Abstract
The p53-family member p73 plays a role in various cellular signaling pathways during development and growth control and it can have tumor suppressor properties. Several isoforms of p73 exist with considerable differences in their function. Whereas the functions of the N-terminal isoforms (TA and ΔNp73) and their opposing pro- and antiapoptotic roles have become evident, the functional differences of the distinct C-terminal splice forms of TAp73 have remained unclear. Here, we characterized the global genomic binding sites for TAp73α and TAp73β by chromatin immunoprecipitation sequencing as well as the transcriptional responses by performing RNA sequencing. We identified a specific p73 consensus binding motif and found a strong enrichment of AP1 motifs in close proximity to binding sites for TAp73α. These AP1 motif-containing target genes are selectively upregulated by TAp73α, while their mRNA expression is repressed upon TAp73β induction. We show that their expression is dependent on endogenous c-Jun and that recruitment of c-Jun to the respective AP1 sites was impaired upon TAp73β expression, in part due to downregulation of c-Jun. Several of these AP1-site containing TAp73α-induced genes impinge on apoptosis induction, suggesting an underlying molecular mechanism for the observed functional differences between TAp73α and TAp73β.
INTRODUCTION
The transcription factors of the p53-family, p53, p63 and p73, maintain the balance between cell survival and induction of apoptosis during development, growth, differentiation and cellular stress. The members of the p53-family thereby display common as well as specific functions (1). They determine the cellular fate dependent on the family member and isoform expressed in a specific tissue. The p73 protein exists in multiple isoforms due to different promoter usage at the N-terminus and to C-terminal splice events. The ΔNp73 isoforms that are derived from an internal promoter, antagonize the growth suppressing, pro-apoptotic functions of p53 and of the full length TAp73 isoforms in a dominant negative way by competing for the respective binding sites (2). Overexpression of ΔNp73 isoforms is found in several tumors (3,4) whereas mutations in the p73 gene are rarely found in human cancers (5).
Under certain conditions, p53 is unable to induce apoptosis in the absence of p73 or p63 (6). Furthermore, mice heterozygous for p53/p73 show a higher tumor burden compared to p53 heterozygous mice (7). Although complete knockout of the p73 gene in mice mainly leads to developmental defects (8), the knockdown of only the TA isoforms induces genomic instability, thus showing tumor suppressor activities of TAp73 (9). TAp73 isoforms have been reported to play a role in DNA damage pathways, since p73 is activated by ionizing irradiation and cisplatin through c-Abl, thereby inducing apoptosis (10–12). Furthermore, TAp73 isoforms are upregulated by different mechanisms through chemotherapeutic drug induced DNA damage (13,14).
The transcriptional function of p73 is complex because of the plethora of p73 isoforms, which have varying transcriptional activity toward target genes. In addition to shared target sites, the p53-family members differ in their ability to transactivate common target genes like p21 (15) or Bax (16,17). Some genes are only induced by specific isoforms, like p57/kip2 by p73β but not by p73α or by p53 (18).
Besides its function as a pro-apoptotic protein, several reports have also described an inhibition of apoptosis or a support of growth by TAp73 in certain cell lines under specific conditions (19,20). It has been shown that a crosstalk between the transcription factor c-Jun and p73 regulates growth and that c-Jun enhances the function of p73 (21,22). However, the exact molecular mechanism of this crosstalk remains unknown.
C-Jun is a member of the AP1 family of heterodimeric transcription factors, regulating growth and apoptosis depending on the cellular environment and on the composition of the respective dimer (23). Dimers containing c-Jun mainly promote growth via G1-progression through the transactivation of Cyclin D1 (24). The fact that a c-Jun null mutation is embryonically lethal and causes retarded growth of cultured cells underscores the importance of c-Jun for cellular growth (25). AP1 dimers can also protect cells from UV-mediated apoptosis by negatively regulating p53 (26) and c-Jun is also required for re-entry of cells into cell cycle after UV-induced p53 mediated growth arrest (27). Due to the complexity of the many p73 isoforms and the varying composition of the Jun/Fos dimers several different interactions between p73 and Jun/Fos might be possible, probably with different consequences for the cellular fate.
To gain insight into the molecular basis for the different physiological function of TAp73α and TAp73β, we identified their binding sites by chromatin immunoprecipitations (ChIPs) coupled with deep sequencing (ChIP-seq) and global expression analysis using RNA sequencing (RNA-seq). This revealed that the two TAp73 isoforms bind to both shared and isoform-specific target sites and distinctly transactivate target genes. We uncovered a p73-consensus motif that is present in a large fraction of the p73 binding sites. The binding sites of TAp73α but not those of TAp73β showed an overrepresentation of AP1 binding sites to which c-Jun can bind simultaneously with TAp73α. The binding of c-Jun to DNA is decreased upon TAp73β expression, which reduces the mRNA and protein levels of c-Jun. The expression of distinct target genes with an AP1 site close to TAp73α binding sites depends on c-Jun and they can influence apoptosis induction by TAp73α, possibly explaining the different physiological responses mediated by the respective TAp73 isoforms.
MATERIAL AND METHODS
Cell culture conditions
The human MDA-MB231-, HCT116-, HEK293 cell lines and the osteosarcoma Saos2 (parental cells or cell lines with inducible p53, TAp73α or TAp73β) (28) were maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum at 37°C. Expression of p53, TAp73α or TAp73β in stably transfected cells was induced with 0.5 µg doxycycline (for TAp73α) or 2.0 µg doxycyclin (for TAp73β and p53). Transfections were performed using the calcium phosphate precipitation method (BES). Transient knockdown of IL1RAP and NEDD4L was achieved by using Dharmacon ON-TARGETplus siRNAs, according to manufacturers manual. Lentiviral transduction was used to deliver pLKO-siRNA constructs for the knockdown of c-Jun and p73 (Sigma), while pLKO-siRNA targeting Luciferase served as a control.
Cell cycle analysis
Saos2 parental cells and Saos cell lines inducible for TAp73α, TAp73β or p53 were induced for 24 h or 48 h, prior to harvest. All cells were collected, resuspended in PBS containing 1% FCS and fixed in ethanol overnight at 4°C. DNA content was stained with propidium iodide (Sigma) for 30 min at room temperature and analyzed by flow cytometry (Becton Dickinson FACScan). The data were analyzed using CellQuest Pro software.
Western blot analysis
Whole cell extract was harvested in 2 × SDS sample buffer, 4 × sample buffer was added to aliquots of chromatin samples (see below for preparation of chromatin), both types of samples were boiled and proteins were separated by SDS–PAGE. After blotting, the following antibodies were used to detect proteins: BL906 (anti-p73; Abcam), DO1 (anti-p53; BD PharMingen), ab13487 (anti-active Caspase3; Abcam), SC45-X (anti-cJun; SantaCruz Biotechnology), SL30 (anti-TATA-box Binding Protein; anti-TBP), M2 (anti-FLAG; Sigma). Secondary antibodies used were either rabbit–anti-mouse or swine–anti-rabbit conjugated to HRP (Dako).
ChIP
ChIP was basically done as described by Denissov et al. (29). To immunoprecipitate p53, DO1 antibody (BD PharMingen) was used and BL906 (Abcam) was used for p73. Immunoprecipitation of c-Jun was done with an anti-c-Jun antibody (Upstate). Real-time qPCR was performed using the SYBR Green mix (Biorad) with the MyIQ machine (Biorad). Primers are listed in Supplemental Table S5.
Prior to the Re-ChIP, p73-antibody (BL906) or c-Jun-antibody (Upstate) was crosslinked to protein-A/G-beads (SantaCruz) using 20 mM dimethylpimelimidate (pH: 8.5). The chromatin complexes from the first ChIP were eluted in 50 μl of elution buffer and the SDS concentration was adjusted with 5 × incubation buffer without SDS to 0.15% in the second ChIP-reaction. The Re-ChIP included an unspecific or a beads only control, respectively, to allow an estimation of leakage and antibody carryover from the first ChIP.
RNA isolation and RT–PCR
Total RNA was extracted using the RNeasy Mini Kit according to protocol (Qiagen). For cDNA synthesis retrotranscription was performed using 1 μg of RNA with random hexamer primers, dNTPs, DTT, buffer and Superscript Retrotranscriptase (Invitrogen). The cDNA was analyzed by real-time qPCR using a MyIQ machine (Biorad). Primers are listed in Supplemental Table S5.
RNA sequencing
To prepare samples for RNA-seq 100 μg of total RNA were subjected to polyA-selection with Oligotex Kit according to protocol (Qiagen). Fragmentation of 100 ng polyA-selected mRNA was done for 105 s at 94°C in 1 × fragmentation buffer (40 mM Tris acetate, pH 8.2; 100 mM potassium acetate; 30 mM magnesium acetate). After purification with RNeasy Mini Kit, according to protocol (Qiagen) first strand cDNA synthesis was performed with random hexamer primers, dNTPs, DTT, buffer and Superscript Retrotranscriptase (Invitrogen). Second strand synthesis was done with Escherichia coli DNA polymerase, E.coli DNA ligase, dNTPs buffer and RNAseH. After purification with Minelute Reaction Cleanup Kit (Qiagen), the obtained material was used to prepare sequencing samples according to the manufacturers protocol (Illumina).
Illumina high-throughput sequencing (ChIP-seq)
Sequencing samples were prepared according to the manufacturers protocol (Illumina). Shortly, adapted sequences were linked to the generated ChIP, the library was size selected (200–250 bp) and amplified by PCR. Clustering and 36-cycle sequencing were performed using an Illumina Cluster Station and Genome Analyzer according to the manufacturer’s instructions. Images acquired from the Genome Analyzer were processed through the bundled image analysis pipeline (Illumina). All 35 bp sequence reads were uniquely mapped to the human genome (NCBI build 36.1, hg18) with zero or one mismatch allowed using ELAND software (Illumina). For visualization purposes, all reads were directionally extended to 200 bp, and the number of overlapping sequence reads was determined for each position in the genome, averaged over a 10 bp window and visualized in the UCSC genome browser (http://genome.ucsc.edu).
