 151 |
| N2—H2⋯Cl1 | 0.93 | 2.37 | 3.289 (4) | 169 |
| N3—H3⋯N5i | 0.93 | 2.22 | 3.034 (9) | 146 |
| N3—H3⋯N5Bi | 0.93 | 2.23 | 3.120 (9) | 160 |
| N4—H4⋯Cl1 | 0.93 | 2.42 | 3.330 (4) | 168 |
Symmetry code: (i) 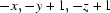 .
.
Acknowledgments
The authors thank the University of California, Davis, for acquisition of the Bruker SMART APEXII diffractometer and the support of the Summer Undergraduate Research Program of the University of California, Davis.
supplementary crystallographic information
Comment
In the title compound (Fig. 1), the MnIII—MnIV centrosymmetric, dinuclear cation is bridged by two oxo ligands. Each manganese atom also binds to a tetradentate cyclam (cyclam = 1,4,8,11-tetraazacyclotetradecane) to achieve a distorted octahedral coordination environment. Due to the center of inversion, there is no distinction between the two different oxidation states in the structure, and it is disordered mixed valent. The dinuclear cation bears a +3 charge, and the charge is balanced by a chloride anion and two tetraphenylborate anions. The chloride resides on a center of symmetry. The configuration of the cyclam ligand is cis-V, in which the N—H bonds alternate above and below the N4 plane (Bosnich et al., 1965).
The structures of similar disordered mixed valent (III/IV) µ-oxo bridged manganese cyclam complexes are reported in Lu, et al., 2001, with two perchlorate and one nitrate anions; Goodson, et al., 1990, with dithionate and thiosulfate anions, and a second structure with three bromides. In the title compound, the Mn2O2 core has Mn—O distances of 1.817 (3) and 1.821 (3) Å. All of the above referenced structures show similar coordination geometry, and the mean Mn—O bond distances in the Mn2O2 core for these complexes is 1.824 Å. A trifluoromethanesulfonate salt (Brewer, et al., 1989) crystallized with discrete MnIII and MnIV metal centers, and these mean Mn—O distances are characteristic of the oxidation states of Mn: MnIII—O, 1.861 Å and MnIV—O, 1.788 Å. Related values were also seen in the structure of a large Mn12 cluster with three units of Mn2O2 core geometry. Average values with average deviations from the mean were MnIII—O, 1.879 (7) Å and MnIV—O, 1.787 (6) Å (Levaton & Olmstead, 2010). Thus, the disordered distances agree well with the average III/IV values.
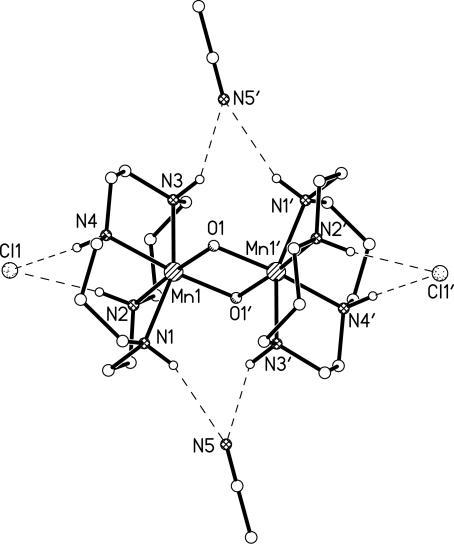
In the structure of the title compound, all of the available hydrogen atoms bonded to the N atoms in the cyclam ligand participate in hydrogen bonding (Fig. 2 and Table 2). The hydrogen atoms bonded to N atoms N2 and N4 are hydrogen bonded to chloride atom. The hydrogen atoms on N atoms N1 and N3 hydrogen bond to the acetonitrile N atoms.
Experimental
The title compound was obtained while attempting to prepare an azido derivative. To a stirred solution of 1,4,8,11-tetraazacyclotetradecane (cyclam), (200 mg, 1 mmol) in 5 ml of methanol was added a solution of MnCl2.4H2O (200 mg, 1 mmol) in 25 ml of methanol. Over the one hour course of the addition, the reaction color progressed from red to dark green. After stirring for 1 hr an excess of NaN3 (200 mg, 2.5 mmol) was added to the reaction mixture in a solution of 9 ml me thanol and 1 ml H2O. No color change was observed upon addition of the azide. After 20 min of stirring NaBPh4 (350 mg, 1 mmol) was added. Addition of the tetraphenylborate salt caused a precipitate to form and the solution turned red-brown. Subsequently, 18 ml of a 5:1 acetonitrile:water and 10 ml of 0.1 M HClO4 were added. The orange-brown solid was collected by filtration. Dichroic, red-green crystals of the product were obtained by slow evaporation of a toluene solution.
Refinement
The C-bound and N-bound H atoms were positioned geometrically with C—H = 0.95—0.98 Å, N—H = 0.88 Å, and allowed to ride on their parent atoms with Uiso(H) = 1.2—1.5 Ueq(C). One of the BPh4- phenyl rings was disordered and was refined in two parts with a rigid group refinement. The acetonitrile molecule was disordered in two parts, each assigned 50% occupancy. Atoms of the disordered acetonitriles were kept isotropic. The atoms of the disordered BPh4- phenyl group were allowed to refine with anisotropic thermal parameters and a similarity restraint (SIMU) of 0.008. The cation displayed large thermal motion and thermal ellipsoids for these atoms were refined with an ISOR 0.01 restraint. An attempt was made to solve and refine the structure in the non-centrosymmetric space group P1 but atoms of the cation became N.P.D.
