Abstract
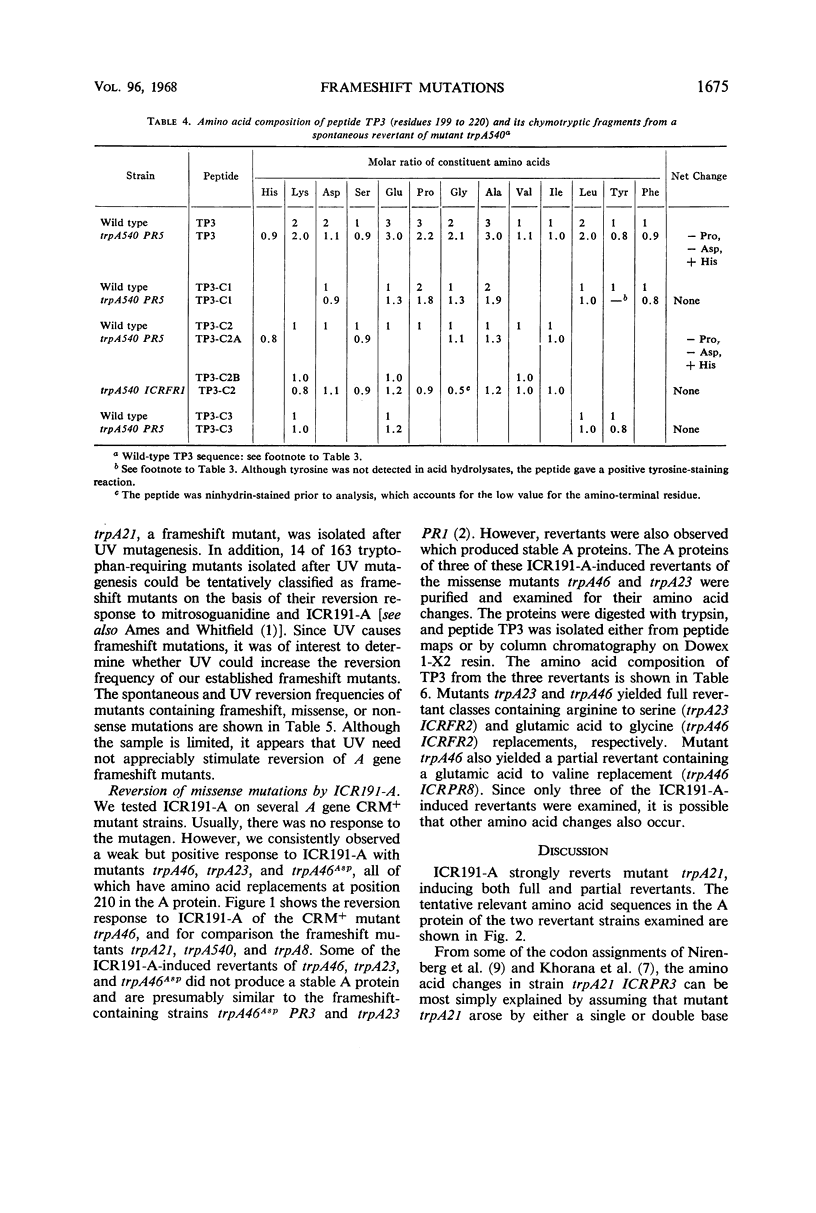
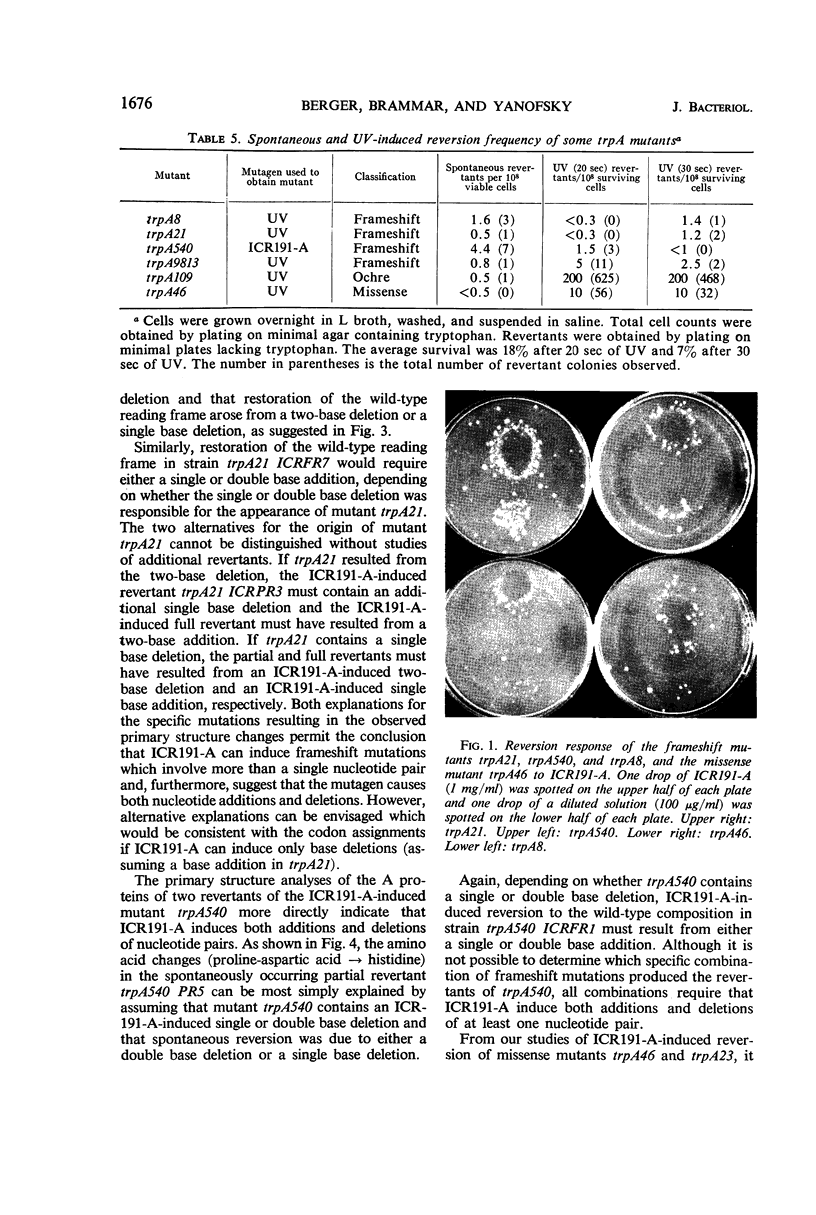
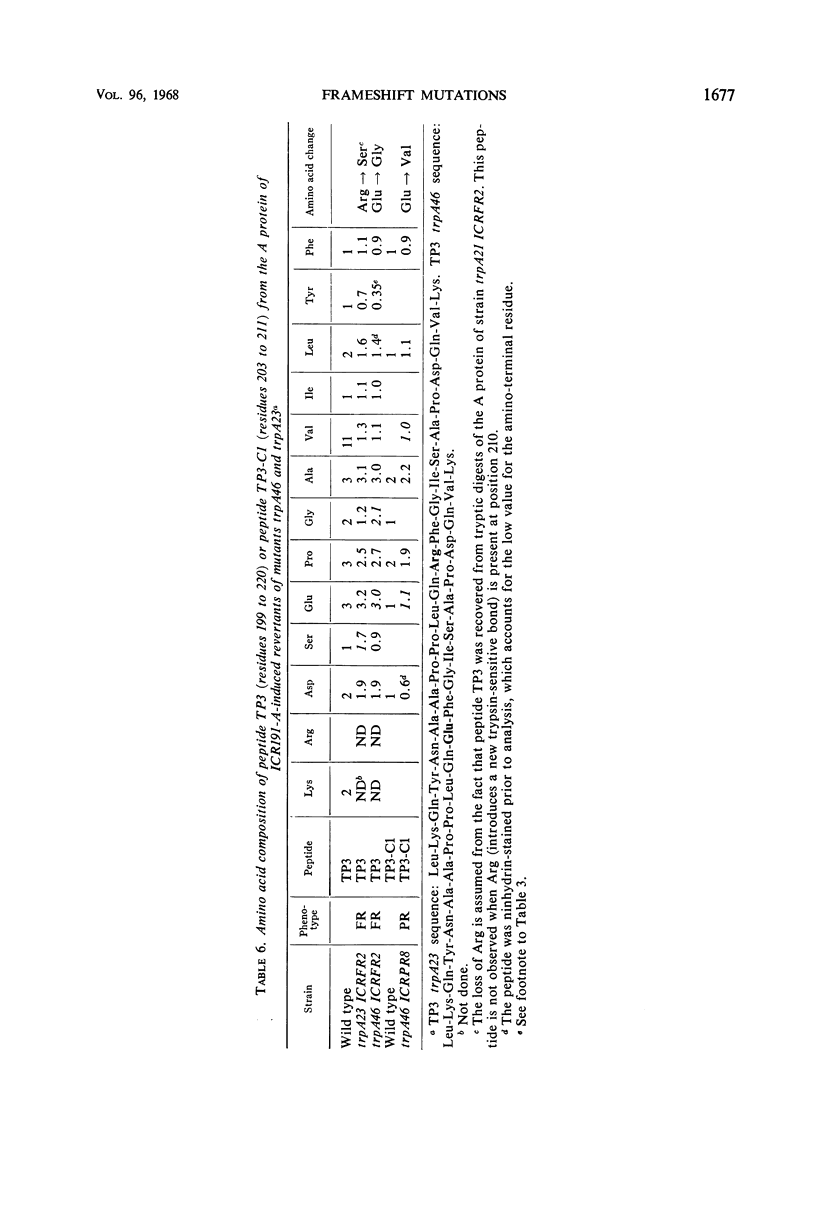
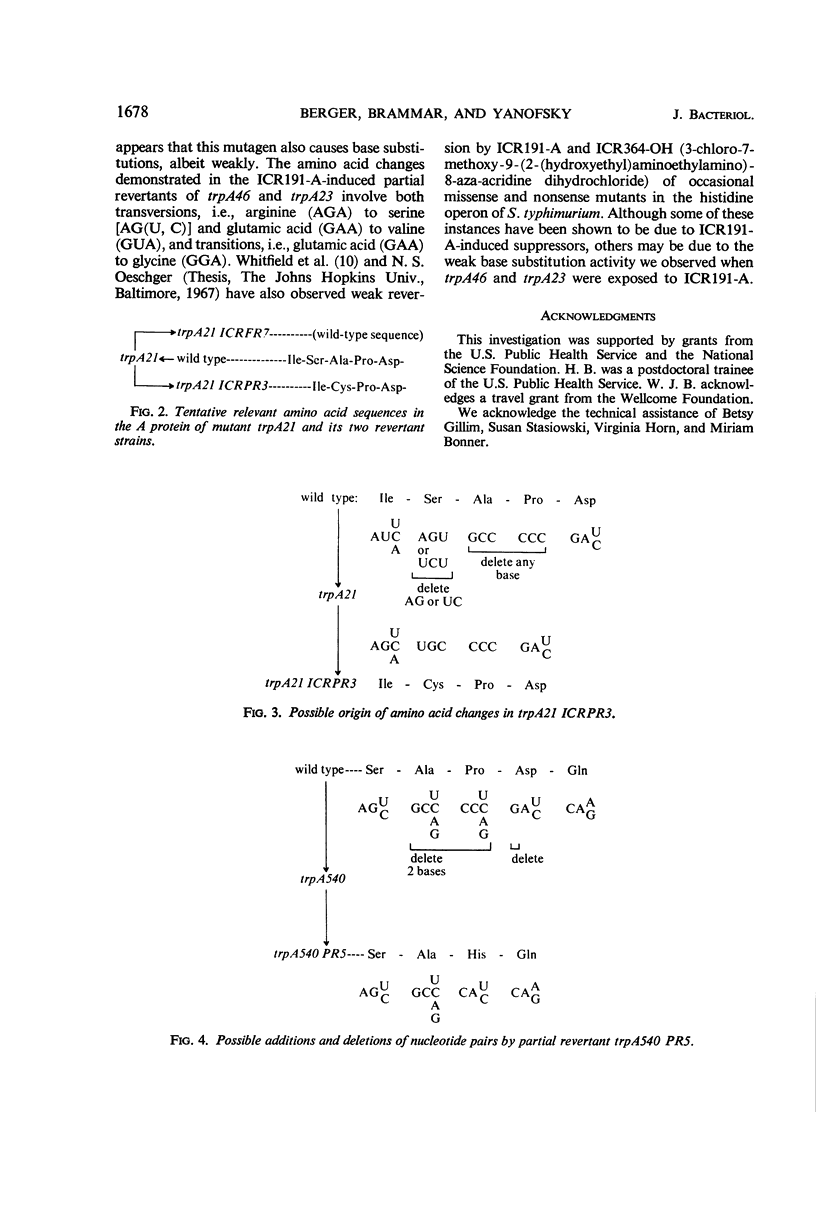
Frameshift mutant trpA21 was isolated after ultraviolet treatment and frameshift mutant trpA540 after ICR191-A (an acridine derivative) treatment of wild-type Escherichia coli K-12. The A proteins of spontaneous and ICR191-A-induced partial revertants of these mutants contained altered amino acid sequences one residue shorter than the comparable sequence in the A protein of wild-type bacteria. The data support the conclusion that ICR191-A causes frameshift mutations. The findings further indicate that both base additions and deletions are elicited by ICR191-A treatment and that mutagenesis by this compound sometimes affects more than one base pair. ICR191-A also weakly reverts some missense mutants. Analyses of the relevant peptides of the