ChIP-seq data analysis
Two biological replicates were sequenced for all ChIP samples. To incorporate the different signal/background ratios of the different biological replicates and generate high-confidence peaks, we called peaks on a set of combined reads randomly sampled from each combination of two replicates. We randomly selected ∼7 million reads from each TAp73α and each Tap73β replicate and 3 million from each p53 replicate. We combined these randomly selected reads (∼14 million for both TAp73α and TAp73β, 6 million for p53) and used this set as input for MACS (30). Peaks were called using default parameters (P-value threshold 1E-5), using a Saos2 input DNA sample as control (∼14 million reads). This random sampling procedure was repeated 10 times, and only the peaks determined in every single analysis were kept. Peaks were mapped to RefSeq genes, downloaded from the UCSC Genome Browser, to determine genomic location.
RNA-seq data analysis
Two biological replicates were sequenced for each sample. Reads were mapped to the genome and all reads mapping within RefSeq genes (downloaded from the UCSC Genome Browser) were counted. The read counts per gene per replicate were used as input for DEGseq (31). This R package was used to call differentially regulated genes using the MARS method, with default parameters. All genes with a significant change (FDR < 0.001) and an absolute log2 fold change of 1 were called as regulated.
Association strength
The continuous association strength per gene was calculated as described in Ouyang et al. (32). The association strength of gene i is calculated as the a weighted sum of intensities of all of the peaks within 2 Mb of the gene, according to the following formula:
where gk is the total normalized number of reads aligned of the kth binding site, dk is the distance (number of nucleotides) between the TSS of gene i and the kth binding site and d0 is 5000. The association strength values are log2 transformed and quantile normalized.
Motif analysis
The location and score of the p53 motif within the 200 bp peaks was determined using p53scan with default settings (www.ncmls.nl/bioinfo/p53scan/) (28).
To determine the p73 motif three motif prediction tools were run on the 200 bp peaks: MotifSampler (33), Weeder (34) and MDmodule (35). A set of 1000 sequences, randomly selected from the highest 5000 peak sequences was used as input to predict motifs. We used the ‘large’ analysis setting for Weeder, and MDmodule and MotifSampler were used to predict 10 motifs for each of the even widths between 6 and 20. The significance of the predicted motifs was determined by scanning the remaining 80% of the 5000 highest peak sequences (4000 sequences) and two different backgrounds: a set of random genomic sequences with a similar genomic distribution as the peak sequences and a set of random sequences generated according to a 1st order Markov model (similar dinucleotide distribution as the peak sequences). P-values were calculated using the hypergeometric distribution with the Benjamini–Hochberg multiple testing correction. All motifs with a P < 0.001 and at least an absolute enrichment >1.5 compared to both backgrounds were determined as significant. The closest matching motif in the JASPAR database (36) was determined using STAMP (37).
The p53scan algorithm was modified to use the best p73-motif matrix found in the motif analysis, hereafter referred to as p73scan. The positional weight matrices (PWM) is included as Supplementary Table S4. All p73scan analyses reported in this study were carried out with a spacer length of 0.
The AP1-motif analyses were carried out with the pwmscan.py program included with p53scan, using the best performing AP1 matrix identified in this study as PWM. This matrix is provided as Supplementary Table S4. As a threshold for the AP1 pwmscan.py 0.98 was used, to select for stringent matches.
Data availability
The data have been deposited in NCBI’s Gene Expression Omnibus (38) and are accessible through GEO Series accession number GSE15780 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15780).
RESULTS
Physiological and molecular differences in the cellular responses to TAp73α and TAp73β
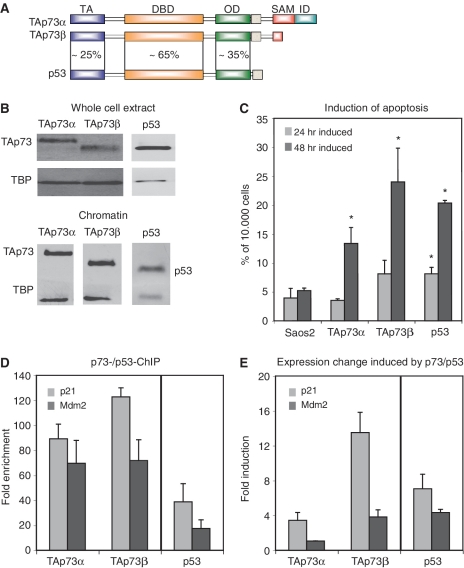
The domain organization of the predominant TAp73α- and TAp73β isoforms shows some similarity with the related tumor suppressor p53, but the three proteins differ completely in their C-terminus (Figure 1A). To characterize their common and distinct functions, we used isogenic Saos cell lines (28) to express TAp73α and TAp73β at comparable protein levels as detected in whole cell lysates as well as in the chromatin-bound fraction (Figure 1B). As a comparison, the effect of p53 on apoptosis induction was included. To quantify the degree of cell death after induction of TAp73α, TAp73β or p53, FACS analysis was performed and apoptosis was measured as the sub-G1 cell population (Figure 1C). Whereas in the parental Saos2 cells only a small proportion of cells undergo programmed cell death, the induction of TAp73α, TAp73β or p53 leads to an increase in the apoptotic population. The levels of apoptosis after TAp73α induction are rather modest. Upon TAp73β induction, on the other hand, apoptosis levels are three to eight times higher than in parental Saos2 cells, thus comparable to p53-induced cell death. Since the different p53-family members displayed distinct effects on the level of apoptosis induction, we examined possible molecular causes for these differences. To analyze the DNA binding capacities of the two p73 isoforms and p53 we used ChIP followed by qPCR (ChIP-qPCR). As shown in Figure 1D binding of p53 and both TAp73 isoforms is observed to the p21 and Mdm2 genes. It is higher for TAp73β at the p21 and Mdm2 binding sites than for TAp73α. Examining the transactivating potential of these p73 isoforms and p53 shows that both isoforms increase the level of p21 mRNA, with a much stronger increase mediated by TAp73β (Figure 1E). The mRNA level of Mdm2 is increased upon TAp73β and p53 expression, but not upon TAp73α expression in this cell system (Figure 1E). Thus, induction of TAp73α and TAp73β had different effects on the transcriptional activation of target genes as well as on the cellular apoptotic response.
Figure 1.
Differential physiological and molecular effects of TAp73α and TAp73β. (A) Domain structure of the p53-family members TAp73α, TAp73β and p53. TA, transactivation domain; DBD, DNA-binding domain; OD, oligomerization domain; SAM, sterile α-motif; ID, inhibitory domain. (B) Expression of TAp73α, TAp73β and p53 in Saos-inducible cell lines. Whole cell extracts or chromatin was harvested after 24 h of induction and protein levels were analyzed by western blot. (C) Induction of apoptosis. Saos cells expressing the indicated member of the p53-family were induced for 24 or 48 h, before the amount of cells in sub-G1 phase, as a measure for apoptotic induction, was counted using FACS. Error bars indicate standard deviation (SD) derived from three independent experiments, asterisks indicate statistical significance as shown by Student’s t-test (P < 0.05). (D) Differential binding of TAp73α and TAp73β to target genes, using p53 as a positive control. Saos TAp73- or p53-inducible cells were induced for 24 h before chromatin was isolated. Complexes of TAp73 or p53 and chromatin were precipitated and qPCR analysis was performed with primers for the putative binding sites and enrichment is shown in fold over an unspecific control (myoglobin). SD results from three independent experiments. (E) Expression changes upon induction of p53-family isoforms. RNA from Saos cells was harvested 24 h after induction of the respective family member. After cDNA synthesis qPCR was performed with the indicated primers and results were normalized against GAPDH expression. The induction was calculated as fold over the normalized expression values from Saos2 parental cells. SD was calculated from three independent experiments.
Global binding profiles reveal common as well as distinct binding sites for TAp73α and TAp73β
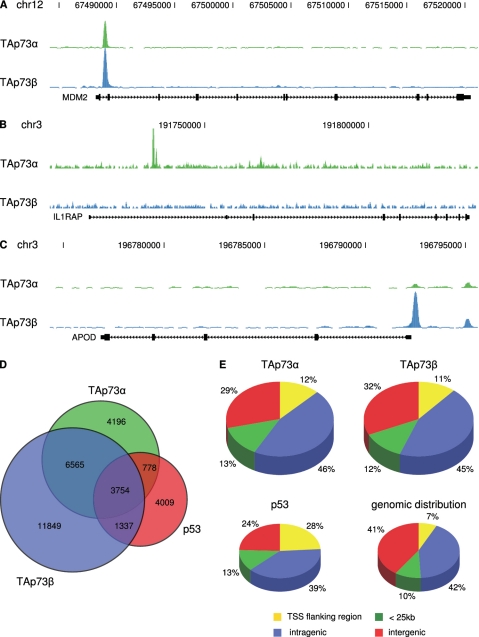
To shed light on the molecular mechanisms for the differential cellular responses induced by the two TAp73 isoforms, we compared their DNA binding site repertoire at a genome-wide scale, performing ChIP-seq of the TAp73 isoforms as well as of p53 (two biological replicates each). To validate the reproducibility of the data sets, we compared the number of reads per peak between two biological replicates and found a high correlation (Supplementary Figure S1). Identifying binding sites by peak calling on the combined data using MACS (30), with an input DNA sample as background control, resulted in 15 293 peaks for TAp73α, 23 505 peaks for TAp73β and 9878 peaks for p53 (Table 1). We identified genome wide binding sites that are shared by both p73 isoforms (10 319 sites) or are preferential for either TAp73α (4196 sites) or TAp73β (11 849 sites) exemplified by the Mdm2, IL1RAP and APOD gene, respectively (Figure 2A–C).
