Abstract
Protein expression in the microbial eukaryotic host Pichia pastoris offers the possibility to generate high amounts of recombinant protein in a fast and easy to use expression system.
As a single-celled microorganism P. pastoris is easy to manipulate and grows rapidly on inexpensive media at high cell densities. Being a eukaryote, P. pastoris is able to perform many of the post-translational modifications performed by higher eukaryotic cells and the obtained recombinant proteins undergo protein folding, proteolytic processing, disulfide bond formation and glycosylation [1].
As a methylotrophic yeast P. pastoris is capable of metabolizing methanol as its sole carbon source. The strong promoter for alcohol oxidase, AOX1, is tightly regulated and induced by methanol and it is used for the expression of the gene of interest. Accordingly, the expression of the foreign protein can be induced by adding methanol to the growth medium [2; 3].
Another important advantage is the secretion of the recombinant protein into the growth medium, using a signal sequence to target the foreign protein to the secretory pathway of P. pastoris. With only low levels of endogenous protein secreted to the media by the yeast itself and no added proteins to the media, a heterologous protein builds the majority of the total protein in the medium and facilitates following protein purification steps [3; 4].
The vector used here (pPICZαA) contains the AOX1 promoter for tightly regulated, methanol-induced expression of the gene of interest; the α-factor secretion signal for secretion of the recombinant protein, a Zeocin resistance gene for selection in both E. coli and Pichia and a C-terminal peptide containing the c-myc epitope and a polyhistidine (6xHis) tag for detection and purification of a recombinant protein. We also show western blot analysis of the recombinant protein using the specific Anti-myc-HRP antibody recognizing the c-myc epitope on the parent vector.
Protocol
Expression of recombinant proteins in the methylotrophic yeast Pichia pastoris
Before you start this protocol you should have your gene of interest cloned in frame in a P. pastoris parent vector and have it sequenced to check for the correct insertion of the gene of interest in the vector.
Step I: Generating electrocompetent yeast cells, linearization of the construct and transformation into P. pastoris
For this step you will need to have the following media and plates at hand:
YPDS plates
600 mL YPD medium
ice-cold sterile water
1 M sorbitol
Amicon Ultra-4 Centrifugal Filter Device
Microcon YM-30 Centrifugal Filter Unit
0.2 cm electroporation cuvette
15 mL sterile glass tube
sterile glass Pasteur Pipette
YPDS plates containing Zeocin
Part 1: Preparation of electrocompetent yeast cells
Four days before the intended transformation streak out P. pastoris cells on a YPDS plate without Zeocin and let them grow a 30°C for 1-2 days or until single colonies form.
Two days before intended transformation grow 5 ml of your P. pastoris strain in YPD medium in a 50 ml Falcon tube at 30°C overnight. Place the Falcon tube in a flask to fix it in the shaker.
The day before the transformation inoculate 500 mL of fresh YPD medium in a 2 liter flask with 0.25 mL of the overnight culture and let grow overnight again in a shaker at 30°C to an OD600 = 1.3-1.5. Note: dilute your sample before measuring on the spectrophotometer so that you get an accurate result.
The day of the transformation have ice-cold sterile water and 1 M sorbitol at hand. Centrifuge the cells at 1500 x g for 5 minutes at 4°C. Resuspend the pellet with 500 mL of ice-cold, sterile water.
Centrifuge the cells again as step 4. Resuspend the pellet with 250 mL of ice-cold, sterile water.
Centrifuge the cells again as in step 4. Resuspend the pellet in 20 mL of ice-cold 1 M sorbitol.
Centrifuge the cells again as in step 4. Resuspend the pellet in 1 mL of ice-cold 1 M sorbitol to a final volume of ~ 1.5 mL. Store cells on ice or at 4°C until further use. Note: we prepare more electrocompetent cells than needed and store them in aliquots of 80 μl in microcentrifuge tubes at -80 °C. We use the stored cells for no longer than one month.
