Abstract
Analysis of Chlamydomonas axonemes revealed that the protein phosphatase, PP2A, is localized to the outer doublet microtubules and is implicated in regulation of dynein-driven motility. We tested the hypothesis that PP2A is localized to the axoneme by a specialized, highly conserved 55-kDa B-type subunit identified in the Chlamydomonas flagellar proteome. The B-subunit gene is defective in the motility mutant pf4. Consistent with our hypothesis, both the B- and C-subunits of PP2A fail to assemble in pf4 axonemes, while the dyneins and other axonemal structures are fully assembled in pf4 axonemes. Two pf4 intragenic revertants were recovered that restore PP2A to the axonemes and re-establish nearly wild-type motility. The revertants confirmed that the slow-swimming Pf4 phenotype is a result of the defective PP2A B-subunit. These results demonstrate that the axonemal B-subunit is, in part, an anchor protein required for PP2A localization and that PP2A is required for normal ciliary motility.
Keywords: Cilia, flagella, dynein, axonemes, protein phosphatases, microtubules
Introduction
Cilia, also called flagella, are responsible for a surprisingly wide range of essential motile and sensory / signaling functions (Berbari et al. 2009; Gerdes et al. 2009; Goetz and Anderson 2010; Satir and Christensen 2007; Smith and Rohatgi 2011). Motile cilia play a central role in the embryonic node where they contribute to normal pattern formation and in the adult where they are required for normal mucociliary transport in the airway, movement of cerebral spinal fluid in the brain ventricles, fluid movement in the oviducts and sperm motility (Brokaw 2009; Lindemann and Lesich 2010; Satir and Christensen 2008; Wallingford 2010). Defects in motile cilia can result in the ciliopathy called primary cilia dyskinesia (PCD), randomization of pattern formation, chronic respiratory infection and infertility (Basu and Brueckner 2008; Hashimoto and Hamada 2010; Hirokawa et al. 2006; Leigh et al. 2009). However, we are only beginning to understand the mechanisms that regulate ciliary motility.
Diverse studies in a number of experimental systems have revealed that ciliary / flagellar motility is regulated by phosphorylation (Elam et al. 2009; Salathe 2007). One of the best studied examples revealed that the inner dynein arm, I1 (or dynein f), is required for control of flagellar waveform (Bayly et al. 2010; Brokaw and Kamiya 1987) and phototaxis (King and Dutcher 1997; Okita et al. 2005). Analysis of isolated Chlamydomonas axonemes demonstrated that dynein-driven microtubule sliding is regulated by phosphorylation of the I1 intermediate chain, IC138 (Porter and Sale, 2000; Wirschell et al. 2007). One of the surprises was that protein kinases and phosphatases are localized to the axoneme and include conserved, ubiquitous kinases (PKA, CK1) and phosphatases (PP1, PP2A) (Gokhale et al. 2009; Habermacher and Sale 1995; Habermacher and Sale 1996; Howard et al. 1994; Yang et al. 2000; Yang and Sale 2000). However, with the exception of the A-kinase anchoring proteins, AKAPs (Gaillard et al. 2001; Gaillard et al. 2006), it is not known how CK1, PP1 or PP2A are anchored in the axoneme.
Biochemical analysis of Chlamydomonas axonemes revealed that PP2A is localized to the outer doublet microtubules (Yang et al. 2000). PP2A is a versatile and ubiquitous serine / threonine phosphatase that plays a role in many vital cellular processes including cell growth, cell cycle, motility and apoptosis (Basu 2010; Eichhorn et al. 2009; Mumby 2007; Shi 2009b; Sontag and Sontag 2006; Virshup and Shenolikar 2009). The holoenzyme is composed of three subunits; the A- scaffolding or structural, C-catalytic and the B-regulatory subunits (Basu 2010). The A- and C-subunits form an obligate heterodimer, which then interacts with a B-type subunit (Janssens et al. 2008; Shi 2009a; Virshup and Shenolikar 2009). In mammals, four different families of B-subunits have been defined (PR55/B, PR61/B’, PR72/B”, and PR93/PR110/B’”). Furthermore, in each family, at least three different genes encode several different B-type subunit isoforms (Eichhorn et al. 2009). The common feature of each family member is the ability to bind to the PP2A A- and C- dimer (Xu et al. 2008): otherwise, there is very little sequence or structural homology among the four families (Eichhorn et al. 2009). However, within each family, the sequences and structures are very similar. One prevalent idea is that B-subunits function to provide specificity by targeting and anchoring PP2A to precise positions, organelles and substrates in the cells (Eichhorn et al. 2009; Lechward et al. 2001). We previously identified A- and C-subunits in Chlamydomonas axonemes (Yang et al. 2000). Thus, we postulated the axoneme contains a specialized B-type subunit for localization of PP2A.
Taking advantage of the Chlamydomonas flagellar proteome (Pazour et al. 2005), we identified a highly conserved axonemal PP2A B-subunit in the B/PR55, WD-repeat family (Supplemental Fig. 1) (Eichhorn et al. 2009). Using an antibody to a polypeptide in the N-terminus, we confirmed the B-subunit is localized to the axoneme. The B-subunit gene maps to the PF4 locus on chromosome I (McVittie 1972) and sequencing confirmed the pf4-1 strain has a mutation in the B-subunit gene. Immunoblots revealed the axonemal B-subunit is missing from pf4 axonemes. Consistent with our hypothesis, the axonemal PP2A C-subunit also fails to assemble in pf4 axonemes. The pf4 cells swim slowly and fail to phototax, a phenotype similar to mutants that lack the inner dynein arm I1 (Brokaw and Kamiya 1987; King and Dutcher 1997; Okita et al. 2005). However, pf4 axonemes show no defect in assembly of the dynein arms or other axonemal structures. Through genetic analysis of pf4, two intragenic revertants were recovered that restore assembly of PP2A in the axoneme, rescue phototaxis and near wild-type swimming, thus confirming the pf4 mutant phenotype is a consequence of the B-subunit mutation. This data demonstrates that the B-subunit is necessary for localization of PP2A in the axoneme and that PP2A is required for normal flagellar motility.
Results and Discussion
A PP2A B-subunit is localized to the axoneme
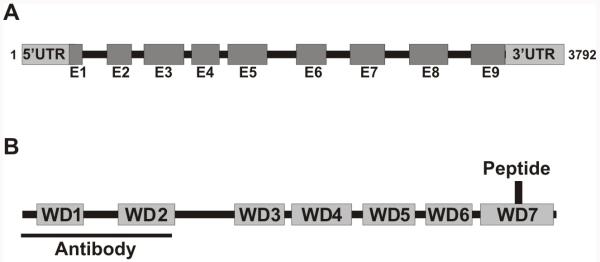
To identify a potential B-type subunit in Chlamydomonas flagella, we analyzed the flagellar proteome database and identified a conserved member of the B/PR55 family of B-subunits (Pazour et al. 2005). The proteome data indicates the B-subunit is an axonemal protein that is solubilized with high salt. The B-subunit gene is 3792 base pairs with nine exons and eight introns (Fig. 1A) and encodes a highly conserved 52.6-kDa protein with seven WD-repeats that form a seven bladed propeller-like structure (Fig. 1B; Sup. Fig. 1, see Xu et al. 2008). While there are other B-type subunits found in the Chlamydomonas genome (Table I), only this B-subunit isoform was identified in the flagellar proteome. Predictably, since PP2A is a ubiquitous and functionally versatile phosphatase (Shi 2009b), the other, non-flagellar B-type subunit proteins are responsible for targeting PP2A to other cellular locations in Chlamydomonas.
