Abstract
In the title molecule, C23H19ClN4OS, the 1,2,4-triazole ring forms dihedral angles of 46.5 (2), 87.4 (2) and 80.9 (2) Å with the three six-membered rings. Weak intermolecular N—H⋯S and C—H⋯O hydrogen bonds consolidate the crystal packing.
Related literature
For the crystal structures of related 1,2,4-triazole-5(4H)-thione derivatives, see: Al-Tamimi et al. (2010 ▶); Fun et al. (2009 ▶); Tan et al. (2010 ▶); Wang et al. (2011 ▶).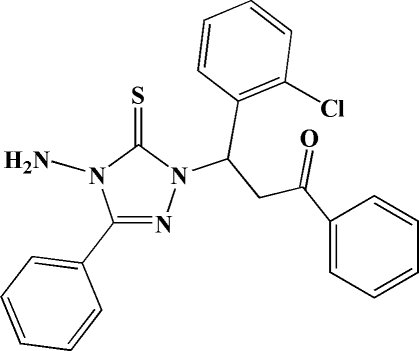
Experimental
Crystal data
C23H19ClN4OS
M r = 434.93
Triclinic,

a = 10.559 (3) Å
b = 10.787 (4) Å
c = 10.835 (3) Å
α = 99.582 (2)°
β = 96.638 (4)°
γ = 115.267 (3)°
V = 1076.2 (6) Å3
Z = 2
Mo Kα radiation
μ = 0.30 mm−1
T = 113 K
0.20 × 0.20 × 0.14 mm
Data collection
Rigaku Saturn CCD area-detector diffractometer
Absorption correction: multi-scan (CrystalClear; Rigaku/MSC, 2005 ▶) T min = 0.943, T max = 0.960
13931 measured reflections
5111 independent reflections
3433 reflections with I > 2σ(I)
R int = 0.038
Refinement
R[F 2 > 2σ(F 2)] = 0.032
wR(F 2) = 0.079
S = 0.95
5111 reflections
279 parameters
H atoms treated by a mixture of independent and constrained refinement
Δρmax = 0.40 e Å−3
Δρmin = −0.29 e Å−3
Data collection: CrystalClear (Rigaku/MSC, 2005 ▶); cell refinement: CrystalClear; data reduction: CrystalClear; program(s) used to solve structure: SHELXS97 (Sheldrick, 2008 ▶); program(s) used to refine structure: SHELXL97 (Sheldrick, 2008 ▶); molecular graphics: SHELXTL (Sheldrick, 2008 ▶); software used to prepare material for publication: SHELXTL.
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811023750/cv5107sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811023750/cv5107Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811023750/cv5107Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N4—H4B⋯S1i | 0.945 (18) | 2.714 (17) | 3.4928 (17) | 140.2 (12) |
| C7—H7⋯O1ii | 0.95 | 2.52 | 3.436 (2) | 161 |
| C21—H21⋯O1iii | 0.95 | 2.53 | 3.415 (2) | 156 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
supplementary crystallographic information
Comment
In continuation of structural study of 1,2,4-triazole-5(4H)-thione derivatives in our group (Wang et al., 2011), we present here the crystal structure of the title compound, (I).
In (I) (Fig.1), all bond lengths and angles are normal and comparable with those observed in related structures (Al-Tamimi et al., 2010; Fun et al., 2009; Tan et al., 2010; Wang et al., 2011). The 1,2,4-triazole ring is planar with an r.m.s. deviation of 0.002 (2) Å. The C1 atom in the triazole ring deviates from the normal Csp2 hybridization state having the bond angles of 102.56 (10)° (N1—C1—N3) and 129.94 (10)° (N1—C1—S1). There are three benzene rings in the molecule. The three benzene rings are inclined with respect to the 1,2,4-triazole ring [dihedral angles of 46.5 (2)° (C18—C23), 87.4 (2)° (C6—C11) and 80.9 (2)% (C12—C17)]. Benzene ring A (C18—C23) attached to the triazole ring makes the dihedral angle of 90.6 (2) and 126.9 (2)° with the benzene rings B (C6—C11) and C (C12—C17), respectively. Ring B and ring C form a dihedral angle 93.6 (2)°.
In the crystal structure, weak intermolecular N—H···S and C—H···O hydrogen bonds (Table 1) consolidate the crystal packing.