Figures
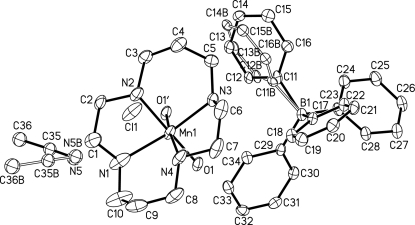
Fig. 1.
The asymmetric unit of the title complex with displacement parameters drawn at the 30% probability level. Disordered components of the acetonitrile and [BPh4]- group are shown.
Fig. 2.
The centrosymmetric dimer of the title cation is shown with H-bonds involving the N—H groups, acetonitriles and chlorides depicted with dashed lines.
Crystal data
| [Mn2O2(C10H24N4)2](C24H20B)2Cl·2C2H3N | Z = 1 |
| Mr = 1298.52 | F(000) = 689 |
| Triclinic, P1 | Dx = 1.280 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 11.437 (2) Å | Cell parameters from 3568 reflections |
| b = 11.713 (2) Å | θ = 2.8–24.6° |
| c = 13.967 (3) Å | µ = 0.47 mm−1 |
| α = 104.136 (3)° | T = 90 K |
| β = 97.697 (3)° | Block, red |
| γ = 107.627 (3)° | 0.15 × 0.11 × 0.08 mm |
| V = 1684.9 (6) Å3 |
Data collection
| Bruker SMART APEXII diffractometer | 6092 independent reflections |
| Radiation source: fine-focus sealed tube | 4083 reflections with I > 2σ(I) |
| graphite | Rint = 0.064 |
| Detector resolution: 8.3 pixels mm-1 | θmax = 25.3°, θmin = 2.8° |
| ω scans | h = −13→13 |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | k = −14→14 |
| Tmin = 0.952, Tmax = 0.972 | l = −16→16 |
| 17931 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.061 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.173 | H-atom parameters constrained |
| S = 1.03 | w = 1/[σ2(Fo2) + (0.080P)2 + 1.3759P] where P = (Fo2 + 2Fc2)/3 |
| 6092 reflections | (Δ/σ)max = 0.007 |
| 433 parameters | Δρmax = 0.54 e Å−3 |
| 258 restraints | Δρmin = −0.81 e Å−3 |
Special details
| Experimental. Crystals were dichroic. |
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Cl1 | 0.5000 | 0.5000 | 0.5000 | 0.0774 (7) | |
| Mn1 | 0.11925 (5) | 0.50038 (5) | 0.50106 (5) | 0.0434 (2) | |
| O1 | −0.0251 (3) | 0.4058 (2) | 0.5257 (2) | 0.0480 (8) | |
| N1 | 0.2187 (3) | 0.6175 (4) | 0.6555 (4) | 0.0738 (14) | |
| H1 | 0.1742 | 0.6709 | 0.6738 | 0.089* | |
| N2 | 0.2773 (3) | 0.6205 (3) | 0.4709 (3) | 0.0509 (10) | |
| H2 | 0.3380 | 0.5826 | 0.4695 | 0.061* | |
| N3 | 0.0891 (3) | 0.3830 (3) | 0.3463 (2) | 0.0357 (7) | |
| H3 | 0.0049 | 0.3316 | 0.3296 | 0.043* | |
| N4 | 0.2090 (3) | 0.3743 (3) | 0.5289 (3) | 0.0579 (11) | |
| H4 | 0.2942 | 0.4107 | 0.5312 | 0.069* | |
| C1 | 0.3437 (4) | 0.7009 (5) | 0.6530 (4) | 0.0686 (15) | |
| H1B | 0.4048 | 0.6565 | 0.6528 | 0.082* | |
| H1C | 0.3751 | 0.7765 | 0.7130 | 0.082* | |
| C2 | 0.3272 (5) | 0.7373 (4) | 0.5578 (4) | 0.0658 (15) | |
| H2A | 0.2677 | 0.7837 | 0.5590 | 0.079* | |
| H2B | 0.4090 | 0.7923 | 0.5518 | 0.079* | |
| C3 | 0.2571 (4) | 0.6510 (4) | 0.3745 (3) | 0.0479 (11) | |
| H3A | 0.3327 | 0.7201 | 0.3744 | 0.058* | |
| H3B | 0.1850 | 0.6811 | 0.3705 | 0.058* | |
| C4 | 0.2318 (4) | 0.5416 (4) | 0.2834 (4) | 0.0547 (12) | |
| H4A | 0.2972 | 0.5034 | 0.2925 | 0.066* | |
| H4B | 0.2384 | 0.5720 | 0.2234 | 0.066* | |
| C5 | 0.1037 (4) | 0.4420 (4) | 0.2635 (3) | 0.0538 (12) | |