Figure 1. TheChlamydomonas PP2A B-subunit.
(A) The B-subunit gene is 3792 base pairs and contains nine exons – E1-E9 - and eight introns. (B) The predicted Chlamydomonas B-subunit protein is 479 amino acids with a mass of 52.6 kDa. The protein contains seven WD-repeats throughout its protein sequence, forming a seven bladed propeller-like structure. Amino acids 1-134 were used to design an antibody to the B-subunit protein. The peptide (amino acids 439-452) identified in the Chlamydomonas Flagellar Proteome resides in the seventh WD repeat domain located in the C-terminus.
Table I. Potential PP2A subunits in the Chlamydomonas Genome Database.
(JGI version 4) reveals several PP2A subunits including 2 A-subunits, 4 C-subunits and 5 B-type subunits. Both A-subunits and one of the C-subunits were found in the Chlamydomonas flagellar proteome (Pazour et al., 2005). Of the five B-type subunits in the genome, only one, PP2A1, was identified in the flagellar proteome. This gene is defective in the pf4 mutant.
| Type | Name1 | Protein ID2 |
Domain/Motif/Family3 | Flagellar Proteome4 |
|---|---|---|---|---|
| A (Scaffold) | PP2A-2r FAP14 |
131550 194683 |
HEAT HEAT |
YES YES |
| B, B’, B”, B’” (Regulatory) |
PP2A-1r (B”) PP2A2 (B’) TAP42-like protein PP2A1/PF4(B) Not annotated (B”) |
112343 127638 148948 185509 405966 |
--- B56 TAP42 WD repeats --- |
NO NO NO YES NO |
| C (Catalytic) | PP2A3 PP2A-c4 PPA1 PP2A-1c |
807 132486 144867 193562 |
Metallophosphatase/PP2Ac Metallophosphatase/PP2Ac Metallophosphatase/PP2Ac Metallophosphatase/PP2Ac |
NO YES NO NO |
Name and/or description in JGI Chlamydomonas reinhardtii v4.
Protein ID in JGI Chlamydomonas reinhardtii v4.
Domain, motif, and family found in SMART and/or pfam analyses.
Presence or absence in flagellar proteome (Pazour et al., 2005).
To confirm that the identified B-subunit localizes to the flagellar axoneme, we generated a B-subunit specific antibody that recognizes a ~50-kDa band on immunoblots of isolated flagella and axonemes, consistent with the predicted size of the B-subunit protein (Fig. 2A; Supplemental Fig. 2). The B-subunit is partially solubilized with detergent (M-M fraction), while the fraction of the B-subunit in the axoneme is solubilized in high salt buffers, confirming the fractionation profile reported in the flagellar proteome and consistent with the solubility of the axonemal A- and C-subunits (Pazour et al. 2005; Yang et al. 2000).
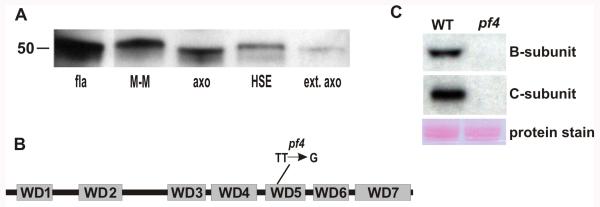
Figure 2. The B-subunit gene is mutated in pf4 cells and the PP2A holoenzyme fails to target to the axoneme.
(A) Immunoblots performed with the B-subunit antibody detect a band in isolated wild-type flagella (fla) with a relative mobility of ~50 kDa, consistent with the predicted size of the B-subunit protein. The B-subunit is partially soluble in detergent (membrane-matrix fraction; M-M); while a fraction of the B-subunit remains associated with the axoneme (axo). The axonemal fraction of the B-subunit is solubilized with high salt buffers (HSE), leaving very little left on the extracted axonemes (ext. axo). Variations in B-subunit migration is a consequence of protein abundance, detergent or salt concentrations in the samples. (B) Schematic of the B-subunit protein shows the location of the pf4 mutation (wild-type sequence is TT; pf4 sequence is G) in the fifth WD repeat. (C) Immunoblots of wild-type and pf4 axonemes demonstrate that the PP2A B-subunit is not present pf4 axonemes. Furthermore, the C-subunit also fails to target to the axoneme in the absence of the B-subunit. A protein stain showing tubulin from these samples is shown below.
The flagellar mutant, pf4, is defective in the B-subunit gene
The B-subunit gene was mapped to Chromosome 1 using BAC clone, 14G12, identified in the Chlamydomonas genome database (http://genome.jgi-psf.org/Chlre4/Chlre4.home.html). The gene maps near the pf4 mutant (Levine and Goodenough 1970; McVittie 1972), a slow-swimming mutant that otherwise had not been characterized. Sequence analysis revealed that pf4 contains a base pair substitution/deletion at positions 2239 and 2240 in exon seven of the B-subunit gene (Fig. 2B; Supplemental fig. 2A, B). This mutation predicts a F306A change in the fifth WD repeat and a frame shift resulting in the coding of a premature stop codon; the mutation predicts a 37-kDa truncated protein product.
To verify the phenotype associated with defects in the B-subunit gene, we backcrossed pf4 to wild-type cells three times and selected one progeny, pf4 9-1, as the representative pf4 mutant strain for all experiments. The pf4 9-1 mutant displayed full-length flagella and reduced swimming speed (see below). From here on, we will refer to pf4 9-1 as pf4.
The B-subunit is required for targeting of PP2A to the axoneme
We predicted that the B-subunit would be missing in pf4 cells and, as a consequence, the entire PP2A complex would fail to assemble in the pf4 axoneme. Immunoblots failed to detect the B-subunit in pf4 axonemes (Fig. 2C) or flagella (Supplemental Fig. 2D). Analysis of additional cell and flagellar fractions from the pf4 mutant demonstrate that pf4 does not express any full length or truncated B-subunit protein. Thus, the pf4 mutant appears to be a null allele for the B-subunit. Notably, the C-subunit also fails to assemble in pf4 axonemes (Fig. 2C) and flagella (Supplemental Fig. 2D). Given that the A- and C-subunits are an obligate heterodimer and consistent with our model, these data indicate that assembly of the B-subunit is necessary for localization of the PP2A C-, and presumably A-, subunits to the outer doublet microtubules. Since in pf4 the PP2A C-subunit fails to assemble in flagella and axonemes, the axonemal B-subunit may be the predominant or sole PP2A B-type subunit in the flagellar compartment.