Experimental
The title compound was synthesized by the reaction of the 3-(2-chlorophenyl)-1- phenyl-2-propen-1-one (2.0 mmol) with 4-amino-3-phenyl-4H-1,2,4-triazole-5- thiol (2.0 mmol) in ethanol. The reaction progress was monitored via TLC. The resulting precipitate was filtered off, washed with cold ethanol, dried and purified to give the target product as colourless solid in 75% yield. Crystals of (I) suitable for single-crystal X-ray analysis were grown by slow evaporation of a solution in chloroform-ethanol (1:1).
Refinement
The H atoms attached to N atoms were located in a difference map and isotropically refined. C-bound H atoms were positioned geometrically (C—H = 0.95–1.00 Å) and refined as riding, with Uiso(H) = 1.2Ueq(C).
Figures
Fig. 1.
View of the molecule of (I) showing the atom-labelling scheme. Displacement ellipsoids are drawn at the 55% probability level.
Crystal data
| C23H19ClN4OS | Z = 2 |
| Mr = 434.93 | F(000) = 452 |
| Triclinic, P1 | Dx = 1.342 Mg m−3 |
| Hall symbol: -P 1 | Mo Kα radiation, λ = 0.71073 Å |
| a = 10.559 (3) Å | Cell parameters from 3478 reflections |
| b = 10.787 (4) Å | θ = 1.9–27.9° |
| c = 10.835 (3) Å | µ = 0.30 mm−1 |
| α = 99.582 (2)° | T = 113 K |
| β = 96.638 (4)° | Prism, colourless |
| γ = 115.267 (3)° | 0.20 × 0.20 × 0.14 mm |
| V = 1076.2 (6) Å3 |
Data collection
| Rigaku Saturn CCD area-detector diffractometer | 5111 independent reflections |
| Radiation source: rotating anode | 3433 reflections with I > 2σ(I) |
| multilayer | Rint = 0.038 |
| Detector resolution: 14.63 pixels mm-1 | θmax = 27.9°, θmin = 2.0° |
| φ and ω scans | h = −13→13 |
| Absorption correction: multi-scan (CrystalClear; Rigaku/MSC, 2005) | k = −14→14 |
| Tmin = 0.943, Tmax = 0.960 | l = −14→14 |
| 13931 measured reflections |
Refinement
| Refinement on F2 | Primary atom site location: structure-invariant direct methods |
| Least-squares matrix: full | Secondary atom site location: difference Fourier map |
| R[F2 > 2σ(F2)] = 0.032 | Hydrogen site location: inferred from neighbouring sites |
| wR(F2) = 0.079 | H atoms treated by a mixture of independent and constrained refinement |
| S = 0.95 | w = 1/[σ2(Fo2) + (0.0349P)2] where P = (Fo2 + 2Fc2)/3 |
| 5111 reflections | (Δ/σ)max = 0.001 |
| 279 parameters | Δρmax = 0.40 e Å−3 |
| 0 restraints | Δρmin = −0.29 e Å−3 |
Special details
| Geometry. All e.s.d.'s (except the e.s.d. in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell e.s.d.'s are taken into account individually in the estimation of e.s.d.'s in distances, angles and torsion angles; correlations between e.s.d.'s in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell e.s.d.'s is used for estimating e.s.d.'s involving l.s. planes. |