| H5A | 0.0381 | 0.4802 | 0.2554 | 0.065* | |
| H5B | 0.0903 | 0.3760 | 0.1991 | 0.065* | |
| C6 | 0.1589 (4) | 0.2975 (4) | 0.3453 (4) | 0.0594 (13) | |
| H6A | 0.2471 | 0.3397 | 0.3428 | 0.071* | |
| H6B | 0.1202 | 0.2226 | 0.2849 | 0.071* | |
| C7 | 0.1555 (4) | 0.2593 (4) | 0.4390 (4) | 0.0676 (15) | |
| H7A | 0.2054 | 0.2038 | 0.4417 | 0.081* | |
| H7B | 0.0676 | 0.2123 | 0.4393 | 0.081* | |
| C8 | 0.1982 (4) | 0.3418 (5) | 0.6219 (4) | 0.0695 (16) | |
| H8A | 0.2278 | 0.2704 | 0.6209 | 0.083* | |
| H8B | 0.1086 | 0.3145 | 0.6256 | 0.083* | |
| C9 | 0.2735 (5) | 0.4502 (5) | 0.7142 (4) | 0.0677 (15) | |
| H9A | 0.3608 | 0.4829 | 0.7057 | 0.081* | |
| H9B | 0.2770 | 0.4181 | 0.7735 | 0.081* | |
| C10 | 0.2253 (6) | 0.5587 (6) | 0.7381 (6) | 0.098 (2) | |
| H10A | 0.1403 | 0.5281 | 0.7515 | 0.117* | |
| H10B | 0.2812 | 0.6235 | 0.8006 | 0.117* | |
| B1 | 0.2591 (4) | −0.1045 (4) | 0.1854 (3) | 0.0326 (9) | |
| C11 | 0.3193 (5) | 0.0254 (4) | 0.1481 (3) | 0.0315 (16) | 0.542 (4) |
| C12 | 0.3289 (5) | 0.1438 (5) | 0.2064 (3) | 0.0257 (15) | 0.542 (4) |
| H12 | 0.3015 | 0.1535 | 0.2682 | 0.031* | 0.542 (4) |
| C13 | 0.3786 (5) | 0.2481 (4) | 0.1743 (4) | 0.0308 (17) | 0.542 (4) |
| H13 | 0.3852 | 0.3291 | 0.2141 | 0.037* | 0.542 (4) |
| C14 | 0.4187 (5) | 0.2339 (4) | 0.0838 (4) | 0.0334 (15) | 0.542 (4) |
| H14 | 0.4527 | 0.3051 | 0.0618 | 0.040* | 0.542 (4) |
| C15 | 0.4090 (4) | 0.1154 (4) | 0.0255 (3) | 0.0401 (16) | 0.542 (4) |
| H15 | 0.4364 | 0.1057 | −0.0363 | 0.048* | 0.542 (4) |
| C16 | 0.3593 (5) | 0.0112 (3) | 0.0577 (3) | 0.0356 (15) | 0.542 (4) |
| H16 | 0.3527 | −0.0698 | 0.0178 | 0.043* | 0.542 (4) |
| C11B | 0.3432 (5) | 0.0282 (5) | 0.1656 (4) | 0.0263 (17) | 0.458 (4) |
| C12B | 0.2979 (5) | 0.1267 (7) | 0.1898 (5) | 0.0306 (19) | 0.458 (4) |
| H12B | 0.2276 | 0.1178 | 0.2203 | 0.037* | 0.458 (4) |
| C13B | 0.3555 (6) | 0.2381 (5) | 0.1693 (5) | 0.0330 (19) | 0.458 (4) |
| H13B | 0.3245 | 0.3054 | 0.1858 | 0.040* | 0.458 (4) |
| C14B | 0.4583 (5) | 0.2511 (4) | 0.1246 (4) | 0.0325 (18) | 0.458 (4) |
| H14B | 0.4977 | 0.3273 | 0.1106 | 0.039* | 0.458 (4) |
| C15B | 0.5037 (5) | 0.1527 (5) | 0.1004 (4) | 0.0371 (17) | 0.458 (4) |
| H15B | 0.5740 | 0.1615 | 0.0698 | 0.045* | 0.458 (4) |
| C16B | 0.4461 (5) | 0.0412 (4) | 0.1209 (4) | 0.0309 (16) | 0.458 (4) |
| H16B | 0.4771 | −0.0261 | 0.1043 | 0.037* | 0.458 (4) |
| C17 | 0.3410 (3) | −0.1988 (3) | 0.1658 (3) | 0.0321 (8) | |
| C18 | 0.4455 (4) | −0.1898 (4) | 0.2359 (3) | 0.0385 (9) | |
| H18 | 0.4718 | −0.1249 | 0.2987 | 0.046* | |
| C19 | 0.5126 (4) | −0.2704 (4) | 0.2186 (3) | 0.0434 (10) | |
| H19 | 0.5823 | −0.2602 | 0.2694 | 0.052* | |
| C20 | 0.4795 (4) | −0.3655 (4) | 0.1283 (3) | 0.0413 (9) | |
| H20 | 0.5247 | −0.4216 | 0.1163 | 0.050* | |
| C21 | 0.3785 (4) | −0.3764 (3) | 0.0563 (3) | 0.0387 (9) | |
| H21 | 0.3543 | −0.4401 | −0.0071 | 0.046* | |
| C22 | 0.3121 (3) | −0.2956 (3) | 0.0752 (3) | 0.0350 (9) | |