The pf4 mutant has defective motility and fails to phototax
The pf4 swimming phenotype is similar to the phenotype of mutant cells lacking I1 dynein (Fig. 3A); pf4 swimming velocities are reduced to velocities exhibited by ida3 mutants (shown here) and other I1-dynein mutants including ida7 (Perrone et al. 1998). Similar to inner dynein arm mutants (Kamiya et al., 1991), flagellar beat frequency is only slightly reduced in pf4 compared to wild-type cells. Thus, like inner dynein arm mutants (Brokaw and Kamiya, 1987; Bayly et al., 2010), reduced swimming speed in pf4 is primarily a consequence of altered flagellar waveform. We performed immunoblot analysis and thin section electron microscopy to determine whether the dynein arms are assembled in pf4 axonemes. Immunoblots determined the dynein arms and radial spokes are fully assembled in pf4 axonemes (Fig. 3B). Transmission electron microscopy also confirmed that there are no obvious structural defects in the dynein arms, radial spokes or central pair apparatus (Fig. 3C). Thus, the motility phenotype in pf4 appears to be a consequence of a failure in PP2A assembly that results in a failure in regulation of dynein motor function.
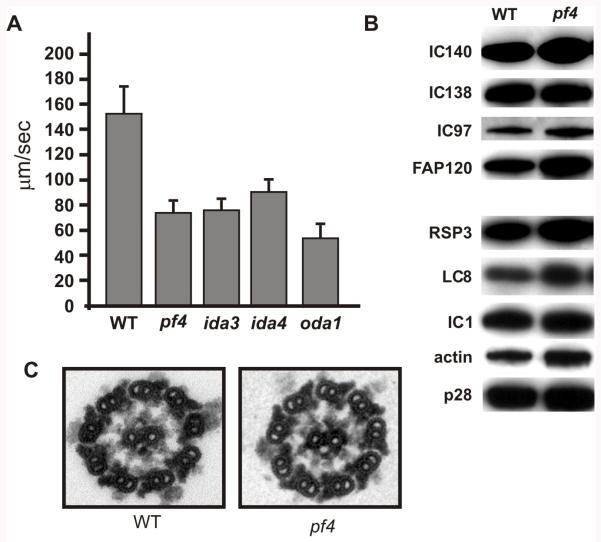
Figure 3. The pf4 axonemes are structurally similar to wild-type.
(A) Swimming velocities of wild-type and mutant cells were analyzed. The pf4 mutant has a reduced swimming velocity that is comparable to mutants lacking I1 dynein (ida3). Also analyzed are ida4, a mutant lacking inner arms a, c, and d, and oda1 lacking the outer dynein arm. (B) Immnoblots of wild-type and pf4 axonemes demonstrate that all of the axonemal dynein motors are assembled in pf4 including the I1 dynein (IC140, IC138, IC97, FAP120), dyneins a, b, c, d, e, and g (actin and p28) and the outer dynein arm (IC1). In addition, there are no defects in assembly of the radial spokes (RSP3). LC8 levels are also unaltered (LC8 is a component of I1, the outer arm and the radial spoke). (C) Transmission electron microscopy of wild-type and pf4 axonemes demonstrates that pf4 assembles all the major axonemal structures including the central pair, the radial spokes, the dynein arms and the outer doublet microtubules.
Diverse genetic and pharmacological evidence has revealed an axonemal phospho-regulatory mechanism that controls dynein activity (Porter and Sale 2000; Wirschell et al. 2007). As indicated above, like the pf4 mutant, mutations that lead to a failure in assembly of I1 dynein results in a slow-swimming phenotype (Fig. 3A; Bower et al. 2009; Hendrickson et al. 2004; Myster et al. 1997; Myster et al. 1999; Perrone et al. 2000; Perrone et al. 1998; Porter et al. 1992; Toba et al. 2011) and defective phototaxis (Fig. 4A; King and Dutcher 1997; Okita et al. 2005). To test whether PP2A may be part of dynein regulatory pathway, we examined phototaxis in pf4 cells. A photo-accumulation assay revealed that pf4 cells do not respond to light; failing to accumulate on one side of a dish, demonstrating that pf4 mutant cells are defective in phototaxis (Fig. 4A). Thus, assembly of PP2A is required for normal motility and phototaxis. One model is that PP2A is part of the phospho-regulatory mechanism that regulates I1 dynein by control of the regulatory phosphoprotein IC138 (discussed below and see Bower et al. 2009; Habermacher and Sale 1997; Hendrickson et al. 2004; King and Dutcher 1997).
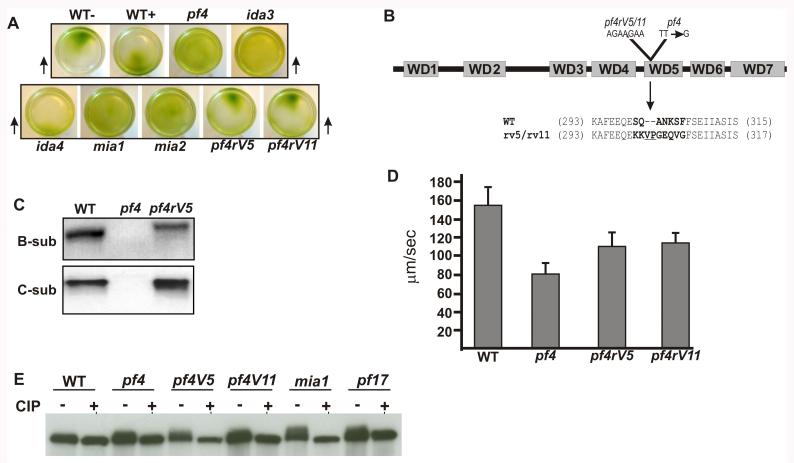
Figure 4. The pf4rV5 and pf4rV11 revertants assemble the PP2A B- and C-subunits in the axoneme and partially rescue the Pf4 phenotype.
(A) Photo-accumulation assays reveal that pf4 is defective in phototaxis. Wild-type cells swim towards (WT+; AGG1) or away (WT−, agg1) from a light source (black arrows) and accumulate on one side of a culture dish, whereas pf4 cells do not. This defect is also present in ida3, mia1 and mia2 mutants—defective in I1 dynein assembly or regulation, but is not a consequence of slow swimming, as another inner arm mutant and an outer arm mutant (ida4 and oda1 respectively) photo-accumulate with wild-type kinetics. These assays revealed that phototaxis is rescued in the pf4 revertants; the revertant cells swim away from the light source and accumulate on one side of the culture dish, similar to wild-type cells (B) Schematic of the B-subunit protein shows the location of the original pf4 mutation and the upstream pf4 revertant mutation—a 7-bp insertion (AGAAGAA) at positions 1802-1808 in exon 6 in the fifth WD repeat domain. (C) Immunoblots of isolated axonemes probed with the B- and C-subunit antibodies demonstrate that PP2A assembly is restored in the revertants (pf4rV5) as compared to wild-type and the pf4 mutant. (D) Analysis of swimming speed demonstrates that the pf4rV5 and pf4rV11 revertant cells swim with a velocity intermediate between wild-type and pf4 cells. The velocity of swimming in the revertants pf4V5 and pf4V11 was significantly faster than the swimming velocity of pf4 (P < 0.001 by t-test). (E) Immunoblots of axonemes treated with buffer alone or CIP were probed with the IC138 antibody as described (Hendrickson et al. 2004; Wirschell et al. 2009). Wild-type axonemes display relatively little IC138 phosphorylation (compare axonemes treated with CIP to untreated axonemes) while axonemes from mutants defective in regulation of I1 dynein (mia1, pf17) show excessive IC138 phosphorylation due to a global inactivation of I1-dynein activity. Axonemes from pf4 and the pf4 revertants also show increased IC138 phosphorylation, consistent with a role for PP2A in regulation of I1 dynein.
