Abstract
Multi-fluorescence recovery after photobleaching is a microscopy technique used to measure the diffusion coefficient (or analogous transport parameters) of macromolecules, and can be applied to both in vitro and in vivo biological systems. Multi-fluorescence recovery after photobleaching is performed by photobleaching a region of interest within a fluorescent sample using an intense laser flash, then attenuating the beam and monitoring the fluorescence as still-fluorescent molecules from outside the region of interest diffuse in to replace the photobleached molecules. We will begin our demonstration by aligning the laser beam through the Pockels Cell (laser modulator) and along the optical path through the laser scan box and objective lens to the sample. For simplicity, we will use a sample of aqueous fluorescent dye. We will then determine the proper experimental parameters for our sample including, monitor and bleaching powers, bleach duration, bin widths (for photon counting), and fluorescence recovery time. Next, we will describe the procedure for taking recovery curves, a process that can be largely automated via LabVIEW (National Instruments, Austin, TX) for enhanced throughput. Finally, the diffusion coefficient is determined by fitting the recovery data to the appropriate mathematical model using a least-squares fitting algorithm, readily programmable using software such as MATLAB (The Mathworks, Natick, MA).
Protocol
1. Align the optics.
The key equipment for an MP-FRAP experiment include: a mode-locked laser source, Pockels Cell (for beam modulation), pulse generator, dichroic, objective lens, fluorescence emission filter, gated photomultiplier tube, and a data recording system (photon counter and multichannel scaler).
2. Determine safe monitor powers.
Attenuate the laser to a low, but reasonable, power for generating fluorescence within the sample.
Focus within the sample.
Set a photon counter to integrate over a time scale much longer than the expected fluorescence recovery. With the focal volume held stationary within the sample (on our microscope software this is achieved by a point scan), record a reasonable number of photon count data.
Increase the power and take another series of photon count data. Repeat until the increase in photon counts appears to diminish significantly with respect to the power increase.
Plot log(photon counts) as a function of log(power). A deviation from a slope of two indicates bleaching during the monitor. Choose as your monitor power a power that is below this cut off, but which allows for a reasonable signal to noise ratio.
3. Determine input parameters to the pulse generator and multichannel scaler.
Determine back-of-the-envelope estimates of the diffusion coefficient and recovery time based on the literature and/or the Stokes-Einstein equation and the diffusion equation.
- Set the necessary parameter values on the pulse generator and the scaler.
- On the pulse generator, set the pre-bleach and bleach times. A good set of rules of thumb is that the pre-bleach should be 1-2 times larger than the expected half recovery time, and the bleach time should be one-tenth the expected half recovery time.
- On the scaler, set the bin width to approximately one half the bleach duration, and set the number of bins per record such that the product of your selected bin width and bins per record is greater than or approximately equal to your expected full recovery plus pre-bleach and bleach times.
- Return to the pulse generator and set the frequency of the pre-bleach/bleach/monitor sequence equal to a value just smaller than the inverse of the product of the bin width times the number of bins per record. *
- Finally, set the number of records/scan on the scaler based on the signal intensity at your chosen monitor power. *It is very important that the time for a complete pre-bleach/bleach/monitor sequence (1/frequency) set on the pulse generator is greater than the data collection time (bin width x bins/record) set on the scaler. If this criteria is not met, multiple bleach triggers may appear within a single record on the scaler.
Set the monitor power to an acceptable level, determined previously. Set the bleach power to 200 mW or so above the bleaching cutoff determined during the monitor bleaching test. These powers are both set as different voltages across the PC crystal.
Seal the microscope, shut off the lights, turn on the PMT, begin a point scan on the microscope software, then start the pulse generator and scaler.
Analyze the resulting recovery curve by plotting the curve and marking three important points: 1) the pre-bleach fluorescence, 2) the fluorescence immediately post-bleach, and 3) the fluorescence at the end of the data set. Use the fluorescence values pre- and immediately post-bleach to better estimate the half-recovery time.