| H22 | 0.2427 | −0.3065 | 0.0238 | 0.042* | |
| C23 | 0.1172 (3) | −0.1789 (3) | 0.1131 (3) | 0.0338 (8) | |
| C24 | 0.0576 (4) | −0.1332 (4) | 0.0454 (3) | 0.0405 (9) | |
| H24 | 0.1015 | −0.0532 | 0.0393 | 0.049* | |
| C25 | −0.0642 (4) | −0.2001 (4) | −0.0140 (3) | 0.0444 (10) | |
| H25 | −0.1010 | −0.1655 | −0.0595 | 0.053* | |
| C26 | −0.1310 (4) | −0.3161 (4) | −0.0067 (3) | 0.0428 (10) | |
| H26 | −0.2145 | −0.3611 | −0.0458 | 0.051* | |
| C27 | −0.0750 (4) | −0.3662 (4) | 0.0581 (3) | 0.0403 (9) | |
| H27 | −0.1190 | −0.4469 | 0.0626 | 0.048* | |
| C28 | 0.0459 (3) | −0.2981 (3) | 0.1165 (3) | 0.0357 (9) | |
| H28 | 0.0822 | −0.3340 | 0.1611 | 0.043* | |
| C29 | 0.2486 (3) | −0.0639 (3) | 0.3049 (3) | 0.0291 (8) | |
| C30 | 0.1384 (3) | −0.0995 (3) | 0.3392 (3) | 0.0341 (8) | |
| H30 | 0.0621 | −0.1500 | 0.2908 | 0.041* | |
| C31 | 0.1344 (4) | −0.0647 (3) | 0.4414 (3) | 0.0393 (9) | |
| H31 | 0.0564 | −0.0909 | 0.4608 | 0.047* | |
| C32 | 0.2429 (4) | 0.0075 (3) | 0.5140 (3) | 0.0380 (9) | |
| H32 | 0.2413 | 0.0289 | 0.5838 | 0.046* | |
| C33 | 0.3534 (4) | 0.0479 (3) | 0.4835 (3) | 0.0373 (9) | |
| H33 | 0.4290 | 0.0997 | 0.5323 | 0.045* | |
| C34 | 0.3548 (3) | 0.0130 (3) | 0.3813 (3) | 0.0353 (9) | |
| H34 | 0.4325 | 0.0432 | 0.3624 | 0.042* | |
| N5 | 0.1329 (9) | 0.8609 (9) | 0.7468 (7) | 0.053 (2)* | 0.50 |
| C35 | 0.1548 (8) | 0.9672 (8) | 0.7751 (6) | 0.0402 (18)* | 0.50 |
| C36 | 0.1803 (8) | 1.0976 (8) | 0.8123 (7) | 0.050 (2)* | 0.50 |
| H36A | 0.1058 | 1.1123 | 0.8322 | 0.076* | 0.50 |
| H36B | 0.2014 | 1.1365 | 0.7593 | 0.076* | 0.50 |
| H36C | 0.2514 | 1.1346 | 0.8713 | 0.076* | 0.50 |
| N5B | 0.1679 (8) | 0.8389 (8) | 0.7379 (6) | 0.047 (2)* | 0.50 |
| C35B | 0.2034 (8) | 0.9323 (8) | 0.7992 (6) | 0.0432 (19)* | 0.50 |
| C36B | 0.2519 (9) | 1.0513 (9) | 0.8797 (7) | 0.059 (2)* | 0.50 |
| H36D | 0.2796 | 1.0372 | 0.9440 | 0.088* | 0.50 |
| H36E | 0.1856 | 1.0877 | 0.8854 | 0.088* | 0.50 |
| H36F | 0.3234 | 1.1091 | 0.8641 | 0.088* | 0.50 |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0257 (8) | 0.0762 (12) | 0.1513 (19) | 0.0152 (8) | 0.0083 (9) | 0.0805 (13) |
| Mn1 | 0.0283 (3) | 0.0300 (3) | 0.0637 (4) | −0.0002 (2) | −0.0153 (3) | 0.0263 (3) |
| O1 | 0.0452 (16) | 0.0273 (13) | 0.0557 (17) | −0.0017 (12) | −0.0218 (13) | 0.0209 (13) |
| N1 | 0.034 (2) | 0.083 (3) | 0.134 (4) | 0.026 (2) | 0.020 (2) | 0.079 (3) |
| N2 | 0.0361 (18) | 0.0423 (19) | 0.070 (2) | 0.0029 (15) | −0.0116 (17) | 0.0352 (18) |
| N3 | 0.0269 (16) | 0.0314 (16) | 0.0460 (19) | 0.0098 (13) | 0.0047 (14) | 0.0092 (14) |
| N4 | 0.0307 (18) | 0.045 (2) | 0.097 (3) | 0.0030 (15) | −0.0096 (18) | 0.045 (2) |
| C1 | 0.032 (2) | 0.086 (4) | 0.081 (3) | 0.002 (2) | −0.006 (2) | 0.047 (3) |
| C2 | 0.052 (3) | 0.049 (3) | 0.065 (3) | −0.020 (2) | −0.023 (2) | 0.030 (2) |
| C3 | 0.043 (2) | 0.041 (2) | 0.065 (3) | 0.0157 (19) | 0.001 (2) | 0.029 (2) |
| C4 | 0.051 (3) | 0.047 (2) | 0.078 (3) | 0.020 (2) | 0.034 (2) | 0.026 (2) |
