Abstract
In the title compound, C20H18N4O2, the imidazopyridine fused ring system is almost perpendicular to the benzene ring [dihedral angle = 87.6 (5)°]. The pyridine ring makes a dihedral angle of 35.5 (5)° with the mean plane of the imidazopyridine fragment. The crystal structure is stabilized by an aromatic π–π stacking interaction between the phenyl rings of neighbouring molecules [centroid–centroid distance = 3.772 (2) Å, interplanar distance = 3.546 (2) Å and slippage = 1.286 (2) Å].
Related literature
For the biological activity of pyridine derivatives, see: Passannanti et al. (1998 ▶); Jiyeon et al. (2010 ▶); Abdel-Alim et al. (2005 ▶); Girgis et al. (2006 ▶); Slominska et al. (2008 ▶); Spanka et al. (2010 ▶). For a related structure, see: Ouzidan et al. (2010 ▶). For sp
3 hybridization, see: Beddoes et al. (1986 ▶).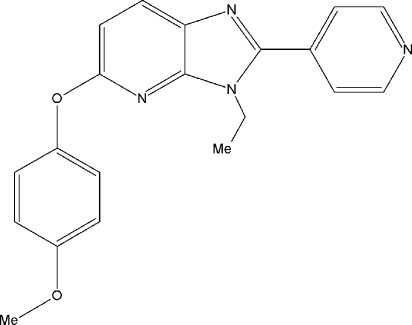
Experimental
Crystal data
C20H18N4O2
M r = 346.38
Monoclinic,

a = 13.6591 (4) Å
b = 13.7104 (4) Å
c = 9.3177 (2) Å
β = 98.940 (1)°
V = 1723.74 (8) Å3
Z = 4
Mo Kα radiation
μ = 0.09 mm−1
T = 293 K
0.25 × 0.22 × 0.19 mm
Data collection
Bruker APEXII CCD area-detector diffractometer
Absorption correction: multi-scan (SADABS; Sheldrick, 1996 ▶) T min = 0.981, T max = 0.985
24827 measured reflections
5887 independent reflections
3938 reflections with I > 2σ(I)
R int = 0.027
Refinement
R[F 2 > 2σ(F 2)] = 0.046
wR(F 2) = 0.140
S = 1.00
5887 reflections
237 parameters
H-atom parameters constrained
Δρmax = 0.30 e Å−3
Δρmin = −0.20 e Å−3
Data collection: APEX2 (Bruker, 2004 ▶); cell refinement: SAINT (Bruker, 2004 ▶); data reduction: SAINT; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: ORTEP-3 (Farrugia, 1997 ▶); software used to prepare material for publication: SHELXL97 and PLATON (Spek, 2009 ▶).
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811023543/lx2189sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811023543/lx2189Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811023543/lx2189Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Acknowledgments
SR and ASP thank Dr Babu Varghese, SAIF, IIT, Chennai, India, for the data collection.
supplementary crystallographic information
Comment
Pyridine derivatives has numerous applications in medicinal chemistry (Passannanti et al., 1998). Furthermore, the imidazo[4,5-b]pyridine moiety is also an important heterocyclic nucleus which has been used extensively in medicinal chemistry. In fact, the heterocycles derived from these intermediates have been tested for their potential as anti-neuroinflammatory (Jiyeon et al., 2010). Pyridine-3-carboxamides have gained attention because of their diverse pharmacological properties such as anti-inflammatory (Abdel-Alim et al., 2005), anticancer (Girgis et al., 2006), cytoprotective (Slominska et al., 2008), and anxiolytic (Spanka et al., 2010) activities. Against this background, and in order to obtain detailed information on molecular conformations in the solid state, an X-ray study of the title compound was carried out.