Intragenic pf4 revertants restore PP2A assembly and rescue phototaxis and near wild-motility
To verify that the defect in the B-subunit gene is responsible for the Pf4 mutant phenotype and to potentially define an axonemal regulatory pathway that includes PP2A, we performed a suppressor screen and recovered two independent intragenic pf4 revertant strains, pf4rV5 and pf4rV11, that harbor an identical second site mutation in the PF4 gene (Fig. 4B, Supplemental Fig. 2A). Mapping analysis revealed that pf4 and the revertants are intragenic based on no recovery of the Pf4 phenotype in 350 tetrads. Sequence analysis revealed a 7-bp insertion (AGAAGAA) upstream of the original pf4 mutation which restores the reading frame. The new sequence in the pf4rV5 and pf4rV11 revertants predicts changes in amino acids 300-308 with the insertion of a valine and a proline, and the alteration of an alanine to a glycine; these amino acids may introduce kinks in the B-sheet structure (Fig. 4B; Supplemental Fig. 2). Thus, the predicted structure of the revertant B-subunit protein should differ from the wild-type PP2A B-subunit.
To determine whether pf4 revertants axonemes assemble PP2A, isolated axonemes were analyzed by immunoblots using the PP2A B- and C-subunit antibodies. The revertant PP2A B-subunit and C-subunit were fully assembled at wild-type levels in pf4 revertant axonemes (Fig. 4C), thus providing additional evidence that the B-subunit is required for localization of the PP2A holoenzyme to the axonemal structure. The revertant B-subunit migrates slightly slower on SDS-PAGE gels compared to the wild-type B-subunit (Fig. 4C). This observation is consistent with the prediction that the revertant protein differs slightly from the wild-type protein, however retains the ability to localize the PP2A holoenzyme in the axoneme.
Since the revertant axonemes assemble PP2A, we examined phototaxis and motility in the pf4 revertant cells. The revertant cells have restored phototaxis (Fig. 4A) and increased swimming velocities relative to the pf4 mutation alone (Fig. 4D). However, the swimming velocities were not fully restored to wild-type levels (Fig. 4D); which indicates that the modified B-subunit expressed in pf4rV5 and pf4rV11 is partially defective. The revertant B-subunit contains a VP insertion, an A302G substitution and several additional amino acid substitutions in the fifth WD repeat which may disrupt the β-sheet structure (Fig. 4B). Perhaps PP2A is accessible only to a subset of its axonemal substrates or the function of PP2A is partially compromised in the revertants. Consistent with this idea, and as an example, a B-type subunit has been shown to interact with Tau to contribute to interaction with the catalytic subunit (Xu et al. 2008). Thus, the inability to fully restore wild-type swimming is most likely a direct result of the modified B-subunit protein. These data also suggest that the phototaxis pathway may be separate from the pathway controlling swimming velocity. Consistent with this interpretation, recent analysis of Chlamydomonas null mutations in the IC138 gene (bop5) revealed that IC138 is required from normal flagellar bending, but not required for phototaxis (See Discussion below and our unpublished data).
The pf4 double mutants display more severe phenotypes
To better understand the role of PP2A in regulation of motility, we generated double mutants between pf4 and dynein arm mutations. As discussed, diverse evidence suggest PP2A may function in a conserved signaling pathway that involves the central pair, radial spokes and at least one ciliary dynein motor, the inner arm I1 dynein (Porter and Sale 2000; Smith and Yang 2004; Wirschell et al. 2007). The Pf4 motility phenotype is consistent with defects in regulation of the inner dynein arms; pf4 and inner dynein arm mutations exhibit slow-smooth swimming phenotypes. If the motility defect in pf4 is due to a mis-regulation of I1 dynein, then we predict the motility phenotype of a pf4, I1 double mutant will be similar to or no worse than the parental motility phenotypes. Furthermore, we predict that double mutants between pf4 and other dynein motors, which are unaffected by PP2A, will display a more severe phenotype.
Table II lists the pf4 double mutants generated in this study and their motility phenotypes. When the pf4 defect is combined with defects in inner arm dyneins a, c, and d (ida4), inner arm dynein e (ida6), the outer dynein arm (oda6) or with defects in regulation of I1 dynein (mia1, mia2), the resulting motility phenotypes were significantly worse than the parental phenotypes. These results indicate that PP2A does not play a unique role in regulation of the single headed inner dynein arms or the outer arms and that axonemal PP2A does not function in regulatory mechanisms unique to the pathways defined by the mia1 and mia2 mutants. In spite of several attempts, we could not isolate double mutants between pf4 and any I1-dynein mutant (ida1, ida7, bop5). The significance of this result is unclear, but is possibly technical or that these combinations are lethal for unknown reasons. It is possible we may be revealing additional, non-ciliary functions for the PP2A B-subunit and I1 dynein.
Table II. Phenotype of pf4 double mutants.
The pf4 mutant phenocopies I1-dynein mutants that are defective in assembly or regulation of I1 dynein. Double mutants generated between pf4 and other dynein deficiencies show more severe phenotypes consistent with the observation that PP2A is involved in a phospho-regulatory pathway that controls I1 dynein.
| Mutant | Defect | Phenotype |
|---|---|---|
| pf4 | PP2A | Slow swimming |
| pf4 x oda6 | PP2A, Outer arm dyneins | Very slow swimming or paralyzed |
| pf4 x ida4 | PP2A, Dynein a, c, and d | Very slow swimming |
| pf4 x ida6 | PP2A, Dynein e, DRC | Sporadic twitching, nearly paralyzed, short flagella |
| pf4 x ida9 | PP2A, Dynein c | Very slow swimming |
| pf4 x mia1 | PP2A, Mia1p | Very slow swimming |
| pf4 x mia2 | PP2A, Mia2p | Sporadic twitching, nearly paralyzed |
Based on pharmacological evidence for a role for PP2A in regulation of I1 dynein and our current results, PP2A may operate, at least in part, in a pathway that includes I1 dynein and possibly IC138 (Bower et al. 2009; Wirschell et al. 2007). Consistent with this hypothesis, we determined that IC138 is highly phosphorylated in pf4 axonemes compared to wild-type axonemes; a phenotype similar to that observed for mutants that are defective in the phospho-regulatory pathway that controls I1 dynein (Fig. 4E). IC138 also appears to be highly phosphorylated in pf4 revertant axonemes (Fig. 4E). As discussed above, this observation may also indicate that the revertant PP2A B-subunit differs in structure from wild-type, and as a consequence, IC138 is abnormally phosphorylated in the revertants resulting in only partial restoration of swimming speed. To date, the key phospho-residues in I1 dynein are not known and, thus, sites of phosphorylation in IC138 may differ in pf4 axonemes compared to the pf4 revertants. Further tests of this model will require identification of specific IC138 phospho-residues and detailed analysis of point mutations in these residues. Nevertheless, these results are consistent with a role for PP2A in regulation of I1 dynein.
What is the role of PP2A in signaling and regulation of motility?