Part 2: Linearization and concentration of the pPICZαA construct
For transformation linearize the vector containing your gene of interest by restriction digest. We use the enzyme PmeI. Other restriction sites are possible. You have to make sure that your insert does not contain the restriction site you want to use to linearize your vector. For transformation into P. pastoris you will need 5-20 μg of linearized DNA in 5-10 μl sterile water.
Also linearize, concentrate and transfer the plain vector (no insert) into P. pastoris. The parent vector without the insert is a control for background intracellular expression and allows you to interpret your expression results.
- Thaw all reagents on ice, combine the following reagents and briefly centrifuge the tube to get all liquid to the bottom, tap the tube and spin again:
- x μl sterile water
- 5.0 μl 10X NEBuffer 4
- 0.5 μl 100X BSA
- up to 2 μg vector DNA (~ 100 ng/μl)
- 2.0 μl PmeI enzyme mix
Incubate at 37°C for 3 hours and heat inactivate the enzyme mix at 65°C for 20 minutes.
In the mean time pre-rinse an Amicon Ultra-4 Centrifugal Filter Device with 1 mL of Milli-Q water, spin the tube at 4000 x g for 6-8 minutes and discard the flow through. Note: do not allow the membrane to dry out once wet. If you are not using the device after pre-rinsing, leave water on the membrane until the device is used.
Transfer the heat-inactivated solution from step 2 to the pre-rinsed Amicon Centrifugal Filter Unit, spin the tube at 4000 x g for 6-8 minutes and discard the flow through. Note: if you have prepared more than one linearization mix, combine all solutions into one Amicon Centrifugal Filter Unit.
Centrifuge until the amount of solution on the Amicon filter is reduced to approximately 100 ~ 150 μl. Add another 1.5 mL of sterile water to the empty tube from above and transfer the solution to the Amicon tube and repeat the centrifugation step. Wash a second time with 1.5 mL of sterile water and discard flow through again. Reduce the final volume to ~ 150 μl. The washing steps remove the remaining salt from the buffer to prevent arching when pulsing the cells. Note: we use a swinging bucket rotor for the centrifugation step; simply place the capped Amicon centrifugal filter unit in a 50 mL Falcon tube to hold it in place during centrifugation. Amicon filter devices retain 50 μl of solution on the filter even after extended centrifugation.
For further concentration of the DNA preparation transfer the remaining DNA solution from the Amicon filter device to a pre-rinsed Microcon YM-30 Centrifugal Filter Unit and rinse the Amicon tube with an additional 50 μl of sterile water to recover all DNA. Centrifuge the Microcon Centrifugal Filter Unit in a microcentrifuge at 10,000 x g until the filter is still slightly covered with liquid. Only spin for a short time and check every minute if only a small amount of liquid is left on the filter. To recover your DNA solution, place the filter upside down in a new microcentrifuge tube and spin in a second centrifugation step at 1000 x g for 3 minutes. The final volume should not exceed 10-15 μl in total, otherwise your DNA might be too diluted for the transformation step. Note: do not spin the filter device too long and let it not dry completely to prevent potential sample loss. If your membrane is dry, add 10-15 μl sterile water onto the membrane, agitate gently for 30 seconds and recover your DNA as described above.
Part 3: Transformation into P. pastoris by electroporation
The linearized and concentrated pPICZαA DNA containing your insert is now ready for transformation into the electrocompetent P. pastoris cells (see step I, part 2). About 15 minutes before the transformation prepare the following reagents and devices: fill a microcentrifuge tube with 1 mL of 1 M sorbitol and place it on ice; place a 0.2 cm electroporation cuvette on ice; label a sterile 15 mL glass tube and have a sterile glass Pasteur pipette at hand.
Transfer 80 μl of the cells from step I, part 2.7 to an ice-cold 0.2 cm electroporation cuvette. Add the concentrated linearized pPICZαA DNA solution from step I, part 3.6 and mix by moving the pipette tip from side-to-side in the electroporation cuvette.Note: when adding the DNA only push pipette to the first stop. After mixing the DNA with the cells push pipette to the second stop and slowly remove from the cuvette.