The bond lengths and angles in (Fig. 1) agree with those observed in other imidazopyridine derivatives (Ouzidan et al., 2010). The imidazopyridine ring system is essentially planar, with maximum deviation of 0.013 (1)° for atom C1. The sum of bond angles around N2[359.2 (9)°] of the imidazole ring is in accordance with sp3 hybridization (Beddoes et al., 1986). The imidazopyridine ring system makes dihedral angles of 35.5 (5) and 87.6 (5)°, respectively, with the pyridine and phenyl rings and also the dihedral angle between the pyridine and phenyl ring is 87.0 (6)°. It shows that the phenyl ring is perpendicular to both the imidazopyridine and pyridine rings. The atoms O1 and O2 are deviated by 0.039 (1) and - 0.021 (1)Å from the leastsquares plane of the phenyl ring.
The molecules lack hydrogen bonding functionality and pack in layers parallel to the (100) planes. The crystal structure is stabilized by an aromatic π–π stacking interaction between the phenyl rings of adjacent molecules, with a Cg···Cg distance of 3.772 (2) Å and an interplanar distance of 3.546 (3) Å resulting in a slippage of 1.286 (3) Å (Cg is the centroid of the C14–C19 phenyl ring).
Experimental
N-ethyl-6-(4-methoxyphenoxy)pyridin-2-amine (0.23 g, 1 mmol) and amide (0.12 g, 1 mmol) successively added to Al3+–Y in xylene at 145°C. After stirring for 16 h, the mixture was diluted with dichloromethane. After removing the catalyst by filtration, followed by solvent evaporation, the resulting crude product was finally purified by column chromatography (silica gel). Single crystals suitable for X-ray diffraction were obtained by slow evaporation of a solution of the title compound in ethylacetate at room temperature.
Refinement
All H atoms were fixed geometrically and allowed to ride on their parent C atoms, with C—H distances fixed in the range 0.93–0.97 Å with Uiso(H) = 1.5Ueq(C) for methyl H 1.2Ueq(C) for other H atoms.
Figures
Fig. 1.
The structure of showing the atom-numbering scheme. The displacement ellipsoids are drawn at the 30% probability level.
Crystal data
| C20H18N4O2 | F(000) = 728 |
| Mr = 346.38 | Dx = 1.335 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| Hall symbol: -P 2ybc | Cell parameters from 5887 reflections |
| a = 13.6591 (4) Å | θ = 1.5–32.0° |
| b = 13.7104 (4) Å | µ = 0.09 mm−1 |
| c = 9.3177 (2) Å | T = 293 K |
| β = 98.940 (1)° | Block, white crystalline |