| C5 | 0.061 (3) | 0.054 (3) | 0.040 (2) | 0.010 (2) | 0.019 (2) | 0.013 (2) |
| C6 | 0.041 (2) | 0.039 (2) | 0.094 (4) | 0.019 (2) | 0.011 (2) | 0.009 (2) |
| C7 | 0.044 (3) | 0.032 (2) | 0.120 (4) | 0.011 (2) | −0.013 (3) | 0.030 (3) |
| C8 | 0.037 (2) | 0.070 (3) | 0.115 (4) | 0.012 (2) | −0.001 (3) | 0.070 (3) |
| C9 | 0.068 (3) | 0.092 (4) | 0.089 (4) | 0.053 (3) | 0.035 (3) | 0.068 (3) |
| C10 | 0.099 (4) | 0.115 (4) | 0.154 (5) | 0.072 (4) | 0.082 (4) | 0.100 (4) |
| B1 | 0.033 (2) | 0.026 (2) | 0.032 (2) | 0.0078 (18) | 0.0034 (18) | 0.0023 (18) |
| C11 | 0.020 (3) | 0.033 (3) | 0.030 (3) | 0.006 (2) | −0.005 (2) | 0.001 (2) |
| C12 | 0.014 (3) | 0.028 (3) | 0.027 (3) | 0.005 (2) | −0.007 (2) | 0.006 (2) |
| C13 | 0.021 (3) | 0.030 (3) | 0.036 (3) | 0.007 (2) | −0.005 (3) | 0.007 (3) |
| C14 | 0.025 (3) | 0.039 (3) | 0.038 (3) | 0.010 (2) | 0.000 (3) | 0.022 (2) |
| C15 | 0.035 (3) | 0.046 (3) | 0.042 (3) | 0.018 (3) | 0.007 (3) | 0.015 (3) |
| C16 | 0.034 (3) | 0.037 (3) | 0.034 (3) | 0.013 (2) | 0.006 (2) | 0.010 (2) |
| C11B | 0.028 (3) | 0.028 (3) | 0.021 (3) | 0.012 (2) | −0.006 (3) | 0.009 (2) |
| C12B | 0.023 (3) | 0.030 (3) | 0.027 (3) | 0.006 (3) | −0.011 (3) | 0.002 (3) |
| C13B | 0.024 (3) | 0.032 (3) | 0.034 (3) | 0.010 (3) | −0.010 (3) | 0.005 (3) |
| C14B | 0.033 (3) | 0.033 (3) | 0.024 (3) | 0.005 (3) | −0.006 (3) | 0.009 (3) |
| C15B | 0.037 (3) | 0.039 (3) | 0.032 (3) | 0.010 (3) | 0.007 (3) | 0.010 (3) |
| C16B | 0.033 (3) | 0.032 (3) | 0.027 (3) | 0.014 (3) | 0.003 (3) | 0.006 (3) |
| C17 | 0.0302 (19) | 0.0253 (18) | 0.035 (2) | 0.0055 (15) | 0.0050 (16) | 0.0053 (15) |
| C18 | 0.034 (2) | 0.036 (2) | 0.038 (2) | 0.0101 (17) | 0.0039 (17) | 0.0038 (17) |
| C19 | 0.038 (2) | 0.044 (2) | 0.047 (2) | 0.0149 (19) | 0.0024 (18) | 0.014 (2) |
| C20 | 0.043 (2) | 0.035 (2) | 0.049 (2) | 0.0177 (18) | 0.0129 (19) | 0.0111 (19) |
| C21 | 0.042 (2) | 0.0259 (19) | 0.043 (2) | 0.0102 (17) | 0.0102 (18) | 0.0023 (17) |
| C22 | 0.032 (2) | 0.0282 (19) | 0.037 (2) | 0.0063 (16) | 0.0026 (16) | 0.0037 (16) |
| C23 | 0.038 (2) | 0.0288 (19) | 0.0294 (19) | 0.0145 (17) | 0.0036 (16) | −0.0010 (15) |
| C24 | 0.045 (2) | 0.032 (2) | 0.038 (2) | 0.0120 (18) | −0.0007 (18) | 0.0047 (17) |
| C25 | 0.045 (2) | 0.042 (2) | 0.039 (2) | 0.018 (2) | −0.0056 (18) | 0.0048 (18) |
| C26 | 0.036 (2) | 0.039 (2) | 0.038 (2) | 0.0102 (18) | −0.0070 (18) | −0.0029 (18) |
| C27 | 0.037 (2) | 0.032 (2) | 0.042 (2) | 0.0108 (17) | −0.0014 (18) | −0.0001 (17) |
| C28 | 0.032 (2) | 0.032 (2) | 0.035 (2) | 0.0112 (16) | −0.0029 (16) | 0.0024 (16) |
| C29 | 0.0282 (19) | 0.0228 (17) | 0.034 (2) | 0.0090 (15) | 0.0036 (15) | 0.0065 (15) |
| C30 | 0.031 (2) | 0.0270 (18) | 0.040 (2) | 0.0086 (16) | 0.0029 (16) | 0.0073 (16) |
| C31 | 0.042 (2) | 0.033 (2) | 0.051 (2) | 0.0133 (18) | 0.0187 (19) | 0.0204 (18) |
| C32 | 0.056 (3) | 0.035 (2) | 0.031 (2) | 0.0222 (19) | 0.0103 (18) | 0.0146 (17) |