With this estimate of the half-recovery time, use the rules of thumb outlined previously to determine new values for the pre-bleach, bleach, and bin width durations. Also, use the pre-bleach fluorescence and fluorescence at the end of the data set to better estimate the time required to achieve full recovery. As a rule of thumb, data should be collected for a time period that allows for the full recovery to be reported for the latter half of the data.
Adjust the parameters on the pulse generator and scaler, take a new curve, and continue tweaking the parameters until your curve exhibits a strong bleach and smooth recovery. Keep in mind that the bleach duration can, and should, be reduced below the rule of thumb, if possible. The bleach power may also be adjusted if too shallow, or too deep, a bleach is achieved.
4. Test for excitation saturation.
Using appropriate monitor and bleach powers, determined previously, take a series of MP-FRAP curves. Analyze each recovery, normalized to the pre-bleach fluorescence, using the appropriate mathematical form and a non-linear least squares fitting algorithm (see "Data Analysis" below). Record the fitted value of the bleach depth parameter.
Continue taking and analyzing successive series of MP-FRAP curves at increasing values of bleach power.
Plot log(bleach depth parameter) as a function of log(power). Any deviation from a slope of two indicates excitation saturation. Choose a bleach power that yields a strong bleach, but does not cause saturation.
5. Continue taking MP-FRAP curves.
Continue taking curves, as above, will all the parameters properly set.
6. Data Analysis.
Remove the pre-bleach and bleach data from the fluorescence data set and adjust the time data such that t = 0 corresponds to the fluorescence value immediately post-bleach.
Normalize the resulting fluorescence recovery with respect to the average pre-bleach fluorescence.
Fit the normalized curve using the appropriate mathematical model and a non-linear, least squares fitting algorithm, such as the lsqcurvefit function in MATLAB. The model presented here accounts for fluorescence recoveries influenced by both diffusion and convective flow:
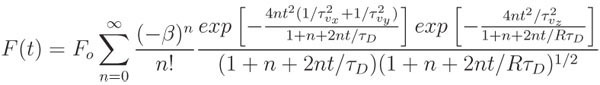 where Fo is the pre-bleach average fluorescence, β is the bleach depth parameter, τD is the characteristic diffusive recovery time, R is the ratio of the square of the axial (wz) to radial (wr) widths of the two-photon focal volume, τvx = wr/vx, τvy = wr/vy, τvz = wz/vz, and v2 = vx2 + vy2 + vz2 is the speed of the convective flow.
For flow confined to the imaging plane, τvz → ∞ and 1/ τvx + 1/ τvy can be replaced with 1/ τvr. In the absence of convective flow, both exponentials in the numerator go to 1 and the expression is greatly reduced, to only two fitting parameters, β and τD.
The diffusion coefficient is easily calculated as D = wr2/8τD.
where Fo is the pre-bleach average fluorescence, β is the bleach depth parameter, τD is the characteristic diffusive recovery time, R is the ratio of the square of the axial (wz) to radial (wr) widths of the two-photon focal volume, τvx = wr/vx, τvy = wr/vy, τvz = wz/vz, and v2 = vx2 + vy2 + vz2 is the speed of the convective flow.
For flow confined to the imaging plane, τvz → ∞ and 1/ τvx + 1/ τvy can be replaced with 1/ τvr. In the absence of convective flow, both exponentials in the numerator go to 1 and the expression is greatly reduced, to only two fitting parameters, β and τD.
The diffusion coefficient is easily calculated as D = wr2/8τD.