Table 1.
ChIP-seq results
| Replicate no. | Uniquely mapped reads (million) | Reads after normalization (million) | Number of peaks | Genes with a peak within 25 kb | |
|---|---|---|---|---|---|
| Saos control | 1 | 14.82 | 14.82 | ||
| Saos TAp73α | 1 | 12.52 | 7.41 | 15 293 | 5405 |
| Saos TAp73α | 2 | 13.77 | 7.41 | ||
| Saos TAp73β | 1 | 12.10 | 7.41 | 23 505 | 6932 |
| Saos TAp73β | 2 | 13.08 | 7.41 | ||
| Saos p53 | 1 | 5.53 | 3.00 | 9878 | 5665 |
| Saos p53 | 2 | 14.71 | 3.00 |
Figure 2.
Global binding site analysis of p53-family members. (A) Common binding site for TAp73α and TAp73β. The lower track displays the location of the Mdm2 proto-oncogene in the UCSC genome browser. The upper track shows the binding site for TAp73α and the middle one the site for TAp73β as determined by ChIP-seq using the Genome Analyzer (Illumina) and visualized with the UCSC genome browser. (B) Preferential binding site for TAp73α. The lower track displays the location of the IL1RAP gene, the other tracks as described in ‘A’. (C) Preferential binding site for TAp73β. The lower track displays the location of the APOD gene, the other tracks as described in ‘A’. (D) Overlap of target genes that were determined by ChIP-seq for TAp73α, TAp73β and p53. (E) The genomic distribution of binding sites for TAp73α, TAp73β and p53 after ChIP-seq is compared with the respective categories within the human genome. Locations of binding sites are divided in transcriptional start site (TSS) flanking region (5 kb upstream of TSS + first exon + first intron), intragenic region (all exons and introns except first), < 25 kb (peaks within 25 kb of the next annotated gene) and intergenic region (peaks > 25 kb away from any annotated gene).
Comparing the binding sites of TAp73α, TAp73β and p53, we found that a large portion of binding sites is bound by all three proteins, but also that a considerable number is preferably bound by only one of the three proteins (Figure 2D). We found that 44% of the TAp73β binding sites and 46% of the p53 binding sites, respectively, overlap with TAp73α (Figure 2D). Thus, while there are many target genes common between TAp73α, TAp73β and p53, there are also isoform and family member-specific binding sites, possibly mediating the differential functions of TAp73α, TAp73β and p53. To gain further insight into the characteristics of the DNA binding sites, we analyzed their genomic distribution (Figure 2E). Both TAp73 isoforms show almost the same genomic binding distribution being significantly enriched in promoter regions, compared to genomic background, but not as pronounced as the p53 binding sites from which almost one third are found close to transcriptional start sites.
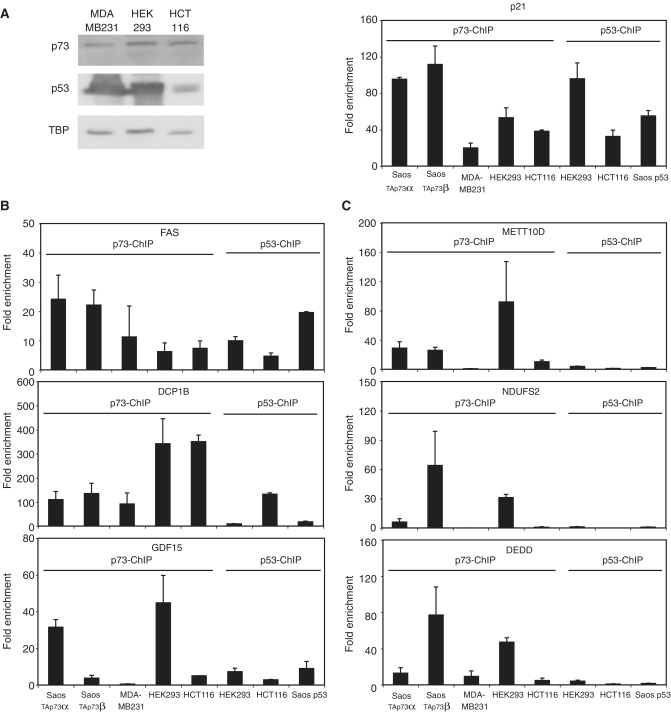
Since we have identified global binding profiles for TAp73α, TAp73β and p53 expressed in isogenic Saos cell lines, we wanted to verify that these binding sites could be physiological target sites of endogenous p73 and p53. Therefore, we have analyzed MDA-MB231-, HEK293- and HCT116-cell lines, which express p53 and p73 (Figure 3A, left), for p73- and p53-DNA binding and compared it with the binding events in the different Saos cells used before. Binding to the positive control p21 occurred in all cases (Figure 3A, right), while some binding sites can be bound by p73 as well as p53 in several different cell lines, e.g. binding sites close to the FAS, DCP1B and GDF15 gene (Figure 3B). Furthermore, we also identified target sites that are selectively bound by p73 only in specific cell lines, such as the METT10D, NDUFS2 and DEDD gene (Figure 3C). Thus, binding sites originally identified in Saos cell lines can also be found in cell lines expressing endogenously p73 or p53. The isoform-specific occupancy appears to be cell type dependent.
Figure 3.
Validation of binding sites in other cell lines. (A) Expression of endogenous p73 and p53 in several human cell lines. Protein samples were harvested from the different cell lines and stained against p73 or p53 as indicated (left). As a positive control for ChIP-qPCR in the respective cell line, binding to the p21-promoter was examined (right). Error bars were derived from two independent experiments. (B) Binding of p73 and p53 to target genes in several cell lines. ChIP-qPCR with a p73-antibody was performed in MDA-MB231, HEK293 and HCT116 cells or against p53 in HCT116 and HEK293 cells. As a comparison, binding to the sites in the different Saos cells is shown. The indicated primers were used to calculate the respective enrichment. Error bars were derived from two independent experiments. (C) Cell line specific binding to target genes. ChIP-qPCR was performed as in ‘B’.
Isoform-specific transcriptional responses
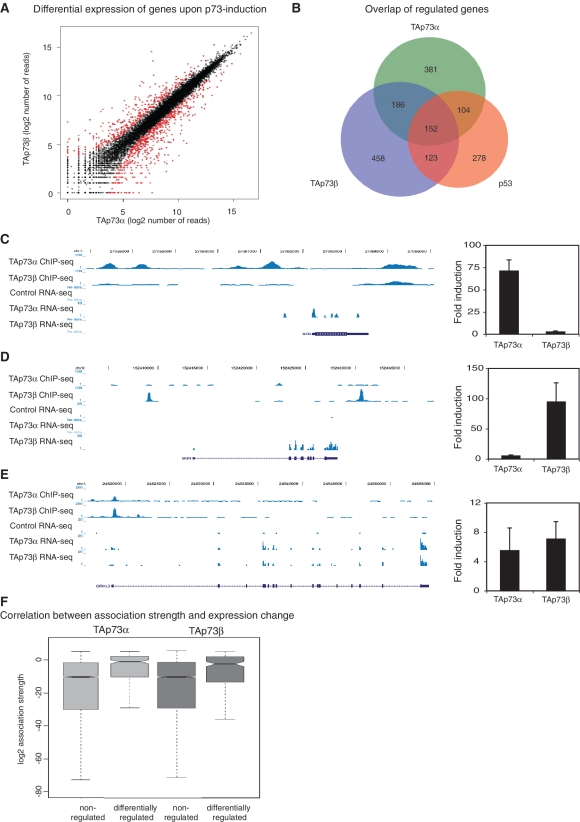
Having identified genome wide binding sites for TAp73α, TAp73β and p53, we were interested in the transcriptional consequences of the induction of the three proteins and to which extent the DNA binding and transcriptional changes could be correlated. Therefore, we performed global transcriptome analysis of TAp73α, TAp73β and p53 expressing Saos cells using RNA-seq. Two biological replicates were sequenced per cell line and the fold change relative to the Saos2 parental control was calculated using DEGseq (31). The two p73 isoforms show distinct expression signatures, with genes specifically regulated after induction of TAp73α (485 genes) or TAp73β (575 genes), as well as genes regulated in both cases (338 genes) (Figure 4A and B). The previously reported, TAp73β-specific target gene p57/Kip2/CDKN1C is also in our genome-wide RNA-seq data and is clearly induced only upon TAp73β expression (Supplementary Figure S2A). Two newly identified examples of isoform-specific regulation are shown in Figure 4C and D: the SFN gene, specifically activated by TAp73α and the BGN gene, specifically activated by TAp73β. Both genes are also differentially bound by one of the p73 isoforms as seen in the ChIP-seq data being in good agreement with the RNA-seq data. The GHRL3 gene on the other hand showed binding and activation by both isoforms (Figure 4E). The differential transcriptional response observed was further validated by RT–qPCR proving good agreement with the RNA-seq results (Figure 4C–E, right).
Figure 4.
