Abstract
In the title compound, C14H11NO3S, the six-membered thiazine ring fused to two benzene rings adopts a distorted boat conformation. The dihedral angle between the mean planes of the two benzene rings is 45.8 (1)°. The crystal packing is stabilized by weak intermolecular C—H⋯O interactions.
Related literature
For synthetic dyes and electroluminescent materials containing phenothiazine, see: Miller et al. (1999 ▶). For antipsychotic drugs, see: Wermuth et al. (2003 ▶). For applications of phenothiazine derivatives in medicine, see: Wang et al. (2008 ▶). For their antitumor activity, see: Lam et al. (2001 ▶). For related structures, see: Harrison et al. (2007 ▶); Jasinski et al. (2011 ▶). For standard bond lengths, see Allen et al. (1987 ▶). For puckering parameters, see: Cremer & Pople (1975 ▶).
Experimental
Crystal data
C14H11NO3S
M r = 273.30
Monoclinic,
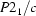
a = 12.5715 (6) Å
b = 8.7648 (4) Å
c = 11.5828 (5) Å
β = 92.142 (4)°
V = 1275.38 (10) Å3
Z = 4
Mo Kα radiation
μ = 0.26 mm−1
T = 173 K
0.35 × 0.15 × 0.15 mm
Data collection
Oxford Diffraction Xcalibur Eos Gemini diffractometer
Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2010 ▶) T min = 0.916, T max = 0.963
5342 measured reflections
2597 independent reflections
2263 reflections with I > 2σ(I)
R int = 0.019
Refinement
R[F 2 > 2σ(F 2)] = 0.036
wR(F 2) = 0.103
S = 1.02
2597 reflections
173 parameters
H-atom parameters constrained
Δρmax = 0.26 e Å−3
Δρmin = −0.40 e Å−3
Data collection: CrysAlis PRO (Oxford Diffraction, 2010 ▶); cell refinement: CrysAlis PRO; data reduction: CrysAlis RED; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536811021854/yk2011sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811021854/yk2011Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811021854/yk2011Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C2—H2B⋯O2i | 0.95 | 2.50 | 3.246 (2) | 135 |
| C8—H8A⋯O1ii | 0.95 | 2.54 | 3.376 (2) | 147 |
| C9—H9A⋯O3ii | 0.95 | 2.56 | 3.322 (2) | 137 |
Symmetry codes: (i) 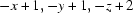 ; (ii)
; (ii) 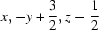 .
.
Acknowledgments
MSS thanks the University of Mysore for the research facilities and R. L. Fine Chem, Bangalore, India, for the gift sample. JPJ acknowledges the NSF–MRI program (grant No. CHE1039027) for funds to purchase the X-ray diffractometer.
supplementary crystallographic information
Comment
Phenothiazine is a well known heterocycle. The phenothiazine structure occurs in many synthetic dyes, electroluminescent materials (Miller et al., 1999) and drugs, especially various antipsychotic drugs, e.g. chlorpromazine and antihistaminic drugs, e.g. promethazine (Wermuth, 2003). Recently, researchers have found some new applications for phenothiazine derivatives in medicine related to antitubercular (Wang et al., 2008) and antitumor activities (Lam et al., 2001). The title compound has been used in the synthesis of oxomemazine, an antihistamine and anticholinergic drug of the phenothiazine chemical class used for the treatment of coughs. The crystal structures of dioxopromethazinium picrate (Harrison et al., 2007) and 1-(10H-phenothiazin-2-yl)ethanone (Jasinski et al., 2011) have been reported. In view of the importance of phenothiazines, this paper reports the crystal structure of the title compound, C14H11NO3S.