Targeting of PP2A to specific sub-cellular domains is an important model for control of substrate specificity. A general role for the B-subunits may be for targeting PP2A to specific sites within the cell for interaction with specific substrates also localized in the same position (Matre et al. 2009; Price et al. 1999; Schmidt et al. 2002; Strack et al. 1998). Based on our current results, we demonstrate that a B/PR55 type B-subunit functions to target PP2A to the flagellar compartment and axoneme where it plays a role in regulating motility. Members of the B/PR55 family are enriched in the ciliated tissues such as the brain and testis (Hatano et al. 1993; Mayer et al. 1991) and play a role in targeting PP2A to microtubules (Nunbhakdi-Craig et al. 2007; Sontag et al. 1999; Tar et al. 2004). An anchoring factor binding PP2A to the microtubules has also been reported (Price et al. 1999). It will now be important to localize PP2A on the outer doublet microtubules and define the axonemal B-subunit interacting proteins that localize PP2A in the axoneme.
Because pharmacological evidence implicates PP2A in control of dynein-driven microtubule sliding (Wirschell et al. 2007) and pf4 mutants strongly phenocopy I1-dynein mutants, our results are consistent with a role of PP2A in regulation of I1 dynein and control of flagellar bending. However, we cannot exclude the possibility that PP2A also regulates additional dyneins. Important questions include identifying specific PP2A substrates in the axoneme and determining if PP2A is directly regulating I1 dynein and IC138.
Materials and methods
Strains, genetic analyses and culture conditions
Chlamydomonas reinhardtii strains include CC124 and CC125 (wild-type), pf4 mt− (CC-880), pf4 mt+ (CC-1027) from the Chlamydomonas Genetics Center (Univ. of MN, St. Paul, MN), pf4 9-1 (recovered from a parental tetrad between wild type and pf4 crosses), pf4rV5 and pf4rV11 (recovered from non-parental tetrad, see below). All experiments utilized the backcrossed progeny pf4 9-1 and referred to as pf4 in this manuscript.
Backcrosses and tetrad dissection were performed as described (Dutcher 1995). Cells harboring the pf4 mutation are palmelloid, which is a phenotype often displayed in mutants with short flagella or a slow-swimming phenotype. Suppressors of pf4 were generated by treating pf4 cells with a UV light dose of 700 μJ/cm2 and screening for individual colonies that hatched. An identical second site mutation in the B-subunit gene was identified in two clones, pf4V5 and pf4V11. In addition to the two intragenic revertants described here, several extragenic suppressor mutations were isolated and will be characterized elsewhere.
Hatching of the palmelloid cells is promoted by resuspending pf4 cells in water or gametic autolysin. Cells were grown in L-medium with aeration on a 14:10 hr light: dark cycle.
Molecular Biology
A restriction fragment containing the first 134 amino acids of the B-subunit was cloned into the pET28A vector (EMD) using BamHI and XhoI to create pCEBsub134 and protein expression induced with 1mM IPTG. The expressed fusion protein was purified using a metal affinity resin (Clontech, Mountain View, CA) and used as an antigen for production of a B-subunit antibody (Spring Valley Labs, Inc, Woodline, MD). B-subunit antibodies were blot purified using the fusion protein. Specific antibodies were eluted using 100 mM citric acid pH 3.0 followed by neutralization with 100 mM Tris pH 9.0.
The pf4 and pf4 revertant mutations (pf4v5 and pf4v11) were identified by sequencing the entire PF4 gene. All primers were designed using Primer3 program (http://frodo.wi.mit.edu/primer3/) and sequencing was performed by Iowa State DNA sequencing facility (http://www.dna.iastate.edu/).
Flagella Isolation
Flagella were isolated from three liters of cells as described (Witman 1986) and resuspended in Na-low buffer (30mM HEPES, pH 7.4; 5mM MgSO4·7H20; 1mM DTT; 0.5mM EDTA; 30Mm NaCl; 1mM PMSF; 0.6 trypsin inhibitory unit [TIU] Aprotinin). NP-40 was added to a final concentration of 1% and axonemes and detergent soluble membrane-matrix fractions separated by centrifugation. To extract the PP2A B-subunit, proteins were extracted from axonemes with 0.6 M NaCl for 30 min and the salt-soluble protein and extracted axonemes were separated by centrifugation. Samples from each step were saved for SDS-PAGE analysis.
SDS-PAGE and immunoblot analysis
Flagellar samples were run on 5%, 7.5% or 10% SDS-PAGE gels and transferred to a nitrocellulose membrane (BioRad, Hercules, CA) for analysis. Equivalent sample loads of each flagellar fraction were loaded. Immunoblots were probed with the following antibodies: affinity purified B-subunit (1:50 – this study), IC140 (1:10,000 – Yang and Sale, 1997), IC138 (1:10,000 – Hendrickson et al., 2004), IC97 (1:10,000 – Wirschell et al., 2009), FAP120 (1:1000 – Ikeda et al., 2009), RSP3 (1:1000 – Williams et al., 1989), LC8 (1:50 – King et al., 1996), IC1 (1:1000 – King et al.,1986), actin (1:100 – Sugase et al., 1996) and p28 (1:250 – LeDizet and Piperno, 1995). For analysis of IC138 phosphorylation, axonemes were treated with calf intestinal phosphatase (CIP) as described (Hendrickson et al. 2004) or buffer alone and resolved on 5% gels and analyzed by immunoblots using the IC138 antibody.
Motility analyses
Swimming velocity measurements were determined as described (Elam et al. 2009). Briefly, cells were placed on a microscope perfusion chamber and visualized using a 10X Plan-Apochromat lens (Carl Zeiss, Inc.) and a darkfield condenser using an Axiovert 35 inverted light microscope (Carl Zeiss, Inc.). Cells were recorded using a silicon intensified camera (VE-1000; Dage-MTI) and converted to a digital format using Labview 7.1 software (National Instruments). Swimming velocity was calculated using manual tracings of twenty cells for each strain from three independent experiments.
Photo-accumulation assays were performed as follows: cells (2.5 × 106 cells/ml in 35 mm petri dishes) were dark adapted for 10 min, then exposed to either ambient or fluorescent light located 30 cm away (2 W/m2) with the same photo-accumulation results. Dishes were photographed after 5 min to determine relative photo-accumulation ability.
Electron Microscopy
Electron microscopy samples were prepared as described (Mitchell and Sale 1999). Embedded axonemes were thin sectioned, stained with lead citrate and uranyl acetate and viewed using a Hitachi H7500 transmission electron microscope. Images were acquired usiing a Gatan 792 BioScan CCD camera with Gatan Digital Micrograph software (Gatan, Pleasanton, CA) and adjustments were made using Adobe Photoshop (Adobe Systems, Mountain View, CA).
Supplementary Material
Supplemental Figure 1: Alignment of the B-subunit was performed using ClustalW and the alignments were color-coded using the percentage composition coloring tool, Colorfy (http://bifrost.wustl.edu/colorfy) (Lin et al. 2010).