Analysis of expression changes induced by TAp73 by RNA-Seq. (A) Differential transcriptional responses upon induction of TAp73α or TAp73β. Upon 24 h induction of TAp73α or TAp73β global expression profiles were obtained in duplicates using RNA-seq. The fold change relative to Saos2 parental control was calculated using DEGseq (31) and differential gene expression of the two p73 isoforms was plotted. (B) Overlap of expression-changing genes after induction of TAp73α, TAp73β or p53 as analyzed by RNA-seq. (C) Preferential binding of TAp73α results in specific transcriptional induction of the SFN gene by TAp73α. The two upper tracks display the ChIP-seq-data from TAp73α or TAp73β, respectively. The three lower tracks show the signals from RNA-seq for Saos2 parental cells or upon induction of either TAp73α or TAp73β. Below the tracks, the respective gene is displayed. On the right the expression change of the respective gene upon TAp73α or TAp73β induction is validated by RT-qPCR. SD was derived from three independent experiments. (D) Preferential binding of TAp73β results in specific transcriptional induction of the BGN-gene by TAp73β. (E) The GRHL3-gene was bound by TAp73α and TAp73β and changes its expression upon induction of both isoforms. (F) Correlation between association strength of TAp73α and TAp73β and expression changes. Non-regulated genes and genes regulated with an absolute log2 > 1 are shown.
The overlap between regulated genes and p73 binding sites is highly significant. Of the TAp73α upregulated genes, 49% have a TAp73α binding site within 25 kb (P = 3.08E-19), and 50% of the TAp73β upregulated genes have a TAp73β binding site (P = 3.56E-7) (Table 2 and Supplementary Table S1). The relation between binding and expression is visualized in Figure 4F, which illustrates that differentially regulated genes have a significantly higher association strength (32) than unregulated genes (TAp73α, P < 2.2E-16; TAp73β, P < 2.2E-16).
Table 2.
Expression changes upon TAp73 induction
| Total number of genes | Genes with a binding site within 25 kb | P-value of overlap between binding and expression change | |
|---|---|---|---|
| Induced by TAp73α | 344 | 170 | 3.08E-019 |
| Repressed by TAp73α | 477 | 100 | 9.99E-001 |
| Induced by TAp73β | 260 | 129 | 3.56E-007 |
| Repressed by TAp73β | 653 | 193 | 9.97E-001 |
| Induced by p53 | 350 | 152 | 6.53E-010 |
| Repressed by p53 | 305 | 66 | 9.97E-001 |
Analyzing the functional annotation of bound and regulated TAp73α and TAp73β target genes by GO analysis using DAVID (39,40), we uncovered a plethora of different physiological aspects that appear to be regulated by TAp73α and TAp73β (Supplementary Table S2). Most strikingly both isoforms regulate the expression of genes involved in different developmental processes such as tissue development, cell differentiation and development of specific anatomical structures. Interestingly, in a KEGG pathway analysis (41) of regulated target genes, the two p73 isoforms appear to regulate different functional subgroups of genes, highlighting that the functions of TAp73α and TAp73β are not merely overlapping, but also distinct (Supplementary Table S2). The most prominent functional link of this KEGG pathway analysis can be seen for TAp73α for metastasis involved processes: TAp73α seems to induce target genes that fall into several functional categories linked to metastasis, such as focal adhesion, ECM-receptor interaction, cell communication and regulation of actin cytoskeleton. For TAp73β, on the other hand, the p53-signalling pathway is the first functional category that appears in the KEGG pathway analysis. Thus, the functional annotation analysis hints at common as well as distinct functions of TAp73α and TAp73β during cellular growth and development.
Characteristics of TAp73α and TAp73β binding sites
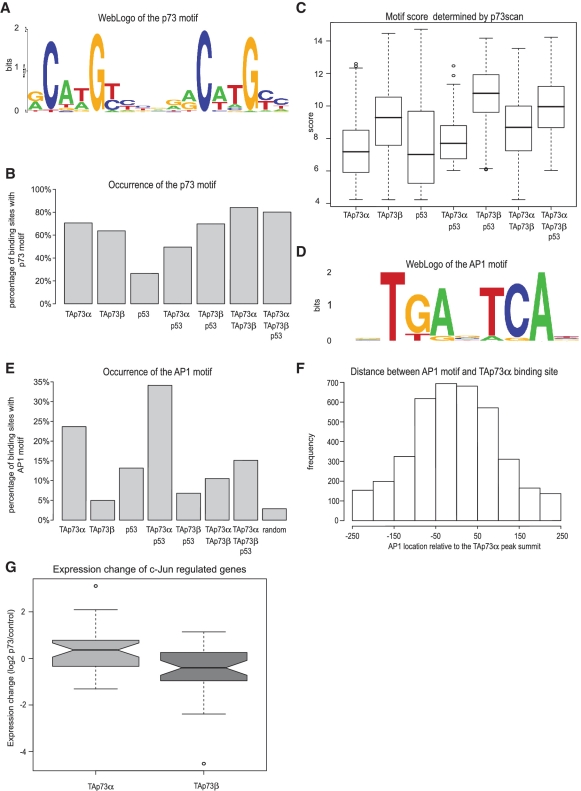
The distinct binding patterns of TAp73α and TAp73β as well as their different transactivating potential led us to analyze the binding sites with respect to their sequence contents and properties. We used a comprehensive discovery approach to predict motifs for the TAp73α as well as the TAp73β binding sites. Three different motif prediction tools (MDmodule, MotifSampler and Weeder) were applied to the binding sites and resulting motifs were tested for their significance compared to a set of background sequences (see ‘Materials and Methods’ for details). After inspection of the significant motifs using STAMP (37), we found the prevalent motif in both TAp73 sets to be a p53-like motif, hereafter referred to as the p73 consensus-binding motif (Figure 5A and Supplementary Table S3). We tested the performance of the discovered motifs by comparing the area under curve (AUC) of the receiver operator curve (ROC), which summarizes the trade-off between sensitivity and specificity when varying the motif score threshold. There was no clear difference in the PWM of the best performing TAp73α and TAp73β motif, but slight differences when comparing it to the PWM we published earlier for p53 (Supplementary Table S4) (28). The best performing p73 consensus motif based on the ROC, AUC is more degenerate than the previously identified p53 motif and clearly shows a better performance for the p73 sequences (Supplementary Figure S3). We used the algorithm p53scan (28) with this PWM to scan all identified binding sites for the new p73 consensus motif. For this purpose, we grouped all binding sites into seven different groups, according to which combinations of p53-family proteins were recruited (Table 3). Subsequently, we determined the fraction of sequences containing a p73 motif for each group (Figure 5B). In the p73 groups, we find that at least 70% of the sequences contain the p73 consensus motif. In contrast, the group of sequences bound only by p53 has a low number of p73 consensus motifs (20%). The score of the p73scan algorithm depends on the conformity of the identified motif in a specific sequence to the ideal consensus motif, the p73 motif in this case. Interestingly, the median motif score for TAp73α and p53 groups of binding sites is lower when compared with groups containing TAp73β binding sites (Figure 5C). The group with the highest score, i.e. with the highest similarity to the ideal consensus p73 motif, is the group of target sites bound by both p53 and TAp73β. We conclude that the p53-family members have distinct requirements toward their binding sites resulting in specific motifs and that a p73 specific binding motif can be extracted from our global analysis.
Figure 5.
Characteristics of TAp73α- and TAp73β binding sites. (A) The newly identified TAp73-binding motif visualized by WebLogo (56). (B) The percentage of binding sites containing the p73 motif as determined by p73scan. Binding sites are divided in seven different groups based on detected binding of TAp73α, TAp73β and/or p53: (i–iii) bound by only one family member, (iv) and (v) by p53 and either TAp73 isoform, (vi) by both TAp73 isoforms and (vii) by both TAp73 isoforms and p53. (C) Boxplot of the TAp73 motif scores, as determined by p73scan, of the groups described in ‘B’. (D) The AP1 sequence motif identified in the TAp73α binding sites visualized by WebLogo. (E) The percentage of binding sites containing the AP1 motif of the groups described in ‘B’. (F) The location of the AP1 motif in TAp73α binding sites, relative to the peak summit. (G) Boxplot of the log2-fold expression change (Saos cells expressing TAp73α or TAp73β versus Saos2 parental control) of the genes identified in Barenco et al. (43) to be c-Jun/NF-kB dependent. Expression of TAp73α cells is shown in light grey, TAp73β is shown in dark grey. The difference in the median fold change between TAp73α and TAp73β is significant (P < 0.001, Wilcoxon rank sum test).
Table 3.
Groups of p53-family specific genes
| Genes bound by different family members | Number of binding sites |
|---|---|
| TAp73α only | 4196 |
| TAp73β only | 11 849 |
| p53 only | 4009 |
| p53 + TAp73α | 4532 |
| p53 + TAp73β | 5091 |
| TAp73α + TAp73β | 10 319 |
| p53 + TAp73α + TAp73β | 3754 |
Besides the p73 motif itself, the AP1 motif (Figure 5D) which can be bound by heterodimers of the Jun/Fos family (42) is significantly overrepresented in the sequences occupied by TAp73α (8.2 times, P < 1E-10) and in the TAp73α and p53 group (11.8 times, P < 1E-10) compared with a random set of background sequences (Figure 5E). Strikingly, whereas this motif is found in 24–35% of the sequences in sites bound by TAp73α or TAp73α and p53, it is not enriched in the TAp73β or the p53 and TAp73β sites. The location of the AP1 motif is plotted relative to the TAp73α peak summit in Figure 5F. The distinct centered distribution indicates that the AP1 motif is located in very close proximity to the actual TAp73α binding site. A similar distribution is also seen for the distance between TAp73β or p53 peak summits and occurring AP1 motifs, although the frequencies of AP1 motif occurrences are lower (Supplementary Figure S4A). While the AP1 and p73 motifs occur close together, there does not seem to be a specific fixed distance (Supplementary Figure S4B).