| Refinement. Refinement of F2 against ALL reflections. The weighted R-factor wR and goodness of fit S are based on F2, conventional R-factors R are based on F, with F set to zero for negative F2. The threshold expression of F2 > σ(F2) is used only for calculating R-factors(gt) etc. and is not relevant to the choice of reflections for refinement. R-factors based on F2 are statistically about twice as large as those based on F, and R- factors based on ALL data will be even larger. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.87508 (4) | 0.30026 (4) | 0.43912 (3) | 0.02271 (9) | |
| Cl1 | 1.14768 (4) | 0.15222 (4) | 0.21807 (4) | 0.04566 (13) | |
| O1 | 0.70047 (10) | 0.13775 (10) | 0.05439 (9) | 0.0298 (2) | |
| N1 | 0.90922 (11) | 0.39665 (11) | 0.22109 (10) | 0.0171 (2) | |
| N2 | 0.88351 (11) | 0.48767 (11) | 0.15864 (10) | 0.0184 (2) | |
| N3 | 0.81314 (11) | 0.49054 (11) | 0.34152 (9) | 0.0175 (2) | |
| N4 | 0.75491 (14) | 0.52876 (15) | 0.44374 (11) | 0.0252 (3) | |
| C1 | 0.86846 (13) | 0.39575 (13) | 0.33424 (11) | 0.0174 (3) | |
| C2 | 0.82420 (13) | 0.54367 (13) | 0.23458 (11) | 0.0172 (3) | |
| C3 | 0.98141 (13) | 0.31632 (13) | 0.17006 (12) | 0.0179 (3) | |
| H3 | 0.9499 | 0.2301 | 0.2049 | 0.021* | |
| C4 | 0.93753 (13) | 0.26682 (13) | 0.02470 (11) | 0.0183 (3) | |
| H4C | 0.9598 | 0.3500 | −0.0121 | 0.022* | |
| H4D | 0.9946 | 0.2203 | −0.0067 | 0.022* | |
| C5 | 0.77970 (14) | 0.16486 (13) | −0.02188 (12) | 0.0201 (3) | |
| C6 | 0.72490 (13) | 0.09573 (13) | −0.16117 (12) | 0.0191 (3) | |
| C7 | 0.58247 (14) | −0.00620 (14) | −0.20344 (13) | 0.0263 (3) | |
| H7 | 0.5211 | −0.0281 | −0.1443 | 0.032* | |
| C8 | 0.53048 (15) | −0.07543 (16) | −0.33125 (14) | 0.0344 (4) | |
| H8 | 0.4331 | −0.1442 | −0.3600 | 0.041* | |
| C9 | 0.61981 (16) | −0.04485 (16) | −0.41761 (13) | 0.0332 (4) | |
| H9 | 0.5838 | −0.0934 | −0.5053 | 0.040* | |
| C10 | 0.76108 (16) | 0.05597 (14) | −0.37670 (13) | 0.0266 (3) | |
| H10 | 0.8221 | 0.0770 | −0.4362 | 0.032* | |
| C11 | 0.81364 (14) | 0.12638 (13) | −0.24900 (12) | 0.0214 (3) | |
| H11 | 0.9108 | 0.1960 | −0.2210 | 0.026* | |
| C12 | 1.14209 (13) | 0.40065 (14) | 0.21643 (11) | 0.0179 (3) | |
| C13 | 1.22678 (15) | 0.33506 (15) | 0.24249 (13) | 0.0249 (3) | |
| C14 | 1.37336 (15) | 0.41100 (17) | 0.28825 (13) | 0.0313 (4) | |
| H14 | 1.4285 | 0.3635 | 0.3066 | 0.038* | |
| C15 | 1.43893 (15) | 0.55614 (17) | 0.30705 (13) | 0.0311 (3) | |