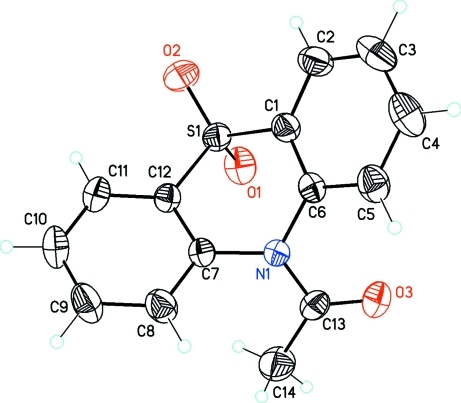
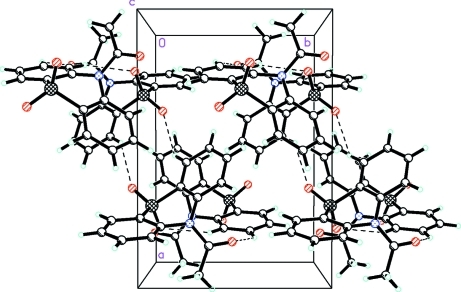
In the title compound the six-membered thiazine ring fused to two benzene rings adopts a distorted boat conformation. (Cremer & Pople, 1975) with puckering parameters Q, θ and φ of 0.6257 (12) Å, 95.85 (13)° and 180.29 (15)°, respectively (Fig. 1). For an ideal boat θ and φ have values of 90.0° and 180°. The dihedral angle between the mean planes of the two benzene rings is 45.8 (1)°. The ethanone group extends away from corner of the boat crease with a -169.76 (14)° (C6/N1/C13/C14) torsion angle. The SO2 group extends away from the opposite corner of the boat crease with a 105.8 (14)° (C2/C1/S1/O1) torsion angle. Bond lengths are in normal ranges (Allenet al., 1987). Crystal packing is stabiized by weak C—H···O (Table 1, Fig. 2) intermolecular interactions.
Experimental
The title compound was obtained as a gift sample from RL Fine Chem, Bangalore, India. X-ray quality crystals were obtained by slow evaporation of solution of a 1:1 mixture of acetone:ethanol (m.p.: 495 K).
Refinement
All of the H atoms were placed in their calculated positions and then refined using the riding model with Atom—H lengths of 0.95Å (CH), or 0.98Å (CH3). Isotropic displacement parameters for these atoms were set to 1.19-1.21 (CH) or 1.49 (CH3) times Ueq of the parent atom.
Figures
Fig. 1.
Molecular structure of the title compound showing the atom labeling scheme and 50% probability displacement ellipsoids.
Fig. 2.
Packing diagram of the title compound viewed down the c axis. Dashed lines indicate weak C—H···O intermolecular interactions.
Crystal data
| C14H11NO3S | F(000) = 568 |
| Mr = 273.30 | Dx = 1.423 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 3700 reflections |
| a = 12.5715 (6) Å | θ = 3.3–32.3° |
| b = 8.7648 (4) Å | µ = 0.26 mm−1 |
| c = 11.5828 (5) Å | T = 173 K |
| β = 92.142 (4)° | Block, colorless |
| V = 1275.38 (10) Å3 | 0.35 × 0.15 × 0.15 mm |
| Z = 4 |
Data collection
| Oxford Diffraction Xcalibur Eos Gemini diffractometer | 2597 independent reflections |
| Radiation source: Enhance (Mo) X-ray Source | 2263 reflections with I > 2σ(I) |
| graphite | Rint = 0.019 |
| Detector resolution: 16.1500 pixels mm-1 | θmax = 26.4°, θmin = 3.3° |
| ω scans | h = −15→15 |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2010) | k = −10→10 |
| Tmin = 0.916, Tmax = 0.963 | l = −14→13 |
| 5342 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.036 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.103 | H-atom parameters constrained |
| S = 1.02 | w = 1/[σ2(Fo2) + (0.0596P)2 + 0.3216P] where P = (Fo2 + 2Fc2)/3 |
| 2597 reflections | (Δ/σ)max = 0.001 |
| 173 parameters | Δρmax = 0.26 e Å−3 |
| 0 restraints | Δρmin = −0.40 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.27860 (3) | 0.53431 (4) | 0.88913 (3) | 0.03458 (15) | |