The relationship between TAp73α and transcription factors binding to AP1 sites was further strengthened when we assessed putative AP1 target genes. Recently, a group of genes predicted to be controlled by c-Jun/NF-ΚB was identified (43). Of those genes, 20% overlap with the genes differentially regulated by TAp73α and TAp73β (P < 0.0001). Strikingly, the median log2 fold expression change for these genes in TAp73α cells is 0.36, while these genes have a median log2 fold change of −0.41 (P < 0.001) in TAp73β cells (Figure 5G). In TAp73α-expressing cells, genes with an AP1 motif are more often upregulated compared to genes lacking this motif (Supplementary Figure S5 and Table S4). This correlation is not observed for TAp73β.
In conclusion, the sequences of the identified p73 binding sites show clear isoform-specific characteristics. In particular, the overrepresentation of the AP1 motif distinguishes the TAp73α from the TAp73β binding sites.
TAp73α activates genes containing an AP1 motif close to a p73 binding site
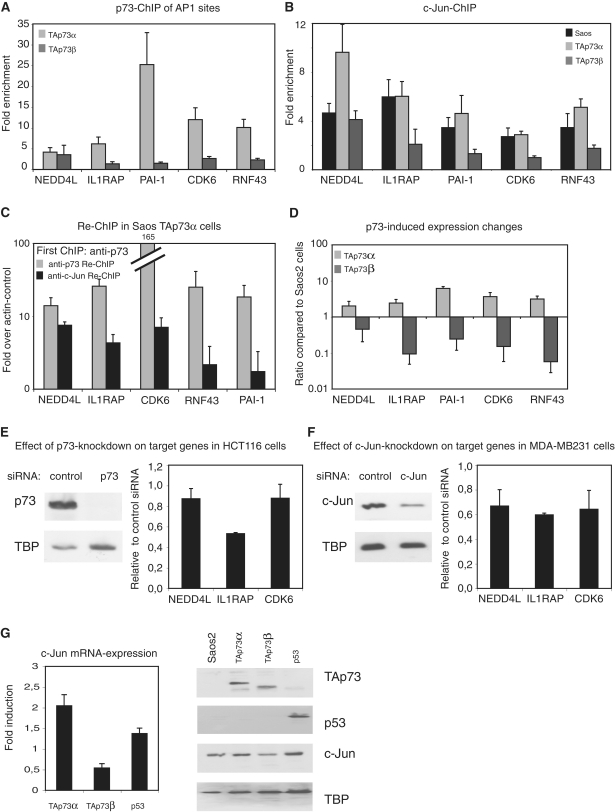
The transcription factors of the AP1 family regulate many growth-related functions; therefore, we wondered whether genes assigned to p73 binding sites co-occurring with AP1 motifs could contribute to the physiological differences we observed between TAp73α and TAp73β. We chose five genes in which an AP1 motif is also present and detected strong enrichment of TAp73α at all tested binding sites, while the binding of TAp73β is not significantly enriched (Figure 6A). Next, we assessed whether c-Jun is differentially recruited in the presence or absence of either TAp73α or TAp73β. ChIP-qPCR of c-Jun revealed clear binding of c-Jun to target sites in parental Saos2 and in TAp73α-expressing cells (Figure 6B). In the cases of NEDD4L and RNF43, the expression of TAp73α increases the binding of c-Jun, compared with parental Saos2 cells. Importantly, in TAp73β expressing cells c-Jun binding to the respective target sites is reduced compared with parental Saos2 cells. We also analyzed whether p53 was bound to these AP1 motif-containing sites and whether it could change the recruitment of c-Jun. While strong binding of p53 was not observed, the binding of c-Jun to these sites appears to remain unchanged in p53-expressing cells compared with parental Saos2 cells (Supplementary Figure S6). To validate that TAp73α and c-Jun are actually bound simultaneously to the same binding regions, we performed ChIP-re-ChIP analysis. For all tested target genes, we could re-ChIP c-Jun at the TAp73α-bound target sites and vice versa, showing that TAp73α and c-Jun can indeed be found on the same binding regions at the same time (Figure 6C and Supplementary Figure S7).
Figure 6.
Opposite effects of TAp73α and TAp73β on target genes close to an AP1 motif. (A) TAp73α but not TAp73β binds to AP1 motif-containing binding sites. ChIP-qPCR was performed in Saos cells expressing TAp73α or TAp73β for 24 h, with primers covering putative AP1 binding sites. SD was derived from three independent experiments. (B) Binding of c-Jun to putative target genes is reduced in TAp73β-expressing cells. ChIP-qPCR of c-Jun was performed in Saos2 cells, either the parental cell line, or cells expressing TAp73α or TAp73β for 24 h. Same primers as in ‘A’ were used and SD was derived from three independent experiments. (C) ChIP-re-ChIP shows binding of TAp73α and c-Jun to the same sites on DNA. After induction of TAp73α chromatin complexes were precipitated first with a p73-antibody. After elution a second round of immunoprecipitation was performed with a c-Jun- or again with a p73-antibody. The second ChIP included an unspecific antibody control to calculate enrichment. SD was derived from three independent experiments. (D) Opposite regulation of AP1 motif-containing target genes by TAp73α and TAp73β. Changes in expression of mRNA of the target genes from ‘A’, upon induction of TAp73α or TAp73β for 24 h. SD was calculated from three independent experiments. (E) Expression of AP1 motif-containing target genes is reduced after knockdown of p73. After knockdown of p73 in HCT116 cells, protein levels of p73 (left) or transcriptional levels of the respective target genes (right) were analyzed. Error bars were derived from two independent experiments. (F) Knockdown of c-Jun reduces the expression of AP1 motif-containing target genes. After knockdown of c-Jun in MDA-MB231 cells, protein levels of c-Jun (left) or transcriptional levels of the respective target genes (right) were analyzed. Error bars were derived from two independent experiments. (G) The c-Jun expression levels are downregulated by TAp73β. Expression of c-Jun mRNA upon induction of TAp73α, TAp73β or p53 for 24 h (left). SD was calculated from three independent experiments. Protein-levels of c-Jun were analyzed in Saos cells after induction of TAp73α, TAp73β or p53 for 24 h before whole cell extract was isolated and the indicated proteins were stained by western blot (right).
In concordance with the binding data, TAp73α induces the mRNA expression of all five genes (Figure 6D). Surprisingly, we found a repression of the level of mRNA of the same genes upon TAp73β induction, in the case of RNF43 or IL1RAP a substantial decrease to 5–10% of the level of expression in the parental Saos2 cells. To show the dependence of these target genes from endogenous p73 and c-Jun, we silenced the expression of p73 and c-Jun in HCT116 and MDA-MB231 cells, respectively, and analyzed the expression of NEDD4L, IL1RAP and CDK6 (Figure 6E and F). The reduction of p73 levels leads to a small decrease of NEDD4L- and CDK6-mRNA and a strong reduction of the IL1RAP expression (Figure 6E). After the reduction of c-Jun expression, the mRNA levels of all three tested genes were reduced more than 30% (Figure 6F).
To further explore the relationship between c-Jun and p73, we analyzed the levels of c-Jun mRNA in TAp73β-expressing cells (Figure 6G, left). Strikingly, upon TAp73β induction the c-Jun mRNA level is strongly reduced, an effect also seen in our RNA-seq data (Supplementary Figure 2C). The protein level of c-Jun is also reduced specifically upon TAp73β induction, while neither TAp73α nor p53 exert a similar effect (Figure 6G, right).
Thus, we have identified a specific set of TAp73α target sites that also contain an AP1 site and that are bound by c-Jun. Very interestingly, these AP1 motif-containing target sites are not bound by TAp73β. Furthermore, while the endogenous expression of these genes seems to depend on p73 and c-Jun, a repression of the associated genes specifically upon TAp73β induction is observed, at least partly due to a negative regulation of c-Jun by TAp73β. Thus, the interplay between c-Jun and TAp73 is isoform dependent, leading to differential binding of c-Jun to its target genes and to a differential transcriptional outcome.
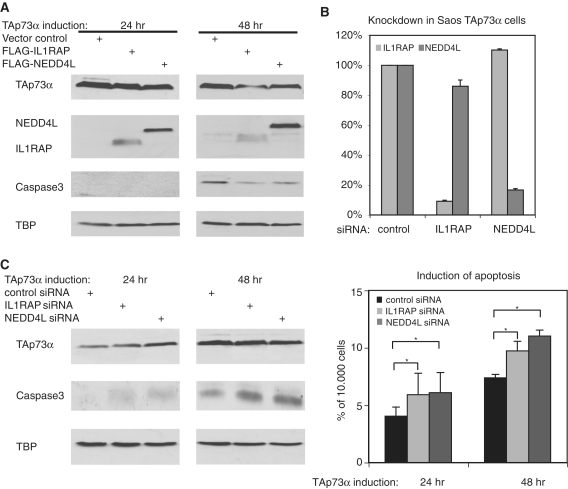
The TAp73α-target genes IL1RAP and NEDD4L can influence apoptosis
Several of the genes that are in close proximity to combined TAp73α/AP1 binding sites are linked to tumorigenesis or malignant growth (Table 4). Because TAp73α can induce these genes, we assessed whether these target genes impede on the induction of programmed cell death. To this end, we overexpressed two of these gene products, NEDD4L and IL1RAP, in Saos cells inducible for TAp73α. The effect of the overexpressed proteins on the TAp73α induced apoptosis was monitored by staining for active Caspase3. TAp73α induces the cleavage of Caspase3 mainly after 48 h (Figure 7A). Upon transfection of IL1RAP, the amount of activated Caspase3 is markedly decreased, especially after 48 h of TAp73α induction. NEDD4L impedes on the accumulation of active Caspase3 in a similar way, mainly after 48 h of TAp73α induction. To examine the physiological role of IL1RAP and NEDD4L in the TAp73α-signaling pathway, we transfected siRNAs directed against IL1RAP and NEDD4L into TAp73α-expressing cells. Analysis of the mRNA after transfection of the respective siRNA and induction of TAp73α for 24 h shows that the knockdown of IL1RAP and NEDD4L has an efficiency of at least 80% (Figure 7B). Strikingly, the knockdown of IL1RAP and NEDD4L increases the induction of active Caspase3 after 24 and 48 h of TAp73α induction (Figure 7C, left). Analyzing the population of cells in sub-G1, we found that the knockdowns lead to a statistically significant increase of TAp73α-induced apoptosis (Figure 7C, right). Thus, we have identified two target genes of TAp73α that seem to impede on the apoptosis induction capability of TAp73α, most likely via a pathway involving c-Jun.