Supplemental Figure 2: A. Chromatograms showing the DNA sequence in pf4 and the pf4 revertants cells. B. The pf4 mutation generates a novel CviKi-1 restriction site in the pf4 mutant strain. PCR products covering the site of the pf4 mutation in exon 7 were digested with CviKi-1. The resulting restriction pattern is diagnostic for the pf4 mutation: wild-type PF4 sequence produces 110-, 38-, 23- and 4-base pair bands, while the pf4 mutation results in 69-, 40-, 38-, 23- and 4-base pair bands (Supplemental figure 2). Digested and undigested PCR products were resolved on 2% agarose gels. C and D. Immunoblots using isolated flagella and axonemes to characterize the antibody to the axonemal PP2A B-subunit and to demonstrate that the PP2A B- and C-subunits fail to assemble in flagella and axonemes from pf4 cells. Protein staining using Ponceau was used to confirm protein loading.
Acknowledgements
We are grateful to Dr. Gregory J. Pazour for aid in analysis of the flagellar proteome database. We wish to acknowledge Rasagnya Viswanadha, Dr. Lea Alford and Ken-ichi Wakabayashi for scientific support, critical discussions of the manuscript and for purification of antibodies used to verify double mutants. This work was supported by NIH grants to WSS (R37GM051173) and SKD (GM-03842). RK is supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science and Technology of Japan (20051007). RY is supported by a long-term fellowship from the TOYOBO Biotechnology Foundation. CE is supported by an NIH NRSA individual predoctoral fellowship (1F31GM087087-01). KY was supported by a fellowship from the CD-BioRap program at Washington University with funds from NSF.
References
- Basu B, Brueckner M. Cilia multifunctional organelles at the center of vertebrate left-right asymmetry. Curr Top Dev Biol. 2008;85:151–74. doi: 10.1016/S0070-2153(08)00806-5. [DOI] [PubMed] [Google Scholar]
- Basu S. PP2A in the Regulation of Cell Motility and Invasion. Curr Protein Pept Sci. 2010;12:3–11. doi: 10.2174/138920311795659443. [DOI] [PubMed] [Google Scholar]
- Bayly PV, Lewis BL, Kemp PS, Pless RB, Dutcher SK. Efficient spatiotemporal analysis of the flagellar waveform of Chlamydomonas reinhardtii. Cytoskeleton (Hoboken) 2010;67(1):56–69. doi: 10.1002/cm.20424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berbari NF, O’Connor AK, Haycraft CJ, Yoder BK. The primary cilium as a complex signaling center. Curr Biol. 2009;19(13):R526–35. doi: 10.1016/j.cub.2009.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bower R, VanderWaal K, O’Toole E, Fox L, Perrone C, Mueller J, Wirschell M, Kamiya R, Sale WS, Porter ME. IC138 defines a subdomain at the base of the I1 dynein that regulates microtubule sliding and flagellar motility. Mol Biol Cell. 2009;20(13):3055–63. doi: 10.1091/mbc.E09-04-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brokaw CJ. Thinking about flagellar oscillation. Cell Motil Cytoskeleton. 2009;66(8):425–36. doi: 10.1002/cm.20313. [DOI] [PubMed] [Google Scholar]
- Brokaw CJ, Kamiya R. Bending patterns of Chlamydomonas flagella: IV. Mutants with defects in inner and outer dynein arms indicate differences in dynein arm function. Cell Motil Cytoskeleton. 1987;8(1):68–75. doi: 10.1002/cm.970080110. [DOI] [PubMed] [Google Scholar]
- Dutcher SK. Mating and tetrad analysis in Chlamydomonas reinhardtii. Methods Cell Biol. 1995;47:531–40. doi: 10.1016/s0091-679x(08)60857-2. [DOI] [PubMed] [Google Scholar]
- Eichhorn PJ, Creyghton MP, Bernards R. Protein phosphatase 2A regulatory subunits and cancer. Biochim Biophys Acta. 2009;1795(1):1–15. doi: 10.1016/j.bbcan.2008.05.005. [DOI] [PubMed] [Google Scholar]
- Elam CA, Sale WS, Wirschell M. The Regulation of Dynein-Driven Microtubule Sliding in Chlamydomonas Flagella by Axonemal Kinases and Phosphatases. In: Stephen MK, Gregory JP, editors. Methods in Cell Biology. Academic Press; 2009. pp. 133–151. [DOI] [PubMed] [Google Scholar]
- Gaillard AR, Diener DR, Rosenbaum JL, Sale WS. Flagellar radial spoke protein 3 is an A-kinase anchoring protein (AKAP) J Cell Biol. 2001;153(2):443–8. doi: 10.1083/jcb.153.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard AR, Fox LA, Rhea JM, Craige B, Sale WS. Disruption of the A-kinase anchoring domain in flagellar radial spoke protein 3 results in unregulated axonemal cAMP-dependent protein kinase activity and abnormal flagellar motility. Mol Biol Cell. 2006;17(6):2626–35. doi: 10.1091/mbc.E06-02-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes JM, Davis EE, Katsanis N. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 2009;137(1):32–45. doi: 10.1016/j.cell.2009.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11(5):331–44. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokhale A, Wirschell M, Sale WS. Regulation of dynein-driven microtubule sliding by the axonemal protein kinase CK1 in Chlamydomonas flagella. J Cell Biol. 2009;186(6):817–24. doi: 10.1083/jcb.200906168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermacher G, Sale WS. Regulation of dynein-driven microtubule sliding by an axonemal kinase and phosphatase in Chlamydomonas flagella. Cell Motil Cytoskeleton. 1995;32(2):106–9. doi: 10.1002/cm.970320207. [DOI] [PubMed] [Google Scholar]
- Habermacher G, Sale WS. Regulation of flagellar dynein by an axonemal type-1 phosphatase in Chlamydomonas. J Cell Sci. 1996;109(Pt 7):1899–907. doi: 10.1242/jcs.109.7.1899. [DOI] [PubMed] [Google Scholar]
- Habermacher G, Sale WS. Regulation of flagellar dynein by phosphorylation of a 138-kD inner arm dynein intermediate chain. J Cell Biol. 1997;136(1):167–76. doi: 10.1083/jcb.136.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto M, Hamada H. Translation of anterior-posterior polarity into left-right polarity in the mouse embryo. Curr Opin Genet Dev. 2010;20(4):433–7. doi: 10.1016/j.gde.2010.04.002. [DOI] [PubMed] [Google Scholar]
- Hatano Y, Shima H, Haneji T, Miura AB, Sugimura T, Nagao M. Expression of PP2A B regulatory subunit beta isotype in rat testis. FEBS Lett. 1993;324(1):71–5. doi: 10.1016/0014-5793(93)81535-8. [DOI] [PubMed] [Google Scholar]