| O1 | 0.18673 (10) | 0.57759 (15) | 0.95043 (10) | 0.0481 (3) | |
| O2 | 0.34587 (11) | 0.41839 (15) | 0.93886 (12) | 0.0552 (4) | |
| O3 | 0.14803 (10) | 0.98466 (14) | 0.75069 (12) | 0.0492 (3) | |
| N1 | 0.22033 (10) | 0.75636 (14) | 0.71011 (11) | 0.0301 (3) | |
| C1 | 0.35385 (12) | 0.69784 (18) | 0.86054 (13) | 0.0330 (3) | |
| C2 | 0.44865 (13) | 0.7260 (2) | 0.92270 (15) | 0.0441 (4) | |
| H2B | 0.4745 | 0.6564 | 0.9801 | 0.053* | |
| C3 | 0.50395 (14) | 0.8569 (2) | 0.89904 (17) | 0.0521 (5) | |
| H3A | 0.5685 | 0.8793 | 0.9410 | 0.063* | |
| C4 | 0.46595 (15) | 0.9559 (2) | 0.81450 (18) | 0.0523 (5) | |
| H4A | 0.5045 | 1.0467 | 0.7999 | 0.063* | |
| C5 | 0.37288 (14) | 0.9262 (2) | 0.75021 (15) | 0.0415 (4) | |
| H5A | 0.3487 | 0.9941 | 0.6909 | 0.050* | |
| C6 | 0.31596 (11) | 0.79526 (17) | 0.77439 (13) | 0.0307 (3) | |
| C7 | 0.21083 (11) | 0.60193 (17) | 0.67173 (13) | 0.0303 (3) | |
| C8 | 0.18008 (14) | 0.5650 (2) | 0.55873 (15) | 0.0413 (4) | |
| H8A | 0.1651 | 0.6434 | 0.5038 | 0.050* | |
| C9 | 0.17142 (16) | 0.4131 (2) | 0.52666 (16) | 0.0486 (5) | |
| H9A | 0.1488 | 0.3881 | 0.4498 | 0.058* | |
| C10 | 0.19506 (15) | 0.2974 (2) | 0.60424 (16) | 0.0470 (4) | |
| H10A | 0.1866 | 0.1939 | 0.5814 | 0.056* | |
| C11 | 0.23104 (13) | 0.33228 (18) | 0.71510 (15) | 0.0388 (4) | |
| H11A | 0.2510 | 0.2536 | 0.7680 | 0.047* | |
| C12 | 0.23754 (12) | 0.48386 (17) | 0.74801 (13) | 0.0301 (3) | |
| C13 | 0.13555 (12) | 0.85896 (18) | 0.70914 (14) | 0.0355 (4) | |
| C14 | 0.03011 (14) | 0.8073 (2) | 0.65925 (19) | 0.0527 (5) | |
| H14A | −0.0254 | 0.8789 | 0.6816 | 0.079* | |
| H14B | 0.0140 | 0.7054 | 0.6886 | 0.079* | |
| H14C | 0.0326 | 0.8039 | 0.5748 | 0.079* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0430 (2) | 0.0320 (2) | 0.0286 (2) | 0.00086 (16) | −0.00171 (16) | 0.00387 (15) |
| O1 | 0.0580 (8) | 0.0502 (7) | 0.0371 (6) | −0.0057 (6) | 0.0143 (6) | −0.0048 (6) |
| O2 | 0.0673 (9) | 0.0431 (7) | 0.0538 (8) | 0.0057 (6) | −0.0169 (7) | 0.0151 (6) |
| O3 | 0.0556 (8) | 0.0313 (6) | 0.0604 (8) | 0.0070 (5) | −0.0022 (6) | −0.0060 (6) |
| N1 | 0.0334 (6) | 0.0253 (6) | 0.0311 (6) | −0.0007 (5) | −0.0050 (5) | 0.0007 (5) |
| C1 | 0.0332 (7) | 0.0339 (8) | 0.0319 (8) | 0.0001 (6) | −0.0005 (6) | −0.0033 (6) |
| C2 | 0.0391 (8) | 0.0518 (10) | 0.0406 (9) | 0.0034 (8) | −0.0086 (7) | −0.0069 (8) |
| C3 | 0.0363 (9) | 0.0687 (13) | 0.0509 (11) | −0.0097 (9) | −0.0056 (8) | −0.0147 (10) |
| C4 | 0.0475 (10) | 0.0565 (12) | 0.0532 (11) | −0.0216 (9) | 0.0068 (9) | −0.0083 (9) |
| C5 | 0.0473 (9) | 0.0389 (9) | 0.0385 (9) | −0.0100 (7) | 0.0036 (7) | 0.0000 (7) |
| C6 | 0.0322 (7) | 0.0300 (7) | 0.0299 (7) | −0.0021 (6) | 0.0011 (6) | −0.0044 (6) |
| C7 | 0.0310 (7) | 0.0268 (7) | 0.0330 (8) | −0.0024 (6) | −0.0006 (6) | −0.0009 (6) |