| H15 | 1.5393 | 0.6090 | 0.3392 | 0.037* | |
| C16 | 1.35845 (15) | 0.62377 (16) | 0.27904 (13) | 0.0292 (3) | |
| H16 | 1.4036 | 0.7233 | 0.2899 | 0.035* | |
| C17 | 1.21141 (14) | 0.54688 (14) | 0.23495 (12) | 0.0231 (3) | |
| H17 | 1.1568 | 0.5950 | 0.2170 | 0.028* | |
| C18 | 0.77955 (13) | 0.65020 (13) | 0.20813 (12) | 0.0194 (3) | |
| C19 | 0.70373 (15) | 0.63013 (15) | 0.08661 (13) | 0.0266 (3) | |
| H19 | 0.6793 | 0.5474 | 0.0224 | 0.032* | |
| C20 | 0.66390 (17) | 0.73115 (18) | 0.05937 (15) | 0.0378 (4) | |
| H20 | 0.6110 | 0.7168 | −0.0233 | 0.045* | |
| C21 | 0.70075 (18) | 0.85272 (17) | 0.15195 (16) | 0.0407 (4) | |
| H21 | 0.6734 | 0.9217 | 0.1327 | 0.049* | |
| C22 | 0.77736 (17) | 0.87375 (16) | 0.27237 (16) | 0.0370 (4) | |
| H22 | 0.8031 | 0.9577 | 0.3356 | 0.044* | |
| C23 | 0.81688 (15) | 0.77314 (14) | 0.30151 (14) | 0.0273 (3) | |
| H23 | 0.8691 | 0.7877 | 0.3846 | 0.033* | |
| H4A | 0.7078 (16) | 0.4471 (16) | 0.4696 (13) | 0.031 (4)* | |
| H4B | 0.8351 (18) | 0.5891 (18) | 0.5104 (15) | 0.052 (5)* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.02278 (18) | 0.02589 (19) | 0.02155 (18) | 0.01071 (15) | 0.00574 (14) | 0.01129 (15) |
| Cl1 | 0.0438 (2) | 0.0299 (2) | 0.0776 (3) | 0.02636 (19) | 0.0153 (2) | 0.0206 (2) |
| O1 | 0.0252 (5) | 0.0299 (6) | 0.0298 (6) | 0.0068 (5) | 0.0135 (5) | 0.0064 (5) |
| N1 | 0.0183 (5) | 0.0172 (5) | 0.0188 (5) | 0.0092 (5) | 0.0061 (4) | 0.0071 (5) |
| N2 | 0.0202 (6) | 0.0184 (6) | 0.0206 (6) | 0.0112 (5) | 0.0057 (5) | 0.0073 (5) |
| N3 | 0.0153 (5) | 0.0228 (6) | 0.0155 (5) | 0.0093 (5) | 0.0052 (4) | 0.0044 (5) |
| N4 | 0.0260 (7) | 0.0372 (8) | 0.0188 (6) | 0.0181 (6) | 0.0116 (5) | 0.0083 (6) |
| C1 | 0.0134 (6) | 0.0189 (6) | 0.0179 (6) | 0.0054 (5) | 0.0037 (5) | 0.0042 (5) |
| C2 | 0.0142 (6) | 0.0194 (6) | 0.0171 (6) | 0.0066 (5) | 0.0039 (5) | 0.0047 (5) |
| C3 | 0.0205 (7) | 0.0162 (6) | 0.0210 (7) | 0.0104 (5) | 0.0068 (5) | 0.0071 (5) |
| C4 | 0.0189 (7) | 0.0163 (6) | 0.0201 (7) | 0.0077 (5) | 0.0066 (5) | 0.0046 (5) |
| C5 | 0.0206 (7) | 0.0159 (6) | 0.0262 (7) | 0.0093 (6) | 0.0073 (6) | 0.0068 (6) |
| C6 | 0.0189 (7) | 0.0143 (6) | 0.0256 (7) | 0.0091 (5) | 0.0036 (6) | 0.0056 (5) |
| C7 | 0.0190 (7) | 0.0251 (8) | 0.0347 (8) | 0.0103 (6) | 0.0047 (6) | 0.0072 (6) |
| C8 | 0.0219 (8) | 0.0313 (9) | 0.0399 (9) | 0.0085 (7) | −0.0069 (7) | 0.0020 (7) |
| C9 | 0.0389 (9) | 0.0312 (8) | 0.0238 (8) | 0.0153 (7) | −0.0066 (7) | 0.0029 (7) |
| C10 | 0.0355 (8) | 0.0227 (7) | 0.0230 (7) | 0.0131 (6) | 0.0053 (6) | 0.0096 (6) |