| C33 | 0.037 (2) | 0.036 (2) | 0.033 (2) | 0.0134 (17) | −0.0016 (17) | 0.0041 (17) |
| C34 | 0.032 (2) | 0.035 (2) | 0.033 (2) | 0.0094 (16) | 0.0041 (16) | 0.0040 (16) |
Geometric parameters (Å, °)
| Mn1—O1 | 1.817 (3) | C15—C16 | 1.3900 |
| Mn1—O1i | 1.821 (3) | C15—H15 | 0.9500 |
| Mn1—N1 | 2.187 (5) | C16—H16 | 0.9500 |
| Mn1—N2 | 2.092 (3) | C11B—C12B | 1.3900 |
| Mn1—N3 | 2.178 (3) | C11B—C16B | 1.3900 |
| Mn1—N4 | 2.116 (3) | C12B—C13B | 1.3900 |
| Mn1—Mn1i | 2.7211 (13) | C12B—H12B | 0.9500 |
| N1—C1 | 1.478 (6) | C13B—C14B | 1.3900 |
| N1—C10 | 1.485 (7) | C13B—H13B | 0.9500 |
| N1—H1 | 0.9300 | C14B—C15B | 1.3900 |
| N2—C3 | 1.482 (5) | C14B—H14B | 0.9500 |
| N2—C2 | 1.484 (6) | C15B—C16B | 1.3900 |
| N2—H2 | 0.9300 | C15B—H15B | 0.9500 |
| N3—C6 | 1.456 (5) | C16B—H16B | 0.9500 |
| N3—C5 | 1.488 (5) | C17—C18 | 1.399 (5) |
| N3—H3 | 0.9300 | C17—C22 | 1.399 (5) |
| N4—C8 | 1.450 (6) | C18—C19 | 1.385 (5) |
| N4—C7 | 1.495 (6) | C18—H18 | 0.9500 |
| N4—H4 | 0.9300 | C19—C20 | 1.380 (6) |
| C1—C2 | 1.497 (6) | C19—H19 | 0.9500 |
| C1—H1B | 0.9900 | C20—C21 | 1.379 (5) |
| C1—H1C | 0.9900 | C20—H20 | 0.9500 |
| C2—H2A | 0.9900 | C21—C22 | 1.382 (5) |
| C2—H2B | 0.9900 | C21—H21 | 0.9500 |
| C3—C4 | 1.492 (6) | C22—H22 | 0.9500 |
| C3—H3A | 0.9900 | C23—C24 | 1.393 (5) |
| C3—H3B | 0.9900 | C23—C28 | 1.404 (5) |
| C4—C5 | 1.511 (6) | C24—C25 | 1.398 (5) |
| C4—H4A | 0.9900 | C24—H24 | 0.9500 |
| C4—H4B | 0.9900 | C25—C26 | 1.379 (6) |
| C5—H5A | 0.9900 | C25—H25 | 0.9500 |
| C5—H5B | 0.9900 | C26—C27 | 1.379 (5) |
| C6—C7 | 1.484 (7) | C26—H26 | 0.9500 |
| C6—H6A | 0.9900 | C27—C28 | 1.387 (5) |
| C6—H6B | 0.9900 | C27—H27 | 0.9500 |
| C7—H7A | 0.9900 | C28—H28 | 0.9500 |
| C7—H7B | 0.9900 | C29—C30 | 1.391 (5) |
| C8—C9 | 1.503 (7) | C29—C34 | 1.395 (5) |
| C8—H8A | 0.9900 | C30—C31 | 1.396 (5) |
| C8—H8B | 0.9900 | C30—H30 | 0.9500 |
| C9—C10 | 1.516 (7) | C31—C32 | 1.375 (5) |
| C9—H9A | 0.9900 | C31—H31 | 0.9500 |
| C9—H9B | 0.9900 | C32—C33 | 1.372 (6) |
| C10—H10A | 0.9900 | C32—H32 | 0.9500 |
| C10—H10B | 0.9900 | C33—C34 | 1.389 (5) |
| B1—C23 | 1.639 (5) | C33—H33 | 0.9500 |
| B1—C17 | 1.649 (5) | C34—H34 | 0.9500 |
| B1—C29 | 1.651 (5) | N5—C35 | 1.146 (11) |
| B1—C11B | 1.672 (6) | C35—C36 | 1.411 (11) |
| B1—C11 | 1.705 (6) | C36—H36A | 0.9800 |
| C11—C12 | 1.3900 | C36—H36B | 0.9800 |
| C11—C16 | 1.3900 | C36—H36C | 0.9800 |
| C12—C13 | 1.3900 | N5B—C35B | 1.129 (11) |
| C12—H12 | 0.9500 | C35B—C36B | 1.454 (12) |
| C13—C14 | 1.3900 | C36B—H36D | 0.9800 |
| C13—H13 | 0.9500 | C36B—H36E | 0.9800 |
| C14—C15 | 1.3900 | C36B—H36F | 0.9800 |
| C14—H14 | 0.9500 | ||
| O1—Mn1—O1i | 83.18 (11) | C17—B1—C11B | 106.0 (4) |
| O1—Mn1—N1 | 100.05 (14) | C29—B1—C11B | 105.6 (3) |
| O1—Mn1—N2 | 174.88 (13) | C23—B1—C11 | 105.4 (3) |
| O1—Mn1—N3 | 95.63 (12) | C17—B1—C11 | 111.8 (3) |
| O1—Mn1—N4 | 90.76 (13) | C29—B1—C11 | 110.7 (3) |
| O1i—Mn1—N1 | 97.81 (13) | C12—C11—C16 | 120.0 |
| O1i—Mn1—N2 | 91.73 (13) | C12—C11—B1 | 121.2 (3) |