Table 4.
Known function of TAp73α target genes with an AP1 motif
| Gene name | Known function | Reference |
|---|---|---|
| RNF43 | Upregulated in colorectal cancers; growth promoting if exogenously expressed | (47,57) |
| NEDD4L | Na+ channel regulation/tumor associated via Wnt-signaling in liver cancer/neuronal survival via Trk neurotrophin receptors | (58,59) |
| CDK6 | Catalytic subunit for G1/S transition/stimulates growth in prostate cancer/overexpressed in medullablastoma; poor prognosis marker | (46,60) |
| IL1RAP | Involved in IL1 action during inflammation/activation of NFkB pathway | (61,62) |
| PAI-1 | Promoter of tumor progression/poor prognosis marker | (63–66) |
| RIPK4 | Can activate NFkappaB/processed during apoptosis/involved in differentiation | (67,68) |
| GAP43 | Growth associated protein 43/highly expressed during neuronal development and axonal regeneration/neurite outgrowth/AP1& TrkA associated | (69) |
| SDC1 | Participates in cell proliferation, cell migration and cell-matrix interactions/Altered (higher) syndecan-1 expression has been detected in several different tumor types | (70,71) |
| NGFR | Tumor suppressor in retinoblastoma/necessary for cell survival in ESCC/Inducer of apoptosis in absence of ligand/Interaction with TrkA/critical regulator of glioma invasion | (72–75) |
Figure 7.
Target genes of TAp73α and c-Jun impede on apoptosis induction. (A) Overexpression of IL1RAP, NEDD4L or an empty vector in Saos cells expressing TAp73α. After induction for 24 or 48 h whole cell extracts were analyzed by western blot. (B) Knockdown of IL1RAP and NEDD4L. TAp73α-expressing Saos cells were transfected twice with a pool of four different siRNAs against each gene or a non-targeting siRNA-pool. After induction for 24 h mRNA was harvested and the efficiency of knockdown was monitored setting the non-targeting sample to 100%. Error bars result from two biological replicas. (C) Knockdown of IL1RAP and NEDD4L increases apoptosis. TAp73α-expressing Saos cells were transfected as in ‘B’, prior to isolation of whole cell extract and western blot analysis (left panel) or preparation for FACS analysis to determine the amount of cells in sub-G1 phase as a measure for apoptotic induction (right). Asterisks indicates P < 0.05. SD was derived from three independent experiments.
We propose that the differences in apoptosis induction by TAp73α and TAp73β are mediated at least partly by the differential collaboration with c-Jun: in the case of TAp73α AP1 binding sites are bound by c-Jun and the respective proliferation related target genes are induced leading to a weaker apoptotic response of TAp73α. On the other hand the TAp73β binding sites are not only devoid of AP1 sites, but TAp73β negatively regulates the c-Jun levels leading to much less c-Jun being bound at the respective AP1 sites which ultimately results in much higher levels of apoptosis.
DISCUSSION
A marked physiological difference between the two most commonly expressed TAp73 isoforms, TAp73α and TAp73β has been reported (16,44), but the molecular mechanism for the observed different cellular response upon TAp73α and TAp73β activation has remained elusive. Here, we analyze the differences of TAp73α and TAp73β target gene binding and regulation and propose that the balance between survival and apoptosis induction by TAp73α and TAp73β is mediated at least partly by the differential interplay with c-Jun. In our global binding site analysis, we identified a consensus binding motif for TAp73α and TAp73β. Although this motif resembles the previously described p53 consensus motif (28), it has several distinct features, e.g. the bases outside the four nucleotide core are less preserved and it is thereby closer related to the binding motif described for p63 (45).
Assessing the molecular differences between TAp73α and TAp73β, we found that an AP1 motif is strongly enriched in the region surrounding many binding sites of TAp73α, but not in TAp73β binding sites. We observed a striking difference in the expression of the target genes with an AP1 motif in the TAp73α- versus TAp73β-expressing cells. While these target genes are selectively bound by TAp73α together with c-Jun and subsequently upregulated, TAp73β represses their mRNA expression. In line with these findings, we observed that the recruitment of c-Jun to the AP1 sites of these genes is impaired upon TAp73β induction probably caused by a downregulation of c-Jun mRNA and protein by TAp73β. Several of the assigned target genes are related to malignant growth, like CDK6 and RNF43 (46,47). We show that two genes specifically upregulated in TAp73α-expressing cells, NEDD4L and IL1RAP, are able to influence the induction of apoptosis upon their overexpression or siRNA-mediated downregulation, thereby showing that TAp73α indeed induces antiapoptotic factors.
Analysis of TAp73α and TAp73β transcriptional activities
It has been reported previously that TAp73β can induce the expression of p53-target genes like p21 or Mdm2 to a higher extent than TAp73α (15,48,49), while also TAp73α-specific target genes have been reported (50). Furthermore, TAp73β has been described to be a much stronger inducer of apoptosis (51,52). In line with this, our data show that known target genes like Mdm2 and p21 are bound with higher affinity and induced to higher levels through TAp73β and that the induction of apoptosis by TAp73β is much stronger. Nevertheless, our study now extends previous analysis by combining ChIP-seq and RNA-seq to compare these two most prevalent TAp73 isoforms with each other as well as with p53 with respect to global target gene binding and expression. Strikingly, besides common target genes bound and regulated by TAp73α, TAp73β and p53, we found target genes that were regulated in an isoform-specific manner. In summary, our data provide a genome-wide analysis of TAp73α, TAp73β and p53 chromatin occupancy and the correlation thereof with transcriptional changes upon TAp73α, TAp73β and p53 induction. This enables a careful examination of the isoform specific as well as overlapping target genes and provides a basis for further functional characterization.
Isoform-specific functional interplay of TAp73 with c-Jun
Our data show that the cellular outcome of the interplay between c-Jun and p73 depends on the presence of the respective TAp73 isoform. First, our motif analysis of genome-wide data showed a striking co-occurrence of c-Jun-binding motifs with TAp73α binding sites, to which both proteins bind simultaneously. Second, induction of TAp73β resulted in a reduction of c-Jun mRNA and protein levels and hence to a lack of recruitment of c-Jun. Third, there is a positive correlation between TAp73α-induced genes and c-Jun target genes, while this correlation is inversed for TAp73β-regulated genes. Thus, the poor induction of cell death by TAp73α could be due to activation of antiapoptotic target genes along with c-Jun, while the stronger apoptosis induced by TAp73β is caused at least partly by a reduction of cellular c-Jun levels and subsequent reduced recruitment of c-Jun to DNA resulting in repression of these target genes. Several of these target genes are linked to malignant growth and their mRNA levels were induced by TAp73α but repressed by TAp73β. The negative regulation of genes related to proliferation by TAp73β and p53 that was reported earlier (53) might, therefore, also be extended to these AP1 motif-containing genes. This AP1 motif can be bound by c-Jun, which has been reported to stabilize p73 and thereby to enhance its function, although no direct interaction was observed (22). More recently, it was reported that c-Jun plays an important role in apoptosis induced by chemotherapeutic treatments mediated by YAP1, a critical regulator of p73 (54,55). On the other hand, c-Jun and p73 have also been shown to co-operate during growth promotion and to synergistically induce transcription from c-Jun response element containing promoter constructs in the absence of additional apoptosis-inducing treatments (21). Based on our findings, we speculate that the differences in cellular outcome are due to the presence of the particular TAp73 isoform in the respective cellular system and its respective interplay with c-Jun which appears to be cell type dependent. The weaker induction of cell death by TAp73α that we observed in our experimental system might, therefore, be mediated by the activation of antiapoptotic target genes together with c-Jun, while the stronger apoptosis induced by TAp73β might be due to the repression of these target genes and a reduction of c-Jun levels. The cell-type, treatment- and isoform-specific differences influencing the cellular fate hint toward several layers of regulation between p73 and c-Jun and might be a very important molecular explanation for the different observed phenotypes induced by the TAp73 isoforms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Dutch Cancer Society (KWF) (KUN 2003-2926 to M.K. and L.S., KUN 2005-3347 to L.S.); Netherlands Organization for Scientific Research (NWO) (Vidi 846.05.002 to S.J.v.H. and M.L.). Funding for open access charge: Georg-Speyer-Haus; Frankfurt
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We would like to thank M. Martin for the pFLAG-mIL1RAcP expression construct and W. Sundquist for the pCl.FLAG.-NEDD4L wt expression construct, as well as F. Galvagni for the c-Jun knockdown constructs.