| C11 | 0.0231 (7) | 0.0153 (6) | 0.0247 (7) | 0.0077 (6) | 0.0028 (6) | 0.0060 (6) |
| C12 | 0.0202 (7) | 0.0224 (7) | 0.0143 (6) | 0.0116 (6) | 0.0060 (5) | 0.0053 (5) |
| C13 | 0.0285 (8) | 0.0271 (8) | 0.0273 (7) | 0.0177 (6) | 0.0100 (6) | 0.0107 (6) |
| C14 | 0.0275 (8) | 0.0485 (10) | 0.0308 (8) | 0.0271 (8) | 0.0091 (7) | 0.0132 (7) |
| C15 | 0.0189 (7) | 0.0444 (10) | 0.0271 (8) | 0.0124 (7) | 0.0048 (6) | 0.0069 (7) |
| C16 | 0.0223 (7) | 0.0271 (8) | 0.0317 (8) | 0.0064 (6) | 0.0059 (6) | 0.0037 (6) |
| C17 | 0.0212 (7) | 0.0234 (7) | 0.0261 (7) | 0.0115 (6) | 0.0042 (6) | 0.0061 (6) |
| C18 | 0.0182 (6) | 0.0215 (7) | 0.0242 (7) | 0.0108 (6) | 0.0123 (6) | 0.0100 (6) |
| C19 | 0.0310 (8) | 0.0337 (8) | 0.0255 (7) | 0.0206 (7) | 0.0130 (6) | 0.0120 (6) |
| C20 | 0.0488 (10) | 0.0559 (11) | 0.0347 (9) | 0.0396 (9) | 0.0199 (8) | 0.0254 (8) |
| C21 | 0.0553 (11) | 0.0414 (10) | 0.0577 (11) | 0.0382 (9) | 0.0363 (10) | 0.0324 (9) |
| C22 | 0.0443 (10) | 0.0237 (8) | 0.0515 (11) | 0.0181 (7) | 0.0276 (9) | 0.0114 (8) |
| C23 | 0.0256 (8) | 0.0253 (8) | 0.0320 (8) | 0.0111 (6) | 0.0122 (6) | 0.0069 (6) |
Geometric parameters (Å, °)
| S1—C1 | 1.6705 (13) | C9—C10 | 1.381 (2) |
| Cl1—C13 | 1.7397 (16) | C9—H9 | 0.9500 |
| O1—C5 | 1.2247 (14) | C10—C11 | 1.3828 (18) |
| N1—C1 | 1.3456 (15) | C10—H10 | 0.9500 |
| N1—N2 | 1.3791 (13) | C11—H11 | 0.9500 |
| N1—C3 | 1.4628 (15) | C12—C13 | 1.3886 (17) |
| N2—C2 | 1.3080 (15) | C12—C17 | 1.3926 (18) |
| N3—C2 | 1.3712 (16) | C13—C14 | 1.3847 (19) |
| N3—C1 | 1.3736 (16) | C14—C15 | 1.381 (2) |
| N3—N4 | 1.4130 (14) | C14—H14 | 0.9500 |
| N4—H4A | 0.918 (15) | C15—C16 | 1.375 (2) |
| N4—H4B | 0.945 (18) | C15—H15 | 0.9500 |
| C2—C18 | 1.4716 (17) | C16—C17 | 1.3871 (18) |
| C3—C12 | 1.5138 (17) | C16—H16 | 0.9500 |
| C3—C4 | 1.5233 (17) | C17—H17 | 0.9500 |
| C3—H3 | 1.0000 | C18—C19 | 1.3910 (18) |
| C4—C5 | 1.5158 (17) | C18—C23 | 1.3974 (18) |
| C4—H4C | 0.9900 | C19—C20 | 1.3861 (19) |
| C4—H4D | 0.9900 | C19—H19 | 0.9500 |
| C5—C6 | 1.4913 (18) | C20—C21 | 1.383 (2) |
| C6—C11 | 1.3926 (17) | C20—H20 | 0.9500 |
| C6—C7 | 1.3935 (18) | C21—C22 | 1.382 (2) |
| C7—C8 | 1.3809 (19) | C21—H21 | 0.9500 |
| C7—H7 | 0.9500 | C22—C23 | 1.3865 (19) |
| C8—C9 | 1.384 (2) | C22—H22 | 0.9500 |
| C8—H8 | 0.9500 | C23—H23 | 0.9500 |
| C1—N1—N2 | 113.49 (10) | C9—C10—C11 | 119.92 (13) |
| C1—N1—C3 | 124.44 (10) | C9—C10—H10 | 120.0 |
| N2—N1—C3 | 121.98 (10) | C11—C10—H10 | 120.0 |
| C2—N2—N1 | 104.42 (10) | C10—C11—C6 | 120.33 (13) |
| C2—N3—C1 | 109.50 (10) | C10—C11—H11 | 119.8 |
| C2—N3—N4 | 125.04 (11) | C6—C11—H11 | 119.8 |