| V = 1723.74 (8) Å3 | 0.25 × 0.22 × 0.19 mm |
| Z = 4 |
Data collection
| Bruker APEXII CCD area-detector diffractometer | 5887 independent reflections |
| Radiation source: fine-focus sealed tube | 3938 reflections with I > 2σ(I) |
| graphite | Rint = 0.027 |
| Detector resolution: 10.0 pixels mm-1 | θmax = 32.1°, θmin = 1.5° |
| ω and φ scans | h = −20→20 |
| Absorption correction: multi-scan (SADABS; Sheldrick, 1996) | k = −20→20 |
| Tmin = 0.981, Tmax = 0.985 | l = −13→12 |
| 24827 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.046 | Hydrogen site location: difference Fourier map |
| wR(F2) = 0.140 | H-atom parameters constrained |
| S = 1.00 | w = 1/[σ2(Fo2) + (0.068P)2 + 0.2334P] where P = (Fo2 + 2Fc2)/3 |
| 5887 reflections | (Δ/σ)max < 0.001 |
| 237 parameters | Δρmax = 0.30 e Å−3 |
| 0 restraints | Δρmin = −0.20 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.34424 (9) | 0.10472 (9) | 0.42615 (13) | 0.0383 (3) | |
| C2 | 0.34457 (9) | 0.18018 (9) | 0.52777 (14) | 0.0406 (3) | |
| H2 | 0.3850 | 0.2344 | 0.5240 | 0.049* | |
| C3 | 0.28465 (9) | 0.17335 (9) | 0.63296 (13) | 0.0389 (3) | |
| H3 | 0.2834 | 0.2222 | 0.7020 | 0.047* | |
| C4 | 0.22585 (8) | 0.09028 (8) | 0.63192 (12) | 0.0335 (2) | |
| C5 | 0.23419 (8) | 0.02078 (8) | 0.52553 (12) | 0.0330 (2) | |
| C6 | 0.12671 (8) | −0.02604 (8) | 0.66320 (12) | 0.0332 (2) | |
| C7 | 0.16889 (10) | −0.14643 (8) | 0.46766 (13) | 0.0403 (3) | |
| H7A | 0.1356 | −0.1952 | 0.5182 | 0.048* | |
| H7B | 0.2363 | −0.1683 | 0.4670 | 0.048* | |
| C8 | 0.11655 (12) | −0.13785 (11) | 0.31346 (15) | 0.0547 (4) | |
| H8A | 0.0490 | −0.1186 | 0.3137 | 0.082* | |
| H8B | 0.1181 | −0.1997 | 0.2655 | 0.082* | |
| H8C | 0.1493 | −0.0897 | 0.2629 | 0.082* | |
| C9 | 0.05197 (8) | −0.08407 (8) | 0.72257 (12) | 0.0341 (2) | |
| C10 | 0.05089 (10) | −0.08248 (9) | 0.87147 (13) | 0.0427 (3) | |
| H10 | 0.0986 | −0.0476 | 0.9329 | 0.051* | |
| C11 | −0.02193 (11) | −0.13343 (10) | 0.92665 (15) | 0.0500 (3) | |
| H11 | −0.0225 | −0.1305 | 1.0262 | 0.060* | |
| C12 | −0.08936 (10) | −0.18810 (10) | 0.70402 (15) | 0.0459 (3) | |
| H12 | −0.1370 | −0.2251 | 0.6457 | 0.055* | |
| C13 | −0.02073 (9) | −0.13820 (9) | 0.63755 (13) | 0.0386 (3) | |
| H13 | −0.0233 | −0.1409 | 0.5373 | 0.046* | |
| C14 | 0.40351 (9) | 0.04175 (9) | 0.22118 (14) | 0.0425 (3) | |
| C15 | 0.33767 (10) | 0.04307 (10) | 0.09494 (15) | 0.0488 (3) | |