- Hendrickson TW, Perrone CA, Griffin P, Wuichet K, Mueller J, Yang P, Porter ME, Sale WS. IC138 is a WD-repeat dynein intermediate chain required for light chain assembly and regulation of flagellar bending. Mol Biol Cell. 2004;15(12):5431–42. doi: 10.1091/mbc.E04-08-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N, Tanaka Y, Okada Y, Takeda S. Nodal flow and the generation of left-right asymmetry. Cell. 2006;125(1):33–45. doi: 10.1016/j.cell.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Howard DR, Habermacher G, Glass DB, Smith EF, Sale WS. Regulation of Chlamydomonas flagellar dynein by an axonemal protein kinase. J Cell Biol. 1994;127(6 Pt 1):1683–92. doi: 10.1083/jcb.127.6.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Yamamoto R, Wirschell M, Yagi T, Bower R, Porter ME, Sale WS, Kamiya R. A novel ankyrin-repeat protein interacts with the regulatory proteins of inner arm dynein f (I1) of Chlamydomonas reinhardtii. Cell Motil Cytoskeleton. 2009;66(8):448–56. doi: 10.1002/cm.20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssens V, Longin S, Goris J. PP2A holoenzyme assembly: in cauda venenum (the sting is in the tail) Trends Biochem Sci. 2008;33(3):113–21. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Kamiya R, Kurimoto E, Muto E. Two types of Chlamydomonas flagellar mutants missing different components of inner-arm dynein. J Cell Biol. 1991;112(3):441–7. doi: 10.1083/jcb.112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SM, Barbarese E, Dillman JF, 3rd, Patel-King RS, Carson JH, Pfister KK. Brain cytoplasmic and flagellar outer arm dyneins share a highly conserved Mr 8,000 light chain. J Biol Chem. 1996;271(32):19358–66. doi: 10.1074/jbc.271.32.19358. [DOI] [PubMed] [Google Scholar]
- King SM, Otter T, Witman GB. Purification and characterization of Chlamydomonas flagellar dyneins. Methods Enzymol. 1986;134:291–306. doi: 10.1016/0076-6879(86)34097-7. [DOI] [PubMed] [Google Scholar]
- King SJ, Dutcher SK. Phosphoregulation of an inner dynein arm complex in Chlamydomonas reinhardtii is altered in phototactic mutant strains. J Cell Biol. 1997;136(1):177–91. doi: 10.1083/jcb.136.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeDizet M, Piperno G. ida4-1, ida4-2, and ida4-3 are intron splicing mutations affecting the locus encoding p28, a light chain of Chlamydomonas axonemal inner dynein arms. Mol Biol Cell. 1995;6(6):713–23. doi: 10.1091/mbc.6.6.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechward K, Awotunde OS, Swiatek W, Muszynska G. Protein phosphatase 2A: variety of forms and diversity of functions. Acta Biochim Pol. 2001;48(4):921–33. [PubMed] [Google Scholar]
- Leigh MW, Pittman JE, Carson JL, Ferkol TW, Dell SD, Davis SD, Knowles MR, Zariwala MA. Clinical and genetic aspects of primary ciliary dyskinesia/Kartagener syndrome. Genet Med. 2009;11(7):473–87. doi: 10.1097/GIM.0b013e3181a53562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine RP, Goodenough UW. The genetics of photosynthesis and the chloroplast in Chlamydomonas reinhardi. Annu. Rev. Genet. 1970;4:397–408. doi: 10.1146/annurev.ge.04.120170.002145. [DOI] [PubMed] [Google Scholar]
- Lin H, Kwan AL, Dutcher SK. Synthesizing and salvaging NAD: lessons learned from Chlamydomonas reinhardtii. PLoS Genet. 2010;6(9) doi: 10.1371/journal.pgen.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemann CB, Lesich KA. Flagellar and ciliary beating: the proven and the possible. J Cell Sci. 2010;123(Pt 4):519–28. doi: 10.1242/jcs.051326. [DOI] [PubMed] [Google Scholar]
- Matre P, Meyer C, Lillo C. Diversity in subcellular targeting of the PP2A B’eta subfamily members. Planta. 2009;230(5):935–45. doi: 10.1007/s00425-009-0998-z. [DOI] [PubMed] [Google Scholar]
- Mayer RE, Hendrix P, Cron P, Matthies R, Stone SR, Goris J, Merlevede W, Hofsteenge J, Hemmings BA. Structure of the 55-kDa regulatory subunit of protein phosphatase 2A: evidence for a neuronal-specific isoform. Biochemistry. 1991;30(15):3589–97. doi: 10.1021/bi00229a001. [DOI] [PubMed] [Google Scholar]
- McVittie A. Flagellum mutants of Chlamydomonas reinhardii. J. Gen. Microbiol. 1972;71(3):525–540. doi: 10.1099/00221287-71-3-525. [DOI] [PubMed] [Google Scholar]
- Mitchell DR, Sale WS. Characterization of a Chlamydomonas insertional mutant that disrupts flagellar central pair microtubule-associated structures. J Cell Biol. 1999;144(2):293–304. doi: 10.1083/jcb.144.2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumby M. The 3D structure of protein phosphatase 2A: new insights into a ubiquitous regulator of cell signaling. ACS Chem Biol. 2007;2(2):99–103. doi: 10.1021/cb700021z. [DOI] [PubMed] [Google Scholar]
- Myster SH, Knott JA, O’Toole E, Porter ME. The Chlamydomonas Dhc1 gene encodes a dynein heavy chain subunit required for assembly of the I1 inner arm complex. Mol Biol Cell. 1997;8(4):607–20. doi: 10.1091/mbc.8.4.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myster SH, Knott JA, Wysocki KM, O’Toole E, Porter ME. Domains in the 1alpha dynein heavy chain required for inner arm assembly and flagellar motility in Chlamydomonas. J Cell Biol. 1999;146(4):801–18. doi: 10.1083/jcb.146.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunbhakdi-Craig V, Schuechner S, Sontag JM, Montgomery L, Pallas DC, Juno C, Mudrak I, Ogris E, Sontag E. Expression of protein phosphatase 2A mutants and silencing of the regulatory B alpha subunit induce a selective loss of acetylated and detyrosinated microtubules. J Neurochem. 2007;101(4):959–71. doi: 10.1111/j.1471-4159.2007.04503.x. [DOI] [PubMed] [Google Scholar]
- Okita N, Isogai N, Hirono M, Kamiya R, Yoshimura K. Phototactic activity in Chlamydomonas ‘non-phototactic’ mutants deficient in Ca2+-dependent control of flagellar dominance or in inner-arm dynein. J Cell Sci. 2005;118(Pt 3):529–37. doi: 10.1242/jcs.01633. [DOI] [PubMed] [Google Scholar]
- Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J Cell Biol. 2005;170(1):103–13. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone CA, Myster SH, Bower R, O’Toole ET, Porter ME. Insights into the structural organization of the I1 inner arm dynein from a domain analysis of the 1beta dynein heavy chain. Mol Biol Cell. 2000;11(7):2297–313. doi: 10.1091/mbc.11.7.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrone CA, Yang P, O’Toole E, Sale WS, Porter ME. The Chlamydomonas IDA7 locus encodes a 140-kDa dynein intermediate chain required to assemble the I1 inner arm complex. Mol Biol Cell. 1998;9(12):3351–65. doi: 10.1091/mbc.9.12.3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Power J, Dutcher SK. Extragenic suppressors of paralyzed flagellar mutations in Chlamydomonas reinhardtii identify loci that alter the inner dynein arms. J Cell Biol. 1992;118(5):1163–76. doi: 10.1083/jcb.118.5.1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter ME, Sale WS. The 9 + 2 axoneme anchors multiple inner arm dyneins and a network of kinases and phosphatases that control motility. J Cell Biol. 2000;151(5):F37–42. doi: 10.1083/jcb.151.5.f37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price NE, Wadzinski B, Mumby MC. An anchoring factor targets protein phosphatase 2A to brain microtubules. Brain Res Mol Brain Res. 1999;73(1-2):68–77. doi: 10.1016/s0169-328x(99)00237-5. [DOI] [PubMed] [Google Scholar]