| C1—N3—N4 | 125.45 (10) | C13—C12—C17 | 117.10 (12) |
| N3—N4—H4A | 105.0 (9) | C13—C12—C3 | 121.22 (12) |
| N3—N4—H4B | 104.9 (9) | C17—C12—C3 | 121.68 (11) |
| H4A—N4—H4B | 106.4 (13) | C14—C13—C12 | 121.95 (13) |
| N1—C1—N3 | 102.56 (10) | C14—C13—Cl1 | 118.52 (11) |
| N1—C1—S1 | 129.94 (10) | C12—C13—Cl1 | 119.53 (11) |
| N3—C1—S1 | 127.46 (9) | C15—C14—C13 | 119.62 (13) |
| N2—C2—N3 | 110.02 (11) | C15—C14—H14 | 120.2 |
| N2—C2—C18 | 124.06 (11) | C13—C14—H14 | 120.2 |
| N3—C2—C18 | 125.91 (10) | C16—C15—C14 | 119.78 (14) |
| N1—C3—C12 | 110.60 (10) | C16—C15—H15 | 120.1 |
| N1—C3—C4 | 111.47 (10) | C14—C15—H15 | 120.1 |
| C12—C3—C4 | 112.37 (10) | C15—C16—C17 | 120.11 (14) |
| N1—C3—H3 | 107.4 | C15—C16—H16 | 119.9 |
| C12—C3—H3 | 107.4 | C17—C16—H16 | 119.9 |
| C4—C3—H3 | 107.4 | C16—C17—C12 | 121.41 (12) |
| C5—C4—C3 | 112.76 (10) | C16—C17—H17 | 119.3 |
| C5—C4—H4C | 109.0 | C12—C17—H17 | 119.3 |
| C3—C4—H4C | 109.0 | C19—C18—C23 | 119.75 (13) |
| C5—C4—H4D | 109.0 | C19—C18—C2 | 119.19 (12) |
| C3—C4—H4D | 109.0 | C23—C18—C2 | 121.02 (12) |
| H4C—C4—H4D | 107.8 | C20—C19—C18 | 119.86 (14) |
| O1—C5—C6 | 121.17 (12) | C20—C19—H19 | 120.1 |
| O1—C5—C4 | 120.25 (12) | C18—C19—H19 | 120.1 |
| C6—C5—C4 | 118.56 (10) | C21—C20—C19 | 120.33 (15) |
| C11—C6—C7 | 119.30 (12) | C21—C20—H20 | 119.8 |
| C11—C6—C5 | 121.73 (12) | C19—C20—H20 | 119.8 |
| C7—C6—C5 | 118.92 (11) | C22—C21—C20 | 120.00 (14) |
| C8—C7—C6 | 120.06 (13) | C22—C21—H21 | 120.0 |
| C8—C7—H7 | 120.0 | C20—C21—H21 | 120.0 |
| C6—C7—H7 | 120.0 | C21—C22—C23 | 120.39 (15) |
| C7—C8—C9 | 120.19 (14) | C21—C22—H22 | 119.8 |
| C7—C8—H8 | 119.9 | C23—C22—H22 | 119.8 |
| C9—C8—H8 | 119.9 | C22—C23—C18 | 119.67 (14) |
| C10—C9—C8 | 120.20 (14) | C22—C23—H23 | 120.2 |
| C10—C9—H9 | 119.9 | C18—C23—H23 | 120.2 |
| C8—C9—H9 | 119.9 | ||
| C1—N1—N2—C2 | −0.53 (14) | C8—C9—C10—C11 | 0.2 (2) |
| C3—N1—N2—C2 | −177.29 (11) | C9—C10—C11—C6 | 0.3 (2) |
| N2—N1—C1—N3 | 0.61 (13) | C7—C6—C11—C10 | −0.26 (19) |
| C3—N1—C1—N3 | 177.27 (11) | C5—C6—C11—C10 | 177.17 (11) |
| N2—N1—C1—S1 | 178.26 (9) | N1—C3—C12—C13 | 145.32 (12) |
| C3—N1—C1—S1 | −5.07 (19) | C4—C3—C12—C13 | −89.41 (14) |
| C2—N3—C1—N1 | −0.45 (13) | N1—C3—C12—C17 | −34.62 (16) |
| N4—N3—C1—N1 | −179.92 (11) | C4—C3—C12—C17 | 90.66 (14) |
| C2—N3—C1—S1 | −178.18 (10) | C17—C12—C13—C14 | 1.71 (19) |
| N4—N3—C1—S1 | 2.35 (19) | C3—C12—C13—C14 | −178.22 (12) |
| N1—N2—C2—N3 | 0.21 (13) | C17—C12—C13—Cl1 | −178.59 (9) |
| N1—N2—C2—C18 | 179.04 (11) | C3—C12—C13—Cl1 | 1.47 (17) |