| H15 | 0.2913 | 0.0930 | 0.0769 | 0.059* | |
| C16 | 0.34062 (10) | −0.03057 (11) | −0.00606 (15) | 0.0508 (3) | |
| H16 | 0.2964 | −0.0299 | −0.0927 | 0.061* | |
| C17 | 0.40900 (10) | −0.10480 (10) | 0.02181 (15) | 0.0479 (3) | |
| C18 | 0.47462 (11) | −0.10467 (11) | 0.14910 (16) | 0.0546 (4) | |
| H18 | 0.5211 | −0.1544 | 0.1679 | 0.066* | |
| C19 | 0.47204 (10) | −0.03162 (11) | 0.24862 (16) | 0.0512 (3) | |
| H19 | 0.5167 | −0.0318 | 0.3348 | 0.061* | |
| C20 | 0.35584 (16) | −0.18189 (16) | −0.2055 (2) | 0.0799 (5) | |
| H20A | 0.2878 | −0.1767 | −0.1920 | 0.120* | |
| H20B | 0.3653 | −0.2416 | −0.2555 | 0.120* | |
| H20C | 0.3727 | −0.1276 | −0.2621 | 0.120* | |
| N1 | 0.29165 (7) | 0.02406 (7) | 0.42238 (11) | 0.0374 (2) | |
| N2 | 0.17080 (7) | −0.05379 (7) | 0.54614 (10) | 0.0340 (2) | |
| N3 | 0.15774 (7) | 0.05932 (7) | 0.71772 (10) | 0.0357 (2) | |
| N4 | −0.09155 (9) | −0.18655 (9) | 0.84645 (13) | 0.0512 (3) | |
| O1 | 0.40353 (7) | 0.11775 (7) | 0.32231 (11) | 0.0511 (2) | |
| O2 | 0.41700 (9) | −0.18162 (9) | −0.06945 (12) | 0.0716 (3) |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0354 (6) | 0.0366 (6) | 0.0433 (6) | −0.0005 (4) | 0.0079 (5) | 0.0034 (5) |
| C2 | 0.0382 (6) | 0.0315 (6) | 0.0517 (7) | −0.0044 (5) | 0.0052 (5) | −0.0009 (5) |
| C3 | 0.0407 (6) | 0.0313 (5) | 0.0433 (6) | −0.0008 (5) | 0.0022 (5) | −0.0056 (5) |
| C4 | 0.0353 (5) | 0.0304 (5) | 0.0338 (5) | 0.0009 (4) | 0.0027 (4) | −0.0008 (4) |
| C5 | 0.0350 (5) | 0.0299 (5) | 0.0338 (5) | 0.0000 (4) | 0.0042 (4) | 0.0004 (4) |
| C6 | 0.0364 (5) | 0.0316 (5) | 0.0318 (5) | 0.0020 (4) | 0.0057 (4) | 0.0008 (4) |
| C7 | 0.0471 (7) | 0.0287 (5) | 0.0468 (7) | −0.0017 (5) | 0.0128 (5) | −0.0070 (5) |
| C8 | 0.0681 (9) | 0.0502 (8) | 0.0454 (7) | −0.0077 (7) | 0.0075 (7) | −0.0135 (6) |
| C9 | 0.0375 (6) | 0.0297 (5) | 0.0360 (5) | 0.0038 (4) | 0.0084 (4) | 0.0031 (4) |
| C10 | 0.0508 (7) | 0.0416 (6) | 0.0365 (6) | −0.0059 (5) | 0.0091 (5) | −0.0021 (5) |
| C11 | 0.0647 (9) | 0.0483 (7) | 0.0403 (7) | −0.0071 (6) | 0.0187 (6) | 0.0014 (6) |
| C12 | 0.0408 (7) | 0.0469 (7) | 0.0496 (7) | −0.0049 (5) | 0.0064 (5) | 0.0004 (6) |
| C13 | 0.0394 (6) | 0.0402 (6) | 0.0357 (6) | 0.0012 (5) | 0.0044 (5) | 0.0025 (5) |
| C14 | 0.0419 (6) | 0.0421 (6) | 0.0470 (7) | −0.0027 (5) | 0.0181 (5) | 0.0038 (5) |
| C15 | 0.0454 (7) | 0.0443 (7) | 0.0571 (8) | 0.0077 (6) | 0.0097 (6) | 0.0095 (6) |