Incubate the cuvette with the cells on ice for 5 minutes.
- Wipe the outside of the cuvette with a tissue and pulse the cells according to the parameters for yeast (Saccharomyces cerevisiae) as suggested by the manufacturer of the specific electroporation device being used.Note: we use a Bio-Rad GenePulser with the following conditions:
- Charging voltage (V): 1500;
- Capacitance (μF): 25;
- Resistance (Ω): 200.
Immediately add 1 mL of ice-cold 1 M sorbitol to the cuvette. Transfer the cuvette content to a sterile 15 mL tube using the sterile glass Pasteur pipette.
Let the tube incubate at 30°C without shaking for 1.5 hours.
Add 5-7 sterilized glass beads on four labeled YPDS plates containing 100 μg/mL Zeocin. Spread 250 μl of the electroporation mix from the 15 mL glass tube on each plate. Shake the plate horizontally to evenly spread out the cells. Let the plate dry for 15 minutes and then remove the beads from the agar by inverting the plate.Note: we use 100 μg/mL Zeocin to select for transformants when using the GS115 Pichia strain. The selection conditions may vary if you are using another Pichia strain.
Incubate plates upside down for 2 to 3 days at 30°C until colonies form. Wrap the plates in a black plastic to prevent degradation of the light sensitive Zeocin. Less amount of antibiotic in the plates can result in false positive clones.
After colonies have formed, pick 12 colonies and purify them by streaking the clones on fresh YPDS plates containing 100 μg/mL of Zeocin.
Step II: Protein expression in Pichia pastoris
For this step you will need to have the following media, plates and reagents at hand:
BMGY medium
glycerol, sterilized
BMMY medium
methanol, sterilized
EPICENTRE Yeast DNA purification kit
You also need the following flasks:
250 mL flask, autoclaved
1 L baffled flask, autoclaved
200 mL beaker, autoclaved
Part 1: Protein expression in Pichiapastoris
All of the following steps are also performed for the transformed control vector (with no insert).
Pick a single colony from the purified Pichia colonies from step I, part 3.9 and inoculate in 25 mL BMGY in a sterile 250 mL flask. Grow at 30°C in a shaking incubator (250-300 rpm) until culture reaches an OD600 = 2-6 (approximately 16-18 hours). Note: dilute your sample before measuring on the spectrophotometer so that you get an accurate result. The cells will be in log-phase growth.
When the cells have reached the appropriate OD600 prepare a glycerol stock. Transfer 800 μl of the 25 mL cell culture to a 2 mL Corning cyrogenic vial and add 200 μl of sterilized glycerol. Freeze the glycerol stock and store at -80°C.
For yeast DNA purification, transfer 1.5 mL of the 25 mL cell culture to a microcentrifuge tube. Harvest the cells by centrifuging at 1300 x g for 1 minute at room temperature. These cells are used for analyzing Pichia integrants to double check if the gene of interest has integrated into the Pichia genome (see step II, part2). Store the cell pellet at 4°C until further analysis.
Transfer the rest of the 25 mL culture to a 50 mL Falcon tube and harvest the cells by centrifuging at 3000 x g for 5 minutes at room temperature. Decant supernatant and place the tubes upside down on a tissue to remove any residual media. To wash the cell pellet resuspend it in 20 mL BMMY to remove any remaining BMGY medium, as glycerol from the BMGY medium can inhibit the later expression. Centrifuge again at 3000 x g for 5 minutes at room temperature and decant the supernatant.
Resuspend cell pellet to an OD600 of 1.0 in BMMY medium to induce expression (approximately 100-200 mL) and transfer the culture in a 1 liter baffled flask. Cover the flask with a 200 mL beaker and return to incubator to continue growth at 30°C.Note: it is important that the temperature does not exceed 30°C. If the temperature of your incubator fluctuates, set the temperature at 28°C. Adequate aeration is also an important parameter for efficient expression during methanol induction (when inducing expression, never exceed a culture volume > 10-30% of your total flask volume). We highly recommend the use of baffled flasks, as they introduce more oxygen in the culture media.