- Salathe M. Regulation of mammalian ciliary beating. Annu Rev Physiol. 2007;69:401–22. doi: 10.1146/annurev.physiol.69.040705.141253. [DOI] [PubMed] [Google Scholar]
- Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2007;69:377–400. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- Satir P, Christensen ST. Structure and function of mammalian cilia. Histochem Cell Biol. 2008;129(6):687–93. doi: 10.1007/s00418-008-0416-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt K, Kins S, Schild A, Nitsch RM, Hemmings BA, Gotz J. Diversity, developmental regulation and distribution of murine PR55/B subunits of protein phosphatase 2A. Eur J Neurosci. 2002;16(11):2039–48. doi: 10.1046/j.1460-9568.2002.02274.x. [DOI] [PubMed] [Google Scholar]
- Shi Y. Assembly and structure of protein phosphatase 2A. Sci China C Life Sci. 2009a;52(2):135–46. doi: 10.1007/s11427-009-0018-3. [DOI] [PubMed] [Google Scholar]
- Shi Y. Serine/threonine phosphatases: mechanism through structure. Cell. 2009b;139(3):468–84. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Smith EF, Rohatgi R. Cilia 2010: the surprise organelle of the decade. Sci Signal. 2011;4(155) doi: 10.1126/scisignal.4155mr1. mr1. [DOI] [PubMed] [Google Scholar]
- Smith EF, Yang P. The radial spokes and central apparatus: mechano-chemical transducers that regulate flagellar motility. Cell Motil Cytoskeleton. 2004;57(1):8–17. doi: 10.1002/cm.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontag E, Nunbhakdi-Craig V, Lee G, Brandt R, Kamibayashi C, Kuret J, White CL, 3rd, Mumby MC, Bloom GS. Molecular interactions among protein phosphatase 2A, tau, and microtubules. Implications for the regulation of tau phosphorylation and the development of tauopathies. J Biol Chem. 1999;274(36):25490–8. doi: 10.1074/jbc.274.36.25490. [DOI] [PubMed] [Google Scholar]
- Sontag JM, Sontag E. Regulation of cell adhesion by PP2A and SV40 small tumor antigen: an important link to cell transformation. Cell Mol Life Sci. 2006;63(24):2979–91. doi: 10.1007/s00018-006-6300-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack S, Zaucha JA, Ebner FF, Colbran RJ, Wadzinski BE. Brain protein phosphatase 2A: developmental regulation and distinct cellular and subcellular localization by B subunits. J Comp Neurol. 1998;392(4):515–27. [PubMed] [Google Scholar]
- Sugase Y, Hirono M, Kindle KL, Kamiya R. Cloning and characterization of the actin-encoding gene of Chlamydomonas reinhardtii. Gene. 1996;168(1):117–21. doi: 10.1016/0378-1119(95)00711-3. [DOI] [PubMed] [Google Scholar]
- Tar K, Birukova AA, Csortos C, Bako E, Garcia JG, Verin AD. Phosphatase 2A is involved in endothelial cell microtubule remodeling and barrier regulation. J Cell Biochem. 2004;92(3):534–46. doi: 10.1002/jcb.20036. [DOI] [PubMed] [Google Scholar]
- Toba S, Fox LA, Sakakibara H, Porter ME, Oiwa K, Sale WS. Distinct roles of 1{alpha} and 1{beta} heavy chains of the inner arm dynein I1 of Chlamydomonas flagella. Mol Biol Cell. 2011;22(3):342–53. doi: 10.1091/mbc.E10-10-0806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup DM, Shenolikar S. From promiscuity to precision: protein phosphatases get a makeover. Mol Cell. 2009;33(5):537–45. doi: 10.1016/j.molcel.2009.02.015. [DOI] [PubMed] [Google Scholar]
- Wallingford JB. Planar cell polarity signaling, cilia and polarized ciliary beating. Curr Opin Cell Biol. 2010;22(5):597–604. doi: 10.1016/j.ceb.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BD, Velleca MA, Curry AM, Rosenbaum JL. Molecular cloning and sequence analysis of the Chlamydomonas gene coding for radial spoke protein 3: flagellar mutation pf-14 is an ochre allele. J Cell Biol. 1989;109(1):235–45. doi: 10.1083/jcb.109.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirschell M, Hendrickson T, Sale WS. Keeping an eye on I1: I1 dynein as a model for flagellar dynein assembly and regulation. Cell Motil Cytoskeleton. 2007;64(8):569–79. doi: 10.1002/cm.20211. [DOI] [PubMed] [Google Scholar]
- Wirschell M, Yang C, Yang P, Fox L, Yanagisawa HA, Kamiya R, Witman GB, Porter ME, Sale WS. IC97 is a novel intermediate chain of I1 dynein that interacts with tubulin and regulates interdoublet sliding. Mol Biol Cell. 2009;20(13):3044–54. doi: 10.1091/mbc.E09-04-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witman GB. Isolation of Chlamydomonas flagella and flagellar axonemes. Methods Enzymol. 1986;134:280–90. doi: 10.1016/0076-6879(86)34096-5. [DOI] [PubMed] [Google Scholar]
- Xu Y, Chen Y, Zhang P, Jeffrey PD, Shi Y. Structure of a protein phosphatase 2A holoenzyme: insights into B55-mediated Tau dephosphorylation. Mol Cell. 2008;31(6):873–85. doi: 10.1016/j.molcel.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Sale WS. The Mr 140,000 intermediate chain of Chlamydomonas flagellar inner arm dynein is a WD-repeat protein implicated in dynein arm anchoring. Mol Biol Cell. 1998;9(12):3335–49. doi: 10.1091/mbc.9.12.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Fox L, Colbran RJ, Sale WS. Protein phosphatases PP1 and PP2A are located in distinct positions in the Chlamydomonas flagellar axoneme. J Cell Sci. 2000;113( Pt 1):91–102. doi: 10.1242/jcs.113.1.91. [DOI] [PubMed] [Google Scholar]
- Yang P, Sale WS. Casein kinase I is anchored on axonemal doublet microtubules and regulates flagellar dynein phosphorylation and activity. J Biol Chem. 2000;275(25):18905–12. doi: 10.1074/jbc.M002134200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1: Alignment of the B-subunit was performed using ClustalW and the alignments were color-coded using the percentage composition coloring tool, Colorfy (http://bifrost.wustl.edu/colorfy) (Lin et al. 2010).
Supplemental Figure 2: A. Chromatograms showing the DNA sequence in pf4 and the pf4 revertants cells. B. The pf4 mutation generates a novel CviKi-1 restriction site in the pf4 mutant strain. PCR products covering the site of the pf4 mutation in exon 7 were digested with CviKi-1. The resulting restriction pattern is diagnostic for the pf4 mutation: wild-type PF4 sequence produces 110-, 38-, 23- and 4-base pair bands, while the pf4 mutation results in 69-, 40-, 38-, 23- and 4-base pair bands (Supplemental figure 2). Digested and undigested PCR products were resolved on 2% agarose gels. C and D. Immunoblots using isolated flagella and axonemes to characterize the antibody to the axonemal PP2A B-subunit and to demonstrate that the PP2A B- and C-subunits fail to assemble in flagella and axonemes from pf4 cells. Protein staining using Ponceau was used to confirm protein loading.