| C16 | 0.0489 (7) | 0.0559 (8) | 0.0462 (7) | 0.0039 (6) | 0.0025 (6) | 0.0070 (6) |
| C17 | 0.0490 (7) | 0.0491 (7) | 0.0473 (7) | 0.0052 (6) | 0.0129 (6) | 0.0019 (6) |
| C18 | 0.0499 (8) | 0.0579 (9) | 0.0554 (8) | 0.0177 (7) | 0.0060 (6) | 0.0030 (7) |
| C19 | 0.0466 (7) | 0.0594 (9) | 0.0468 (7) | 0.0080 (6) | 0.0045 (6) | 0.0042 (6) |
| C20 | 0.0881 (13) | 0.0854 (13) | 0.0627 (11) | 0.0002 (11) | 0.0005 (9) | −0.0198 (10) |
| N1 | 0.0399 (5) | 0.0342 (5) | 0.0396 (5) | −0.0011 (4) | 0.0109 (4) | −0.0010 (4) |
| N2 | 0.0402 (5) | 0.0285 (4) | 0.0342 (5) | −0.0025 (4) | 0.0085 (4) | −0.0030 (4) |
| N3 | 0.0396 (5) | 0.0329 (5) | 0.0346 (5) | 0.0001 (4) | 0.0060 (4) | −0.0017 (4) |
| N4 | 0.0538 (7) | 0.0511 (7) | 0.0517 (7) | −0.0089 (5) | 0.0174 (5) | 0.0016 (5) |
| O1 | 0.0557 (6) | 0.0447 (5) | 0.0585 (6) | −0.0112 (4) | 0.0262 (5) | −0.0036 (4) |
| O2 | 0.0796 (8) | 0.0689 (7) | 0.0634 (7) | 0.0212 (6) | 0.0021 (6) | −0.0142 (6) |
Geometric parameters (Å, °)
| C1—N1 | 1.3161 (15) | C10—H10 | 0.9300 |
| C1—O1 | 1.3667 (14) | C11—N4 | 1.3312 (18) |
| C1—C2 | 1.4020 (17) | C11—H11 | 0.9300 |
| C2—C3 | 1.3744 (17) | C12—N4 | 1.3322 (17) |
| C2—H2 | 0.9300 | C12—C13 | 1.3823 (17) |
| C3—C4 | 1.3928 (16) | C12—H12 | 0.9300 |
| C3—H3 | 0.9300 | C13—H13 | 0.9300 |
| C4—N3 | 1.3842 (14) | C14—C15 | 1.365 (2) |
| C4—C5 | 1.3924 (15) | C14—C19 | 1.3708 (19) |
| C5—N1 | 1.3328 (14) | C14—O1 | 1.4048 (16) |
| C5—N2 | 1.3724 (14) | C15—C16 | 1.385 (2) |
| C6—N3 | 1.3192 (14) | C15—H15 | 0.9300 |
| C6—N2 | 1.3788 (13) | C16—C17 | 1.3784 (19) |
| C6—C9 | 1.4686 (15) | C16—H16 | 0.9300 |
| C7—N2 | 1.4638 (14) | C17—O2 | 1.3687 (17) |
| C7—C8 | 1.5066 (19) | C17—C18 | 1.371 (2) |
| C7—H7A | 0.9700 | C18—C19 | 1.369 (2) |
| C7—H7B | 0.9700 | C18—H18 | 0.9300 |
| C8—H8A | 0.9600 | C19—H19 | 0.9300 |
| C8—H8B | 0.9600 | C20—O2 | 1.406 (2) |
| C8—H8C | 0.9600 | C20—H20A | 0.9600 |
| C9—C13 | 1.3857 (17) | C20—H20B | 0.9600 |
| C9—C10 | 1.3899 (16) | C20—H20C | 0.9600 |
| C10—C11 | 1.3786 (18) | ||
| N1—C1—O1 | 118.16 (10) | N4—C12—C13 | 124.07 (13) |
| N1—C1—C2 | 125.63 (11) | N4—C12—H12 | 118.0 |
| O1—C1—C2 | 116.21 (10) | C13—C12—H12 | 118.0 |
| C3—C2—C1 | 119.44 (11) | C12—C13—C9 | 118.98 (11) |
| C3—C2—H2 | 120.3 | C12—C13—H13 | 120.5 |
| C1—C2—H2 | 120.3 | C9—C13—H13 | 120.5 |
| C2—C3—C4 | 117.22 (11) | C15—C14—C19 | 120.64 (13) |
| C2—C3—H3 | 121.4 | C15—C14—O1 | 119.97 (12) |