REFERENCES
- 1.Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene. 2007;26:5169–5183. doi: 10.1038/sj.onc.1210337. [DOI] [PubMed] [Google Scholar]
- 2.Moll U, Slade N. p63 and p73: roles in development and tumor formation. Mol. Cancer Res. 2004;2:371–386. [PubMed] [Google Scholar]
- 3.Zaika AI, Slade N, Erster SH, Sansome C, Joseph TW, Pearl M, Chalas E, Moll UM. DeltaNp73, a dominant-negative inhibitor of wild-type p53 and TAp73, is up-regulated in human tumors. J. Exp. Med. 2002;196:765–780. doi: 10.1084/jem.20020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Concin N, Becker K, Slade N, Erster S, Mueller-Holzner E, Ulmer H, Daxenbichler G, Zeimet A, Zeillinger R, Marth C, et al. Transdominant DeltaTAp73 isoforms are frequently up-regulated in ovarian cancer. Evidence for their role as epigenetic p53 inhibitors in vivo. Cancer Res. 2004;64:2449–2460. doi: 10.1158/0008-5472.can-03-1060. [DOI] [PubMed] [Google Scholar]
- 5.Melino G, De Laurenzi V, Vousden KH. p73: Friend or foe in tumorigenesis. Nat. Rev. Cancer. 2002;2:605–615. doi: 10.1038/nrc861. [DOI] [PubMed] [Google Scholar]
- 6.Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 7.Flores ER, Sengupta S, Miller JB, Newman JJ, Bronson R, Crowley D, Yang A, McKeon F, Jacks T. Tumor predisposition in mice mutant for p63 and p73: evidence for broader tumor suppressor functions for the p53 family. Cancer Cell. 2005;7:363–373. doi: 10.1016/j.ccr.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 8.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 9.Tomasini R, Tsuchihara K, Wilhelm M, Fujitani M, Rufini A, Cheung C, Khan F, Itie-Youten A, Wakeham A, Tsao M, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agami R, Blandino G, Oren M, Shaul Y. Interaction of c-Abl and p73alpha and their collaboration to induce apoptosis. Nature. 1999;399:809–813. doi: 10.1038/21697. [DOI] [PubMed] [Google Scholar]
- 11.Yuan ZM, Shioya H, Ishiko T, Sun X, Gu J, Huang YY, Lu H, Kharbanda S, Weichselbaum R, Kufe D. p73 is regulated by tyrosine kinase c-Abl in the apoptotic response to DNA damage. Nature. 1999;399:814–817. doi: 10.1038/21704. [DOI] [PubMed] [Google Scholar]
- 12.Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin WG, Levrero M, Wang JY. The tyrosine kinase c-Abl regulates p73 in apoptotic response to cisplatin-induced DNA damage. Nature. 1999;399:806–809. doi: 10.1038/21690. [DOI] [PubMed] [Google Scholar]
- 13.Pediconi N, Ianari A, Costanzo A, Belloni L, Gallo R, Cimino L, Porcellini A, Screpanti I, Balsano C, Alesse E, et al. Differential regulation of E2F1 apoptotic target genes in response to DNA damage. Nat. Cell. Biol. 2003;5:552–558. doi: 10.1038/ncb998. [DOI] [PubMed] [Google Scholar]
- 14.Irwin MS, Kondo K, Marin MC, Cheng LS, Hahn WC, Kaelin WG. Chemosensitivity linked to p73 function. Cancer Cell. 2003;3:403–410. doi: 10.1016/s1535-6108(03)00078-3. [DOI] [PubMed] [Google Scholar]
- 15.De Laurenzi V, Costanzo A, Barcaroli D, Terrinoni A, Falco M, Annicchiarico-Petruzzelli M, Levrero M, Melino G. Two new p73 splice variants, gamma and delta, with different transcriptional activity. J. Exp. Med. 1998;188:1763–1768. doi: 10.1084/jem.188.9.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Melino G, Bernassola F, Ranalli M, Yee K, Zong WX, Corazzari M, Knight RA, Green DR, Thompson C, Vousden KH. p73 Induces apoptosis via PUMA transactivation and Bax mitochondrial translocation. J. Biol. Chem. 2004;279:8076–8083. doi: 10.1074/jbc.M307469200. [DOI] [PubMed] [Google Scholar]
- 17.Flinterman M, Guelen L, Ezzati-Nik S, Killick R, Melino G, Tominaga K, Mymryk JS, Gaken J, Tavassoli M. E1A activates transcription of p73 and Noxa to induce apoptosis. J. Biol. Chem. 2005;280:5945–5959. doi: 10.1074/jbc.M406661200. [DOI] [PubMed] [Google Scholar]
- 18.Blint E, Phillips AC, Kozlov S, Stewart CL, Vousden KH. Induction of p57(KIP2) expression by p73beta. Proc. Natl Acad. Sci. USA. 2002;99:3529–3534. doi: 10.1073/pnas.062491899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toh WH, Logette E, Corcos L, Sabapathy K. TAp73beta and DNp73beta activate the expression of the pro-survival caspase-2S. Nucleic Acids Res. 2008;36:4498–4509. doi: 10.1093/nar/gkn414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nyman U, Sobczak-Pluta A, Vlachos P, Perlmann T, Zhivotovsky B, Joseph B. Full-length p73alpha represses drug-induced apoptosis in small cell lung carcinoma cells. J. Biol. Chem. 2005;280:34159–34169. doi: 10.1074/jbc.M500394200. [DOI] [PubMed] [Google Scholar]
- 21.Vikhanskaya F, Toh W, Dulloo I, Wu Q, Boominathan L, Ng H, Vousden K, Sabapathy K. p73 supports cellular growth through c-Jun-dependent AP-1 transactivation. Nat. Cell. Biol. 2007;9:698–706. doi: 10.1038/ncb1598. [DOI] [PubMed] [Google Scholar]
- 22.Toh WH, Siddique MM, Boominathan L, Lin KW, Sabapathy K. c-Jun regulates the stability and activity of the p53 homologue, p73. J. Biol. Chem. 2004;279:44713–44722. doi: 10.1074/jbc.M407672200. [DOI] [PubMed] [Google Scholar]
- 23.Shaulian E, Karin M. AP-1 as a regulator of cell life and death. Nat. Cell. Biol. 2002;4:E131–E136. doi: 10.1038/ncb0502-e131. [DOI] [PubMed] [Google Scholar]
- 24.Wisdom R, Johnson RS, Moore C. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 1999;18:188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson RS, van Lingen B, Papaioannou VE, Spiegelman BM. A null mutation at the c-jun locus causes embryonic lethality and retarded cell growth in culture. Genes Dev. 1993;7:1309–1317. doi: 10.1101/gad.7.7b.1309. [DOI] [PubMed] [Google Scholar]
- 26.Schreiber M, Kolbus A, Piu F, Szabowski A, Möhle-Steinlein U, Tian J, Karin M, Angel P, Wagner EF. Control of cell cycle progression by c-Jun is p53 dependent. Genes Dev. 1999;13:607–619. doi: 10.1101/gad.13.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shaulian E, Schreiber M, Piu F, Beeche M, Wagner EF, Karin M. The mammalian UV response: c-Jun induction is required for exit from p53-imposed growth arrest. Cell. 2000;103:897–907. doi: 10.1016/s0092-8674(00)00193-8. [DOI] [PubMed] [Google Scholar]
- 28.Smeenk L, Van Heeringen S, Koeppel M, Van Driel M, Bartels S, Akkers R, Denissov S, Stunnenberg H, Lohrum M. Characterization of genome-wide p53-binding sites upon stress response. Nucleic Acids Res. 2008;36:3639–3654. doi: 10.1093/nar/gkn232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denissov S, van Driel M, Voit R, Hekkelman M, Hulsen T, Hernandez N, Grummt I, Wehrens R, Stunnenberg H. Identification of novel functional TBP-binding sites and general factor repertoires. EMBO J. 2007;26:944–954. doi: 10.1038/sj.emboj.7601550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Bio. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 32.Ouyang Z, Zhou Q, Wong WH. ChIP-Seq of transcription factors predicts absolute and differential gene expression in embryonic stem cells. Proc. Natl Acad. Sci. USA. 2009;22:21521–21526. doi: 10.1073/pnas.0904863106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thijs G, Lescot M, Marchal K, Rombauts S, De Moor B, Rouze P, Moreau Y. A higher-order background model improves the detection of promoter regulatory elements by Gibbs sampling. Bioinformatics. 2001;17:1113–1122. doi: 10.1093/bioinformatics/17.12.1113. [DOI] [PubMed] [Google Scholar]
- 34.Pavesi G, Mereghetti P, Mauri G, Pesole G. Weeder Web: discovery of transcription factor binding sites in a set of sequences from co-regulated genes. Nucleic Acids Res. 2004;32:W199–203. doi: 10.1093/nar/gkh465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu X, Brutlag D, Liu J. An algorithm for finding protein–DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat. Biotechnol. 2002;5:835–839. doi: 10.1038/nbt717. [DOI] [PubMed] [Google Scholar]
- 36.Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. 2006;34:D95–D97. doi: 10.1093/nar/gkj115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007;35:W253–W258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar R, Domrachev M, Lash AE. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Bio. 2003;4:P3. [PubMed] [Google Scholar]
- 40.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 41.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakabeppu Y, Ryder K, Nathans D. DNA binding activities of three murine Jun proteins: stimulation by Fos. Cell. 1988;55:907–915. doi: 10.1016/0092-8674(88)90146-8. [DOI] [PubMed] [Google Scholar]