| C8 | 0.0534 (10) | 0.0380 (9) | 0.0321 (8) | −0.0081 (8) | −0.0040 (7) | 0.0014 (7) |
| C9 | 0.0667 (12) | 0.0443 (10) | 0.0346 (9) | −0.0121 (9) | −0.0004 (8) | −0.0097 (8) |
| C10 | 0.0620 (11) | 0.0310 (9) | 0.0485 (10) | −0.0071 (8) | 0.0104 (9) | −0.0115 (8) |
| C11 | 0.0467 (9) | 0.0269 (8) | 0.0432 (9) | 0.0013 (7) | 0.0080 (7) | −0.0001 (7) |
| C12 | 0.0326 (7) | 0.0280 (7) | 0.0297 (7) | 0.0006 (6) | 0.0031 (6) | −0.0008 (6) |
| C13 | 0.0401 (8) | 0.0311 (8) | 0.0351 (8) | 0.0023 (6) | 0.0018 (6) | 0.0060 (7) |
| C14 | 0.0363 (9) | 0.0519 (11) | 0.0697 (13) | 0.0035 (8) | −0.0019 (9) | 0.0010 (10) |
Geometric parameters (Å, °)
| S1—O2 | 1.4292 (13) | C5—C6 | 1.387 (2) |
| S1—O1 | 1.4294 (13) | C5—H5A | 0.9500 |
| S1—C12 | 1.7524 (16) | C7—C8 | 1.389 (2) |
| S1—C1 | 1.7557 (16) | C7—C12 | 1.394 (2) |
| O3—C13 | 1.210 (2) | C8—C9 | 1.386 (2) |
| N1—C13 | 1.394 (2) | C8—H8A | 0.9500 |
| N1—C7 | 1.4283 (19) | C9—C10 | 1.380 (3) |
| N1—C6 | 1.4314 (19) | C9—H9A | 0.9500 |
| C1—C6 | 1.384 (2) | C10—C11 | 1.380 (2) |
| C1—C2 | 1.391 (2) | C10—H10A | 0.9500 |
| C2—C3 | 1.374 (3) | C11—C12 | 1.384 (2) |
| C2—H2B | 0.9500 | C11—H11A | 0.9500 |
| C3—C4 | 1.380 (3) | C13—C14 | 1.496 (2) |
| C3—H3A | 0.9500 | C14—H14A | 0.9800 |
| C4—C5 | 1.388 (3) | C14—H14B | 0.9800 |
| C4—H4A | 0.9500 | C14—H14C | 0.9800 |
| O2—S1—O1 | 117.73 (8) | C8—C7—C12 | 118.45 (14) |
| O2—S1—C12 | 110.23 (8) | C8—C7—N1 | 122.08 (14) |
| O1—S1—C12 | 108.36 (7) | C12—C7—N1 | 119.41 (13) |
| O2—S1—C1 | 109.97 (8) | C9—C8—C7 | 119.52 (16) |
| O1—S1—C1 | 109.16 (7) | C9—C8—H8A | 120.2 |
| C12—S1—C1 | 99.91 (7) | C7—C8—H8A | 120.2 |
| C13—N1—C7 | 123.62 (12) | C10—C9—C8 | 121.26 (16) |
| C13—N1—C6 | 118.52 (13) | C10—C9—H9A | 119.4 |
| C7—N1—C6 | 116.52 (12) | C8—C9—H9A | 119.4 |
| C6—C1—C2 | 121.92 (15) | C11—C10—C9 | 119.88 (16) |
| C6—C1—S1 | 117.79 (11) | C11—C10—H10A | 120.1 |
| C2—C1—S1 | 120.29 (13) | C9—C10—H10A | 120.1 |
| C3—C2—C1 | 118.35 (17) | C10—C11—C12 | 118.89 (16) |
| C3—C2—H2B | 120.8 | C10—C11—H11A | 120.6 |
| C1—C2—H2B | 120.8 | C12—C11—H11A | 120.6 |
| C2—C3—C4 | 120.13 (17) | C11—C12—C7 | 121.87 (15) |
| C2—C3—H3A | 119.9 | C11—C12—S1 | 120.77 (12) |
| C4—C3—H3A | 119.9 | C7—C12—S1 | 117.36 (11) |
| C3—C4—C5 | 121.69 (17) | O3—C13—N1 | 119.75 (15) |
| C3—C4—H4A | 119.2 | O3—C13—C14 | 121.93 (15) |
| C5—C4—H4A | 119.2 | N1—C13—C14 | 118.29 (14) |
| C6—C5—C4 | 118.54 (17) | C13—C14—H14A | 109.5 |
| C6—C5—H5A | 120.7 | C13—C14—H14B | 109.5 |
| C4—C5—H5A | 120.7 | H14A—C14—H14B | 109.5 |
| C1—C6—C5 | 119.33 (14) | C13—C14—H14C | 109.5 |
| C1—C6—N1 | 119.19 (13) | H14A—C14—H14C | 109.5 |
| C5—C6—N1 | 121.45 (14) | H14B—C14—H14C | 109.5 |
| O2—S1—C1—C6 | 154.78 (12) | C13—N1—C7—C12 | −121.42 (16) |
| O1—S1—C1—C6 | −74.65 (14) | C6—N1—C7—C12 | 45.13 (19) |