| O1i—Mn1—N3 | 97.79 (12) | C16—C11—B1 | 118.8 (3) |
| O1i—Mn1—N4 | 173.47 (12) | C13—C12—C11 | 120.0 |
| N1—Mn1—N2 | 80.14 (14) | C13—C12—H12 | 120.0 |
| N1—Mn1—N3 | 159.04 (14) | C11—C12—H12 | 120.0 |
| N1—Mn1—N4 | 85.59 (15) | C12—C13—C14 | 120.0 |
| N2—Mn1—N3 | 85.49 (13) | C12—C13—H13 | 120.0 |
| N2—Mn1—N4 | 94.35 (14) | C14—C13—H13 | 120.0 |
| N3—Mn1—N4 | 80.34 (14) | C15—C14—C13 | 120.0 |
| Mn1—O1—Mn1i | 96.82 (11) | C15—C14—H14 | 120.0 |
| C1—N1—C10 | 112.3 (4) | C13—C14—H14 | 120.0 |
| C1—N1—Mn1 | 108.9 (3) | C14—C15—C16 | 120.0 |
| C10—N1—Mn1 | 119.8 (4) | C14—C15—H15 | 120.0 |
| C1—N1—H1 | 104.8 | C16—C15—H15 | 120.0 |
| C10—N1—H1 | 104.8 | C15—C16—C11 | 120.0 |
| Mn1—N1—H1 | 104.8 | C15—C16—H16 | 120.0 |
| C3—N2—C2 | 110.2 (3) | C11—C16—H16 | 120.0 |
| C3—N2—Mn1 | 116.2 (2) | C12B—C11B—C16B | 120.0 |
| C2—N2—Mn1 | 107.2 (3) | C12B—C11B—B1 | 115.4 (4) |
| C3—N2—H2 | 107.7 | C16B—C11B—B1 | 124.5 (4) |
| C2—N2—H2 | 107.7 | C13B—C12B—C11B | 120.0 |
| Mn1—N2—H2 | 107.7 | C13B—C12B—H12B | 120.0 |
| C6—N3—C5 | 112.2 (3) | C11B—C12B—H12B | 120.0 |
| C6—N3—Mn1 | 109.4 (3) | C12B—C13B—C14B | 120.0 |
| C5—N3—Mn1 | 119.7 (2) | C12B—C13B—H13B | 120.0 |
| C6—N3—H3 | 104.7 | C14B—C13B—H13B | 120.0 |
| C5—N3—H3 | 104.7 | C15B—C14B—C13B | 120.0 |
| Mn1—N3—H3 | 104.7 | C15B—C14B—H14B | 120.0 |
| C8—N4—C7 | 110.7 (4) | C13B—C14B—H14B | 120.0 |
| C8—N4—Mn1 | 115.8 (3) | C14B—C15B—C16B | 120.0 |
| C7—N4—Mn1 | 106.5 (2) | C14B—C15B—H15B | 120.0 |
| C8—N4—H4 | 107.8 | C16B—C15B—H15B | 120.0 |
| C7—N4—H4 | 107.8 | C15B—C16B—C11B | 120.0 |
| Mn1—N4—H4 | 107.8 | C15B—C16B—H16B | 120.0 |
| N1—C1—C2 | 106.8 (4) | C11B—C16B—H16B | 120.0 |
| N1—C1—H1B | 110.4 | C18—C17—C22 | 113.6 (3) |
| C2—C1—H1B | 110.4 | C18—C17—B1 | 124.2 (3) |
| N1—C1—H1C | 110.4 | C22—C17—B1 | 122.2 (3) |
| C2—C1—H1C | 110.4 | C19—C18—C17 | 123.4 (4) |
| H1B—C1—H1C | 108.6 | C19—C18—H18 | 118.3 |
| N2—C2—C1 | 107.9 (4) | C17—C18—H18 | 118.3 |
| N2—C2—H2A | 110.1 | C20—C19—C18 | 120.8 (4) |
| C1—C2—H2A | 110.1 | C20—C19—H19 | 119.6 |
| N2—C2—H2B | 110.1 | C18—C19—H19 | 119.6 |
| C1—C2—H2B | 110.1 | C21—C20—C19 | 117.7 (4) |
| H2A—C2—H2B | 108.4 | C21—C20—H20 | 121.1 |
| N2—C3—C4 | 113.0 (3) | C19—C20—H20 | 121.1 |
| N2—C3—H3A | 109.0 | C20—C21—C22 | 120.7 (4) |
| C4—C3—H3A | 109.0 | C20—C21—H21 | 119.7 |
| N2—C3—H3B | 109.0 | C22—C21—H21 | 119.7 |
| C4—C3—H3B | 109.0 | C21—C22—C17 | 123.8 (3) |
| H3A—C3—H3B | 107.8 | C21—C22—H22 | 118.1 |
| C3—C4—C5 | 113.3 (4) | C17—C22—H22 | 118.1 |
| C3—C4—H4A | 108.9 | C24—C23—C28 | 114.8 (3) |
| C5—C4—H4A | 108.9 | C24—C23—B1 | 125.1 (3) |
| C3—C4—H4B | 108.9 | C28—C23—B1 | 120.2 (3) |
| C5—C4—H4B | 108.9 | C23—C24—C25 | 122.8 (4) |
| H4A—C4—H4B | 107.7 | C23—C24—H24 | 118.6 |
| N3—C5—C4 | 112.7 (4) | C25—C24—H24 | 118.6 |
| N3—C5—H5A | 109.1 | C26—C25—C24 | 120.1 (4) |
| C4—C5—H5A | 109.1 | C26—C25—H25 | 119.9 |
| N3—C5—H5B | 109.1 | C24—C25—H25 | 119.9 |
| C4—C5—H5B | 109.1 | C25—C26—C27 | 119.2 (4) |