| C4—C3—H3 | 121.4 | C19—C14—O1 | 119.35 (13) |
| N3—C4—C5 | 109.76 (10) | C14—C15—C16 | 119.40 (13) |
| N3—C4—C3 | 133.26 (10) | C14—C15—H15 | 120.3 |
| C5—C4—C3 | 116.98 (10) | C16—C15—H15 | 120.3 |
| N1—C5—N2 | 125.53 (10) | C17—C16—C15 | 120.04 (13) |
| N1—C5—C4 | 127.79 (10) | C17—C16—H16 | 120.0 |
| N2—C5—C4 | 106.68 (9) | C15—C16—H16 | 120.0 |
| N3—C6—N2 | 113.32 (9) | O2—C17—C18 | 115.72 (12) |
| N3—C6—C9 | 122.41 (10) | O2—C17—C16 | 124.63 (13) |
| N2—C6—C9 | 124.27 (10) | C18—C17—C16 | 119.66 (13) |
| N2—C7—C8 | 112.19 (10) | C19—C18—C17 | 120.31 (13) |
| N2—C7—H7A | 109.2 | C19—C18—H18 | 119.8 |
| C8—C7—H7A | 109.2 | C17—C18—H18 | 119.8 |
| N2—C7—H7B | 109.2 | C18—C19—C14 | 119.94 (13) |
| C8—C7—H7B | 109.2 | C18—C19—H19 | 120.0 |
| H7A—C7—H7B | 107.9 | C14—C19—H19 | 120.0 |
| C7—C8—H8A | 109.5 | O2—C20—H20A | 109.5 |
| C7—C8—H8B | 109.5 | O2—C20—H20B | 109.5 |
| H8A—C8—H8B | 109.5 | H20A—C20—H20B | 109.5 |
| C7—C8—H8C | 109.5 | O2—C20—H20C | 109.5 |
| H8A—C8—H8C | 109.5 | H20A—C20—H20C | 109.5 |
| H8B—C8—H8C | 109.5 | H20B—C20—H20C | 109.5 |
| C13—C9—C10 | 117.46 (11) | C1—N1—C5 | 112.91 (10) |
| C13—C9—C6 | 123.59 (10) | C5—N2—C6 | 105.53 (9) |
| C10—C9—C6 | 118.91 (11) | C5—N2—C7 | 122.62 (9) |
| C11—C10—C9 | 118.99 (12) | C6—N2—C7 | 131.19 (9) |
| C11—C10—H10 | 120.5 | C6—N3—C4 | 104.72 (9) |
| C9—C10—H10 | 120.5 | C11—N4—C12 | 116.31 (11) |
| N4—C11—C10 | 124.17 (12) | C1—O1—C14 | 116.08 (9) |
| N4—C11—H11 | 117.9 | C17—O2—C20 | 117.96 (13) |
| C10—C11—H11 | 117.9 | ||
| N1—C1—C2—C3 | −1.3 (2) | O1—C14—C19—C18 | −178.17 (12) |
| O1—C1—C2—C3 | 178.21 (11) | O1—C1—N1—C5 | −177.83 (11) |
| C1—C2—C3—C4 | −0.17 (18) | C2—C1—N1—C5 | 1.63 (18) |
| C2—C3—C4—N3 | −179.45 (12) | N2—C5—N1—C1 | 179.15 (11) |
| C2—C3—C4—C5 | 0.96 (16) | C4—C5—N1—C1 | −0.73 (17) |
| N3—C4—C5—N1 | 179.77 (11) | N1—C5—N2—C6 | −179.62 (11) |
| C3—C4—C5—N1 | −0.54 (18) | C4—C5—N2—C6 | 0.28 (12) |
| N3—C4—C5—N2 | −0.12 (13) | N1—C5—N2—C7 | 8.72 (18) |
| C3—C4—C5—N2 | 179.56 (10) | C4—C5—N2—C7 | −171.38 (10) |
| N3—C6—C9—C13 | 142.45 (12) | N3—C6—N2—C5 | −0.36 (13) |
| N2—C6—C9—C13 | −37.09 (17) | C9—C6—N2—C5 | 179.21 (10) |
| N3—C6—C9—C10 | −35.08 (17) | N3—C6—N2—C7 | 170.30 (11) |
| N2—C6—C9—C10 | 145.39 (12) | C9—C6—N2—C7 | −10.13 (19) |
| C13—C9—C10—C11 | −0.54 (19) | C8—C7—N2—C5 | −76.72 (15) |
| C6—C9—C10—C11 | 177.14 (12) | C8—C7—N2—C6 | 113.98 (14) |