| C1—N3—C2—N2 | 0.15 (14) | C12—C13—C14—C15 | −1.0 (2) |
| N4—N3—C2—N2 | 179.62 (11) | Cl1—C13—C14—C15 | 179.27 (10) |
| C1—N3—C2—C18 | −178.65 (12) | C13—C14—C15—C16 | −0.6 (2) |
| N4—N3—C2—C18 | 0.8 (2) | C14—C15—C16—C17 | 1.5 (2) |
| C1—N1—C3—C12 | −87.95 (14) | C15—C16—C17—C12 | −0.8 (2) |
| N2—N1—C3—C12 | 88.45 (13) | C13—C12—C17—C16 | −0.78 (19) |
| C1—N1—C3—C4 | 146.26 (11) | C3—C12—C17—C16 | 179.15 (11) |
| N2—N1—C3—C4 | −37.34 (15) | N2—C2—C18—C19 | 45.75 (18) |
| N1—C3—C4—C5 | −63.58 (13) | N3—C2—C18—C19 | −135.61 (13) |
| C12—C3—C4—C5 | 171.62 (10) | N2—C2—C18—C23 | −131.78 (13) |
| C3—C4—C5—O1 | 4.68 (17) | N3—C2—C18—C23 | 46.86 (18) |
| C3—C4—C5—C6 | −173.49 (10) | C23—C18—C19—C20 | −0.91 (19) |
| O1—C5—C6—C11 | 179.54 (12) | C2—C18—C19—C20 | −178.47 (12) |
| C4—C5—C6—C11 | −2.31 (17) | C18—C19—C20—C21 | 0.8 (2) |
| O1—C5—C6—C7 | −3.03 (18) | C19—C20—C21—C22 | −0.1 (2) |
| C4—C5—C6—C7 | 175.12 (11) | C20—C21—C22—C23 | −0.4 (2) |
| C11—C6—C7—C8 | −0.16 (19) | C21—C22—C23—C18 | 0.4 (2) |
| C5—C6—C7—C8 | −177.66 (12) | C19—C18—C23—C22 | 0.32 (19) |
| C6—C7—C8—C9 | 0.6 (2) | C2—C18—C23—C22 | 177.84 (11) |
| C7—C8—C9—C10 | −0.6 (2) |
Hydrogen-bond geometry (Å, °)
| D—H···A | D—H | H···A | D···A | D—H···A |
| N4—H4B···S1i | 0.945 (18) | 2.714 (17) | 3.4928 (17) | 140.2 (12) |
| C7—H7···O1ii | 0.95 | 2.52 | 3.436 (2) | 161 |
| C21—H21···O1iii | 0.95 | 2.53 | 3.415 (2) | 156 |
Symmetry codes: (i) −x+2, −y+1, −z+1; (ii) −x+1, −y, −z; (iii) x, y+1, z.
Footnotes
Supplementary data and figures for this paper are available from the IUCr electronic archives (Reference: CV5107).
References
- Al-Tamimi, A.-M. S., Bari, A., Al-Omar, M. A., Alrashood, K. A. & El-Emam, A. A. (2010). Acta Cryst. E66, o1756. [DOI] [PMC free article] [PubMed]
- Fun, H.-K., Chantrapromma, S., Sujith, K. V. & Kalluraya, B. (2009). Acta Cryst. E65, o495–o496. [DOI] [PMC free article] [PubMed]
- Rigaku/MSC (2005). CrystalClear Molecular Structure Corporation, The Woodlands, Texas, USA, and Rigaku Corporation, Tokyo, Japan.
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Tan, K. W., Maah, M. J. & Ng, S. W. (2010). Acta Cryst. E66, o2224. [DOI] [PMC free article] [PubMed]
- Wang, W., Gao, Y., Xiao, Z., Yao, H. & Zhang, J. (2011). Acta Cryst. E67, o269. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S1600536811023750/cv5107sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S1600536811023750/cv5107Isup2.hkl
Supplementary material file. DOI: 10.1107/S1600536811023750/cv5107Isup3.cml
Additional supplementary materials: crystallographic information; 3D view; checkCIF report