Add sterilized pure methanol to a final concentration of 0.5% methanol every 24 hours to maintain induction.
At certain time points after the start of the expression transfer 1 mL of the culture to a 1.5 mL microcentrifuge tube. Centrifuge at 1300 x g for 2.5 minutes at room temperature. Transfer the supernatant to a separate 1.5 mL microcentrifuge tube. Store the supernatant and cell pellets at -80°C until further analysis. The samples from different time points will be analyzed to establish the optimal time period for protein expression after induction.Note: we choose 6h, 12h, 24h, 36h and 48h as the time points for taking the samples. Further growth up to 4 days is possible. The optimal time points vary between different expressed proteins.
Analyze the supernatants and cell pellets for protein expression by Coomassie-stained SDS-PAGE and western blot (not described in this protocol).
Part 2: Yeast DNA purification and PCR analysis of the Pichia integrant
All reagents necessary for the yeast DNA purification step of the Pichia culture in BMGY are included in the MasterPure Yeast DNA Purification Kit from EPICENTRE. Note: this analysis is additional and is performed to determine if the gene of interest has integrated into the Pichia genome. The cell pellet from a 1.5 mL sample taken from the culture in BMGY (see step II, part 1.3) is used.
Follow the instructions of the Epicentre MasterPure Yeast DNA Purification manual and run a PCR with the yeast DNA with specific primers for the insert. Analyze 3 μl of your PCR product on an agarose gel containing 1 μl ethidium bromide. Include a 1 Kb+ size marker in one well to interpret the size of your insert and visualize the DNA using a gel documentation station.
The recombinant protein can be analyzed by western blot analysis. Protein purification can be performed by His-tag purification on metal-charged resin (not described in this protocol).
Appendix, list of recipes
Step I:
YPD medium:To prepare 600 mL of YPD medium (Yeast Extract Peptone Dextrose Medium) dissolve 5 g yeast extract and 12 g peptone in 540 mL of water. Autoclave for 20 minutes on liquid cycle.In the mean time prepare 70 mL of 20% dextrose and filter-sterilize before use. Let the autoclaved solution cool to ~60°C and add 60 mL of filter-sterilized 20% dextrose.
YPDS (+ Zeocin) plates:To prepare 500 mL of YPDS + Zeocin agar (Yeast Extract Peptone Dextrose Medium with Sorbitol) dissolve 5 g yeast extract, 91.1 g sorbitol and 10 g peptone in 450 mL of water. Add 10 g of agar and a magnetic stir bar and autoclave for 20 minutes on liquid cycle. In the mean time prepare 60 mL of 20% dextrose and filter-sterilize before use. Let the autoclaved solution cool to ~ 60°C, add 50 mL of filter-sterilized 20% dextrose. Pour a few plates before adding the antibiotic for YPDS plates without Zeocin. Then add 500 μl Zeocin from a 100 mg/ml stock solution to obtain a final concentration of 100 μg/ml Zeocin in the agar. Let stir on a magnetic plate when adding the antibiotic and let stir for about an additional 2 minutes to equally mix.Pour media into Petri dishes and cover with black plastic. Let plates dry on bench overnight. Store YPD plates containing Zeocin at 4°C. The shelf life is one to two weeks. Note: the coverage with the plastic is necessary because Zeocin is light sensitive.