| C9—C10—C11—N4 | 1.3 (2) | N2—C6—N3—C4 | 0.28 (13) |
| N4—C12—C13—C9 | 1.1 (2) | C9—C6—N3—C4 | −179.30 (10) |
| C10—C9—C13—C12 | −0.60 (17) | C5—C4—N3—C6 | −0.09 (13) |
| C6—C9—C13—C12 | −178.16 (11) | C3—C4—N3—C6 | −179.70 (13) |
| C19—C14—C15—C16 | −0.1 (2) | C10—C11—N4—C12 | −0.8 (2) |
| O1—C14—C15—C16 | 177.83 (11) | C13—C12—N4—C11 | −0.4 (2) |
| C14—C15—C16—C17 | 0.6 (2) | N1—C1—O1—C14 | −0.86 (17) |
| C15—C16—C17—O2 | 178.89 (13) | C2—C1—O1—C14 | 179.63 (11) |
| C15—C16—C17—C18 | −0.7 (2) | C15—C14—O1—C1 | 90.64 (14) |
| O2—C17—C18—C19 | −179.25 (14) | C19—C14—O1—C1 | −91.40 (14) |
| C16—C17—C18—C19 | 0.4 (2) | C18—C17—O2—C20 | −175.90 (15) |
| C17—C18—C19—C14 | 0.1 (2) | C16—C17—O2—C20 | 4.5 (2) |
| C15—C14—C19—C18 | −0.2 (2) |
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: LX2189).
References
- Abdel-Alim, A. M., El-Shorbagi, A. A., Abdel-Mothy, S. G. & Abdel-Allah, H. H. M. (2005). Arch. Pharm. Res. 28, 637–647. [DOI] [PubMed]
- Beddoes, R. L., Dalton, L., Joule, T. A., Mills, O. S., Street, J. D. & Watt, C. I. F. (1986). J. Chem. Soc. Perkin Trans. 2, pp. 787–797.
- Bruker (2004). APEX2 and SAINT Bruker AXS Inc., Madison, Wisconsin, USA.
- Farrugia, L. J. (1997). J. Appl. Cryst. 30, 565.
- Girgis, A. S., Hosni, H. M. & Barsoum, F. F. (2006). Bioorg. Med. Chem. 14, 4466-4476. [DOI] [PubMed]
- Jiyeon, O., Sangseop, K., Kyu-Yang, Y., Nak-Jung, K., Hyung, S. H., Je-Yoel, C. & Kyoungho, S. (2010). Biochem. Pharmacol. 79, 596–609.
- Ouzidan, Y., Obbade, S., Capet, F., Essassi, E. M. & Ng, S. W. (2010). Acta Cryst. E66, o946. [DOI] [PMC free article] [PubMed]
- Passannanti, A., Diana, P., Barraja, P., Mingoia, F., Lauria, A. & Cirrincione, G. (1998). Heterocycles, 48, 1229–1235.
- Sheldrick, G. M. (1996). SADABS University of Göttingen, Germany.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Slominska, E. M., Yuen, A., Osman, L., Gebicki, J., Yacoub, M. H. & Smolenski, R. T. (2008). Nucleoside Nucleotides Nucleic Acids, 27, 863–866. [DOI] [PubMed]
- Spanka, C., Glatthar, R., Desrayaud, S., Fendt, M., Orain, D., Troxler, T. & Vranesic, I. (2010). Bioorg. Med. Chem. Lett. 20, 184–188. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811023543/lx2189sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811023543/lx2189Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811023543/lx2189Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