purified A protein show single amino acid replacements compatible with single base-pair changes. In addition, we found that some spontaneously revertible ICR191-A- and ultraviolet light-induced frameshift mutants are not further stimulated to revert by exposure to ultraviolet light.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ames B. N., Whitfield H. J., Jr Frameshift mutagenesis in Salmonella. Cold Spring Harb Symp Quant Biol. 1966;31:221–225. doi: 10.1101/sqb.1966.031.01.030. [DOI] [PubMed] [Google Scholar]
- Berger H., Brammar W. J., Yanofsky C. Analysis of amino acid replacements resulting from frameshift and missense mutations in the tryptophan synthetase A gene of Escherichia coli. J Mol Biol. 1968 Jul 14;34(2):219–238. doi: 10.1016/0022-2836(68)90248-9. [DOI] [PubMed] [Google Scholar]
- Brammar W. J., Berger H., Yanofsky C. Altered amino acid sequences produced by reversion of frameshift mutants of tryptophan synthetase A gene of E. coli. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1499–1506. doi: 10.1073/pnas.58.4.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guest J. R., Yanofsky C. Amino acid sequences surrounding the sulfhydryl groups of the A protein subunit of the Escherichia coli tryptophan synthetase. J Biol Chem. 1966 Jan 10;241(1):1–16. [PubMed] [Google Scholar]
- HELINSKI D. R., YANOFSKY C. Peptide pattern studies on the wild-type A protein of the tryptophan synthetase of Escherichia coli. Biochim Biophys Acta. 1962 Sep 10;63:10–19. doi: 10.1016/0006-3002(62)90333-5. [DOI] [PubMed] [Google Scholar]
- HENNING U., YANOFSKY C. Amino acid replacements associated with reversion and recombination within the A gene. Proc Natl Acad Sci U S A. 1962 Sep 15;48:1497–1504. doi: 10.1073/pnas.48.9.