- 43.Barenco M, Brewer D, Papouli E, Tomescu D, Callard R, Stark J, Hubank M. Dissection of a complex transcriptional response using genome-wide transcriptional modelling. Mol. Syst. Bio. 2009;5:327. doi: 10.1038/msb.2009.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Klanrit P, Flinterman MB, Odell EW, Melino G, Killick R, Norris JS, Tavassoli M. Specific isoforms of p73 control the induction of cell death induced by the viral proteins, E1A or apoptin. Cell Cycle. 2008;7:205–215. doi: 10.4161/cc.7.2.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ortt K, Sinha S. Derivation of the consensus DNA-binding sequence for p63 reveals unique requirements that are distinct from p53. FEBS Lett. 2006;580:4544–4550. doi: 10.1016/j.febslet.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 46.Lim JT, Mansukhani M, Weinstein IB. Cyclin-dependent kinase 6 associates with the androgen receptor and enhances its transcriptional activity in prostate cancer cells. Proc. Natl Acad. Sci. USA. 2005;102:5156–5161. doi: 10.1073/pnas.0501203102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sugiura T, Yamaguchi A, Miyamoto K. A cancer-associated RING finger protein, RNF43, is a ubiquitin ligase that interacts with a nuclear protein, HAP95. Exp. Cell Res. 2008;314:1519–1528. doi: 10.1016/j.yexcr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 48.Lee CW, La Thangue NB. Promoter specificity and stability control of the p53-related protein p73. Oncogene. 1999;18:4171–4181. doi: 10.1038/sj.onc.1202793. [DOI] [PubMed] [Google Scholar]
- 49.Ueda Y, Hijikata M, Takagi S, Chiba T, Shimotohno K. New p73 variants with altered C-terminal structures have varied transcriptional activities. Oncogene. 1999;18:4993–4998. doi: 10.1038/sj.onc.1202817. [DOI] [PubMed] [Google Scholar]
- 50.Fontemaggi G, Kela I, Amariglio N, Rechavi G, Krishnamurthy J, Strano S, Sacchi A, Givol D, Blandino G. Identification of direct p73 target genes combining DNA microarray and chromatin immunoprecipitation analyses. J. Biol. Chem. 2002;277:43359–43368. doi: 10.1074/jbc.M205573200. [DOI] [PubMed] [Google Scholar]
- 51.Oshima Y, Sasaki Y, Negishi H, Idogawa M, Toyota M, Yamashita T, Wada T, Nagoya S, Kawaguchi S, Yamashita T, et al. Antitumor effect of adenovirus-mediated p53 family gene transfer on osteosarcoma cell lines. Cancer Biol. Ther. 2007;6:1058–1066. doi: 10.4161/cbt.6.7.4320. [DOI] [PubMed] [Google Scholar]
- 52.Gonzalez S, Perez-Perez MM, Hernando E, Serrano M, Cordon-Cardo C. p73beta-Mediated apoptosis requires p57kip2 induction and IEX-1 inhibition. Cancer Res. 2005;65:2186–2192. doi: 10.1158/0008-5472.CAN-04-3047. [DOI] [PubMed] [Google Scholar]
- 53.Scian M, Carchman E, Mohanraj L, Stagliano K, Anderson M, Deb D, Crane B, Kiyono T, Windle B, Deb S, et al. Wild-type p53 and p73 negatively regulate expression of proliferation related genes. Oncogene. 2008;27:2583–2593. doi: 10.1038/sj.onc.1210898. [DOI] [PubMed] [Google Scholar]
- 54.Danovi SA, Rossi M, Gudmundsdottir K, Yuan M, Melino G, Basu S. Yes-associated protein (YAP) is a critical mediator of c-Jun-dependent apoptosis. Cell Death Differ. 2008;15:217–219. doi: 10.1038/sj.cdd.4402226. [DOI] [PubMed] [Google Scholar]
- 55.Strano S, Monti O, Pediconi N, Baccarini A, Fontemaggi G, Lapi E, Mantovani F, Damalas A, Citro G, Sacchi A. The transcriptional coactivator yes-associated protein drives p73 gene-target specificity in response to DNA damage. Mol. Cell. 2005;18:447–459. doi: 10.1016/j.molcel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 56.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yagyu R, Furukawa Y, Lin YM, Shimokawa T, Yamamura T, Nakamura Y. A novel oncoprotein RNF43 functions in an autocrine manner in colorectal cancer. Int. J. Oncol. 2004;25:1343–1348. [PubMed] [Google Scholar]
- 58.Arévalo JC, Waite J, Rajagopal R, Beyna M, Chen ZY, Lee FS, Chao MV. Cell survival through Trk neurotrophin receptors is differentially regulated by ubiquitination. Neuron. 2006;50:549–559. doi: 10.1016/j.neuron.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 59.Lee HS, Park MH, Yang SJ, Park KC, Kim NS, Kim YS, Kim DI, Yoo HS, Choi EJ, Yeom YI. Novel candidate targets of Wnt/beta-catenin signaling in hepatoma cells. Life Sci. 2007;80:690–698. doi: 10.1016/j.lfs.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 60.Mendrzyk F, Radlwimmer B, Joos S, Kokocinski F, Benner A, Stange DE, Neben K, Fiegler H, Carter NP, Reifenberger G, et al. Genomic and protein expression profiling identifies CDK6 as novel independent prognostic marker in medulloblastoma. J. Clin. Oncol. 2005;23:8853–8862. doi: 10.1200/JCO.2005.02.8589. [DOI] [PubMed] [Google Scholar]
- 61.Cullinan EB, Kwee L, Nunes P, Shuster DJ, Ju G, McIntyre KW, Chizzonite RA, Labow MA. IL-1 receptor accessory protein is an essential component of the IL-1 receptor. J. Immunol. 1998;161:5614–5620. [PubMed] [Google Scholar]
- 62.Towne JE, Garka KE, Renshaw BR, Virca GD, Sims JE. Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-kappaB and MAPKs. J. Biol. Chem. 2004;279:13677–13688. doi: 10.1074/jbc.M400117200. [DOI] [PubMed] [Google Scholar]
- 63.Bajou K, Noel A, Gerard RD, Masson V, Brunner N, Holst-Hansen C, Skobe M, Fusenig NE, Carmeliet P, Collen D, et al. Absence of host plasminogen activator inhibitor 1 prevents cancer invasion and vascularization. Nature Med. 1998;4:923–928. doi: 10.1038/nm0898-923. [DOI] [PubMed] [Google Scholar]
- 64.Gutierrez LS, Schulman A, Brito-Robinson T, Noria F, Ploplis VA, Castellino FJ. Tumor development is retarded in mice lacking the gene for urokinase-type plasminogen activator or its inhibitor, plasminogen activator inhibitor-1. Cancer Res. 2000;60:5839–5847. [PubMed] [Google Scholar]
- 65.Kortlever RM, Bernards R. Senescence, wound healing and cancer: the PAI-1 connection. Cell Cycle. 2006;5:2697–2703. doi: 10.4161/cc.5.23.3510. [DOI] [PubMed] [Google Scholar]
- 66.Nekarda H, Siewert JR, Schmitt M, Ulm K. Tumour-associated proteolytic factors uPA and PAI-1 and survival in totally resected gastric cancer. Lancet. 1994;343:117. doi: 10.1016/s0140-6736(94)90845-1. [DOI] [PubMed] [Google Scholar]
- 67.Adams S, Pankow S, Werner S, Munz B. Regulation of NF-kappaB activity and keratinocyte differentiation by the RIP4 protein: implications for cutaneous wound repair. J. Invest. Dermatol. 2007;127:538–544. doi: 10.1038/sj.jid.5700588. [DOI] [PubMed] [Google Scholar]
- 68.Meylan E, Martinon F, Thome M, Gschwendt M, Tschopp J. RIP4 (DIK/PKK), a novel member of the RIP kinase family, activates NF-kappa B and is processed during apoptosis. EMBO Rep. 2002;3:1201–1208. doi: 10.1093/embo-reports/kvf236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Diolaiti D, Bernardoni R, Trazzi S, Papa A, Porro A, Bono F, Herbert JM, Perini G, Della Valle G. Functional cooperation between TrkA and p75(NTR) accelerates neuronal differentiation by increased transcription of GAP-43 and p21(CIP/WAF) genes via ERK1/2 and AP-1 activities. Exp. Cell. Res. 2007;313:2980–2992. doi: 10.1016/j.yexcr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 70.Choi D, Kim J, Ryu H, Kim H, Han J, Lee J, Min C. Syndecan-1, a key regulator of cell viability in endometrial cancer. Int. J. Cancer. 2007;121:741–750. doi: 10.1002/ijc.22713. [DOI] [PubMed] [Google Scholar]
- 71.Maeda T, Desouky J, Friedl A. Syndecan-1 expression by stromal fibroblasts promotes breast carcinoma growth in vivo and stimulates tumor angiogenesis. Oncogene. 2006;25:1408–1412. doi: 10.1038/sj.onc.1209168. [DOI] [PubMed] [Google Scholar]
- 72.Dimaras H, Gallie BL. The p75 NTR neurotrophin receptor is a tumor suppressor in human and murine retinoblastoma development. Int. J. Cancer. 2008;122:2023–2029. doi: 10.1002/ijc.23356. [DOI] [PubMed] [Google Scholar]
- 73.Johnston AL, Lun X, Rahn JJ, Liacini A, Wang L, Hamilton MG, Parney IF, Hempstead BL, Robbins SM, Forsyth PA, et al. The p75 neurotrophin receptor is a central regulator of glioma invasion. PLoS Biol. 2007;5:e212. doi: 10.1371/journal.pbio.0050212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khwaja F, Tabassum A, Allen J, Djakiew D. The p75(NTR) tumor suppressor induces cell cycle arrest facilitating caspase mediated apoptosis in prostate tumor cells. Biochem. Biophys. Res. Commun. 2006;341:1184–1192. doi: 10.1016/j.bbrc.2006.01.073. [DOI] [PubMed] [Google Scholar]
- 75.Okumura T, Tsunoda S, Mori Y, Ito T, Kikuchi K, Wang TC, Yasumoto S, Shimada Y. The biological role of the low-affinity p75 neurotrophin receptor in esophageal squamous cell carcinoma. Clin. Cancer Res. 2006;12:5096–5103. doi: 10.1158/1078-0432.CCR-05-2852. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data have been deposited in NCBI’s Gene Expression Omnibus (38) and are accessible through GEO Series accession number GSE15780 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE15780).