| H5A—C5—H5B | 107.8 | C25—C26—H26 | 120.4 |
| N3—C6—C7 | 108.2 (4) | C27—C26—H26 | 120.4 |
| N3—C6—H6A | 110.1 | C26—C27—C28 | 119.8 (4) |
| C7—C6—H6A | 110.1 | C26—C27—H27 | 120.1 |
| N3—C6—H6B | 110.1 | C28—C27—H27 | 120.1 |
| C7—C6—H6B | 110.1 | C27—C28—C23 | 123.4 (4) |
| H6A—C6—H6B | 108.4 | C27—C28—H28 | 118.3 |
| C6—C7—N4 | 109.0 (3) | C23—C28—H28 | 118.3 |
| C6—C7—H7A | 109.9 | C30—C29—C34 | 114.1 (3) |
| N4—C7—H7A | 109.9 | C30—C29—B1 | 125.2 (3) |
| C6—C7—H7B | 109.9 | C34—C29—B1 | 120.7 (3) |
| N4—C7—H7B | 109.9 | C29—C30—C31 | 123.2 (3) |
| H7A—C7—H7B | 108.3 | C29—C30—H30 | 118.4 |
| N4—C8—C9 | 112.1 (4) | C31—C30—H30 | 118.4 |
| N4—C8—H8A | 109.2 | C32—C31—C30 | 120.2 (4) |
| C9—C8—H8A | 109.2 | C32—C31—H31 | 119.9 |
| N4—C8—H8B | 109.2 | C30—C31—H31 | 119.9 |
| C9—C8—H8B | 109.2 | C33—C32—C31 | 118.7 (4) |
| H8A—C8—H8B | 107.9 | C33—C32—H32 | 120.7 |
| C8—C9—C10 | 116.1 (5) | C31—C32—H32 | 120.7 |
| C8—C9—H9A | 108.3 | C32—C33—C34 | 120.0 (4) |
| C10—C9—H9A | 108.3 | C32—C33—H33 | 120.0 |
| C8—C9—H9B | 108.3 | C34—C33—H33 | 120.0 |
| C10—C9—H9B | 108.3 | C33—C34—C29 | 123.7 (4) |
| H9A—C9—H9B | 107.4 | C33—C34—H34 | 118.1 |
| N1—C10—C9 | 113.5 (5) | C29—C34—H34 | 118.1 |
| N1—C10—H10A | 108.9 | N5—C35—C36 | 178.4 (10) |
| C9—C10—H10A | 108.9 | N5B—C35B—C36B | 178.2 (11) |
| N1—C10—H10B | 108.9 | C35B—C36B—H36D | 109.5 |
| C9—C10—H10B | 108.9 | C35B—C36B—H36E | 109.5 |
| H10A—C10—H10B | 107.7 | H36D—C36B—H36E | 109.5 |
| C23—B1—C17 | 108.1 (3) | C35B—C36B—H36F | 109.5 |
| C23—B1—C29 | 109.1 (3) | H36D—C36B—H36F | 109.5 |
| C17—B1—C29 | 111.6 (3) | H36E—C36B—H36F | 109.5 |
| C23—B1—C11B | 116.5 (4) |
Symmetry codes: (i) −x, −y+1, −z+1.
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1···N5 | 0.93 | 2.42 | 3.313 (11) | 160 |
| N1—H1···N5B | 0.93 | 1.98 | 2.832 (10) | 151 |
| N2—H2···Cl1 | 0.93 | 2.37 | 3.289 (4) | 169 |
| N3—H3···N5i | 0.93 | 2.22 | 3.034 (9) | 146 |
| N3—H3···N5Bi | 0.93 | 2.23 | 3.120 (9) | 160 |
| N4—H4···Cl1 | 0.93 | 2.42 | 3.330 (4) | 168 |
Symmetry codes: (i) −x, −y+1, −z+1.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: HG5039).
References
- Bosnich, B., Poon, C. K. & Tobe, M. L. (1965). Inorg. Chem. 4, 1102–1108.
- Brewer, K. J., Calvin, M., Lumpkin, R. S., Otvos, J. W. & Spreer, L. O. (1989). Inorg. Chem. 28, 4446–4451.
- Bruker (2007). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Goodson, P. A., Hodgson, D. J. & Michelsen, K. (1990). Inorg. Chim. Acta, 172, 49–57.
- Levaton, B. B. & Olmstead, M. M. (2010). Acta Cryst. E66, m1226–m1227. [DOI] [PMC free article] [PubMed]
- Lu, Y.-H., Fun, H.-K., Chantrapromma, S., Razak, I. A., Shen, Z., Zuo, J.-L. & You, X.-Z. (2001). Acta Cryst. C57, 911–913. [DOI] [PubMed]
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811019829/hg5039sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811019829/hg5039Isup2.hkl
Additional supplementary materials: crystallographic information; 3D view; checkCIF report