| C12—S1—C1—C6 | 38.88 (13) | C12—C7—C8—C9 | 3.4 (2) |
| O2—S1—C1—C2 | −24.71 (16) | N1—C7—C8—C9 | −179.49 (16) |
| O1—S1—C1—C2 | 105.85 (14) | C7—C8—C9—C10 | −1.5 (3) |
| C12—S1—C1—C2 | −140.62 (14) | C8—C9—C10—C11 | −1.9 (3) |
| C6—C1—C2—C3 | 1.8 (3) | C9—C10—C11—C12 | 3.3 (3) |
| S1—C1—C2—C3 | −178.75 (14) | C10—C11—C12—C7 | −1.3 (2) |
| C1—C2—C3—C4 | −0.8 (3) | C10—C11—C12—S1 | 177.93 (13) |
| C2—C3—C4—C5 | −1.0 (3) | C8—C7—C12—C11 | −2.0 (2) |
| C3—C4—C5—C6 | 1.7 (3) | N1—C7—C12—C11 | −179.24 (14) |
| C2—C1—C6—C5 | −1.1 (2) | C8—C7—C12—S1 | 178.73 (12) |
| S1—C1—C6—C5 | 179.43 (12) | N1—C7—C12—S1 | 1.50 (18) |
| C2—C1—C6—N1 | 177.23 (14) | O2—S1—C12—C11 | 26.68 (16) |
| S1—C1—C6—N1 | −2.26 (19) | O1—S1—C12—C11 | −103.47 (14) |
| C4—C5—C6—C1 | −0.6 (2) | C1—S1—C12—C11 | 142.39 (13) |
| C4—C5—C6—N1 | −178.90 (15) | O2—S1—C12—C7 | −154.06 (12) |
| C13—N1—C6—C1 | 122.59 (15) | O1—S1—C12—C7 | 75.79 (13) |
| C7—N1—C6—C1 | −44.68 (19) | C1—S1—C12—C7 | −38.35 (13) |
| C13—N1—C6—C5 | −59.1 (2) | C7—N1—C13—O3 | 174.72 (15) |
| C7—N1—C6—C5 | 133.59 (15) | C6—N1—C13—O3 | 8.4 (2) |
| C13—N1—C7—C8 | 61.5 (2) | C7—N1—C13—C14 | −3.5 (2) |
| C6—N1—C7—C8 | −131.99 (15) | C6—N1—C13—C14 | −169.76 (14) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C2—H2B···O2i | 0.95 | 2.50 | 3.246 (2) | 135 |
| C8—H8A···O1ii | 0.95 | 2.54 | 3.376 (2) | 147 |
| C9—H9A···O3ii | 0.95 | 2.56 | 3.322 (2) | 137 |
Symmetry codes: (i) −x+1, −y+1, −z+2; (ii) x, −y+3/2, z−1/2.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: YK2011).
References
- Allen, F. H., Kennard, O., Watson, D. G., Brammer, L., Orpen, A. G. & Taylor, R. (1987). J. Chem. Soc. Perkin Trans. 2, pp. S1–19.
- Cremer, D. & Pople, J. A. (1975). J. Am. Chem. Soc. 97, 1354–1358.
- Harrison, W. T. A., Ashok, M. A., Yathirajan, H. S. & Narayana Achar, B. (2007). Acta Cryst. E63, o3277.
- Jasinski, J. P., Pek, A. E., Nayak, P. S., Narayana, B. & Yathirajan, H. S. (2011). Acta Cryst. E67, o430–o431. [DOI] [PMC free article] [PubMed]
- Lam, M., Oleinick, N. L. & Nieminen, A. L. (2001). J. Biol. Chem. 276, 47379–47386. [DOI] [PubMed]
- Miller, M. T., Gantzel, P. K. & Karpishin, T. B. (1999). J. Am. Chem. Soc. 121, 4292–4293.
- Oxford Diffraction (2010). CrysAlis PRO and CrysAlis RED Oxford Diffraction Ltd, Yarnton, England.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Wang, J., Dong, M., Liang, J., Chang, Z., Feng, S., Wang, H. & Ding, N. (2008). Chin. J. Lab. Diagn. 12, 381–382.
- Wermuth, C. G. (2003). The Practice of Medicinal Chemistry, 2nd ed. London: Academic Press.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S1600536811021854/yk2011sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811021854/yk2011Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811021854/yk2011Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report