7. Representative Results.
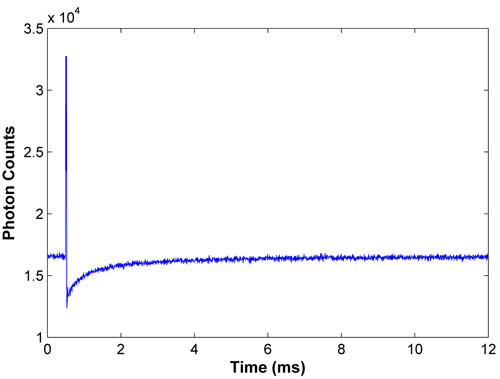 Figure 1. Representative bleach and recovery curve for FITC-BSA. A good recovery curve exhibits a strong bleach and smooth recovery, with the fluorescence at full recovery being reported for the latter half of the data.
Figure 1. Representative bleach and recovery curve for FITC-BSA. A good recovery curve exhibits a strong bleach and smooth recovery, with the fluorescence at full recovery being reported for the latter half of the data.
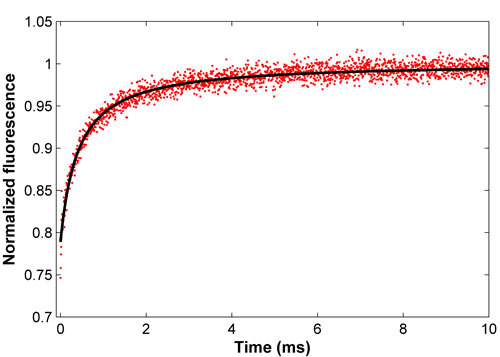 Figure 2. Normalized recovery curve for FITC-BSA (red) and the associated least-squares fit (black). This fit yields a diffusion coefficient of 52.9 um2/s, consistent with the literature.
Figure 2. Normalized recovery curve for FITC-BSA (red) and the associated least-squares fit (black). This fit yields a diffusion coefficient of 52.9 um2/s, consistent with the literature.
Discussion
The power of multi-photon fluorescence recovery after photobleaching lies in its ability to probe thick samples with 3D resolution. Since its development in the 1990's, MP-FRAP has been used to determine the diffusion coefficient (or analogous transport parameters) in cell bodies, ex vivo thick tissue slices, and in vivo tissue and interstitium. In this article, we presented the equipment necessary to run an MP-FRAP experiment, as well as the proper procedure for aligning the beam path, setting experimental parameters, taking data, and analyzing recovery curves.
The choices of an appropriate excitation wavelength and emission filter should be guided by the two-photon cross-sections and emission spectra. This information is often included with the technical data for various dyes. Also, in aligning the laser beam, it is important that proper overfilling of the back lens of the objective be achieved. This is often accomplished by adding a beam expander to the optical system. Proper overfilling can be verified by scanning sub-resolution fixed fluorescent beads in both the axial and radial directions and then plotting and fitting the fluorescence profiles to Gaussians to find the 1/e2 widths for comparison with literature values.
It is also important to choose appropriate monitoring and bleaching powers. The power used to monitor the fluorescence pre- and post-bleach should be low enough not to cause appreciable bleaching, but high enough to allow for a good signal-to-noise ratio. The bleaching power must avoid excitation saturation. In an MP-FRAP experiment there is an upper limit to the fluorescence excitation rate, which is proportional to the square of the power incident on the sample. This limit marks the start of the excitation saturation regime. Fluorescence recovery curves produced using bleach powers operating in the excitation saturation regime will yield erroneously low diffusion coefficients.
Acknowledgments
This work was funded by a Department of Defense Era of Hope Scholar Award (No. W81XWH-05-0396) and a Pew Scholar in the Biomedical Sciences Award to Edward B. Brown III.
References
- Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Biophys J. 1990;60:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- Brown EB, Wu ES, Zipfel W, Webb WW. Measurement of molecular diffusion in solution by multiphoton fluorescence photobleaching recovery. Biophys J. 1999;77:2837–2849. doi: 10.1016/S0006-3495(99)77115-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan KD, Sipprell WHBrown, Jr EB, Brown EB. Improved model of fluorescence recovery expands the application of multiphoton fluorescence recover after photobleaching in vivo. Biophys J. 2009;96:5082–5094. doi: 10.1016/j.bpj.2009.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]