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorana H. G., Büchi H., Ghosh H., Gupta N., Jacob T. M., Kössel H., Morgan R., Narang S. A., Ohtsuka E., Wells R. D. Polynucleotide synthesis and the genetic code. Cold Spring Harb Symp Quant Biol. 1966;31:39–49. doi: 10.1101/sqb.1966.031.01.010. [DOI] [PubMed] [Google Scholar]
- Martin R. G. Frameshift mutants in the histidine operon of Salmonella typhimurium. J Mol Biol. 1967 Jun 14;26(2):311–328. doi: 10.1016/0022-2836(67)90300-2. [DOI] [PubMed] [Google Scholar]
- Nirenberg M., Caskey T., Marshall R., Brimacombe R., Kellogg D., Doctor B., Hatfield D., Levin J., Rottman F., Pestka S. The RNA code and protein synthesis. Cold Spring Harb Symp Quant Biol. 1966;31:11–24. doi: 10.1101/sqb.1966.031.01.008. [DOI] [PubMed] [Google Scholar]
- Whitfield H. J., Jr, Martin R. G., Ames B. N. Classification of aminotransferase (C gene) mutants in the histidine operon. J Mol Biol. 1966 Nov 14;21(2):335–355. doi: 10.1016/0022-2836(66)90103-3. [DOI] [PubMed] [Google Scholar]