Step II:
- BMGY (Buffered Glycerol-complex Medium) and BMMY (Buffered Methanol-complex Medium):Dissolve for each medium 8 g of yeast extract and 16 g peptone in 560 mL water. Autoclave for 20 minutes on liquid cycle.In the mean time prepare the following solutions:
- 1 M potassium phosphate buffer, pH 6.0:
- 10X YNB (13.4% Yeast Nitrogen Base with Ammonium Sulfate without amino acids):
- 500X B (0.02% Biotin):
- 10X M (5% Methanol):
- 10X GY (10% Glycerol):
- 80 mL 1 M potassium phosphate buffer, pH 6.0;
- 80 mL 10X YNB
- 0.16 mL 500X B
- 80 mL 10X GY
Representative Results
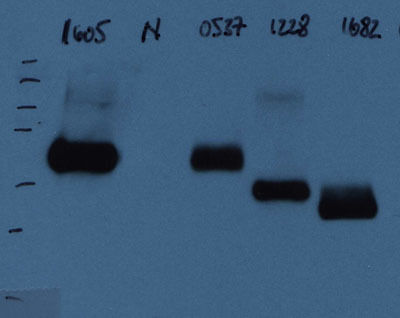 Figure 1. This western blot image shows four expressed proteins (labeled as 1605, 0537, 1228, and 1682) after 24 hours of expression. The antibody used for immunoblotting was a c-myc-HRP antibody. The second lane (labeled N) is a negative control and was loaded with supernatant from cells that do not express a recombinant protein because they were transformed with the parent (plain) vector.
Figure 1. This western blot image shows four expressed proteins (labeled as 1605, 0537, 1228, and 1682) after 24 hours of expression. The antibody used for immunoblotting was a c-myc-HRP antibody. The second lane (labeled N) is a negative control and was loaded with supernatant from cells that do not express a recombinant protein because they were transformed with the parent (plain) vector.
Discussion
Protein expression using the methylotrophic yeast Pichia pastoris as a host system is described in this protocol. The protein can be secreted into the medium depending on the used cloning vector. The secretion of the recombinant protein makes the subsequent purification easier. However, the shown conditions may have to be optimized for the expression of different proteins and changes in the conditions and expression times may result in increased protein levels. The vector used in this protocol has the feature of a c-myc epitope and an antibody for this epitope can be purchased. This allows the analysis of proteins for which there are no antibodies yet available. The His-tag feature of the parent vector facilitates the protein purification on metal-chelating resin and there is also an antibody for the His-tag section of the vector available.
As described in this protocol, protein expression in Pichia pastoris is a multistep-process which needs to be well planned and prepared. A time of two to three weeks is required to perform all steps but excluding the cloning of the construct with the gene of interest. The basis for successful transformation into P. pastoris are high transformation efficient competent yeast cells and a linearized construct which can integrate into the yeast genome. Western blot analysis and His-tag purification are applications to determine the level of recombinant protein expression and are the next steps to perform after this protocol.
Acknowledgments
We would like to thank the Canadian Foundation for Innovation, the British Columbia Knowledge Development Fund, and the Canadian Institutes of Health Research (CIHR) for supporting this work. M.T was supported by a fellowship from the TULA foundation funded Centre for Microbial Diversity and Evolution (CMDE).
References
- Cereghino GP, Cregg JM. Applications of yeast in biotechnology: protein production and genetic analysis. Curr Opin Biotechnol. 1999;10:422–427. doi: 10.1016/s0958-1669(99)00004-x. [DOI] [PubMed] [Google Scholar]
- Cregg JM, Barringer KJ, Hessler AY. Pichia pastoris as a Host System for Transformations. Mol. Cell. Biol. 1985;5:3376–3385. doi: 10.1128/mcb.5.12.3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cregg JM, Cereghino JL, Shi J, Higgins DR. Recombinant protein expression in Pichia pastoris. Mol Biotechnol. 2000;16:23–52. doi: 10.1385/MB:16:1:23. [DOI] [PubMed] [Google Scholar]
- Cereghino GP, Cereghino JL, Ilgen C, Cregg JM. Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr Opin Biotechnol. 2002;4:329–332. doi: 10.1016/s0958-1669(02)00330-0. [DOI] [PubMed] [Google Scholar]


